Abstract
In this study, we investigated the impact of soil pH on the diversity and abundance of archaeal ammonia oxidizers in 27 different forest soils across Germany. DNA was extracted from topsoil samples, the amoA gene, encoding ammonia monooxygenase, was amplified; and the amplicons were sequenced using a 454-based pyrosequencing approach. As expected, the ratio of archaeal (AOA) to bacterial (AOB) ammonia oxidizers’ amoA genes increased sharply with decreasing soil pH. The diversity of AOA differed significantly between sites with ultra-acidic soil pH (<3.5) and sites with higher pH values. The major OTUs from soil samples with low pH could be detected at each site with a soil pH <3.5 but not at sites with pH >4.5, regardless of geographic position and vegetation. These OTUs could be related to the Nitrosotalea group 1.1 and the Nitrososphaera subcluster 7.2, respectively, and showed significant similarities to OTUs described from other acidic environments. Conversely, none of the major OTUs typical of sites with a soil pH >4.6 could be found in the ultra- and extreme acidic soils. Based on a comparison with the amoA gene sequence data from a previous study performed on agricultural soils, we could clearly show that the development of AOA communities in soils with ultra-acidic pH (<3.5) is mainly triggered by soil pH and is not influenced significantly by the type of land use, the soil type, or the geographic position of the site, which was observed for sites with acido-neutral soil pH.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The amount and quality of available nitrogen determine both productivity and biodiversity in most natural and semi-natural ecosystems [1]. In forest ecosystems, for example, the distribution patterns of ammonia and nitrate in soil are strongly influenced by the plant community composition and the growth kinetics of trees [2]. In this respect both the subsequent delivery of inorganic nitrogen via nitrogen fixation or mineralization and the conversion of ammonia to nitrate via nitrification are of interest. Previously, many studies have investigated agricultural soils to identify environmental characteristics that influence the abundance and activity of ammonia oxidizers and nitrite oxidizers, the two major groups of microorganisms that perform nitrification [3]. However, despite the importance of nitrification in the productivity of woodlands, datasets from these ecosystems are rare [4], though they could be useful in improving our understanding of the role of abiotic and biotic parameters that influence nitrifiers in forest soils. In addition, the role of heterotrophic nitrification in such ecosystems cannot be ruled out, but has been reported in relatively few studies [5, 6].
We recently identified soil pH as a key driver of the composition of ammonia-oxidizing microbes in soils, potentially through its influence on the ammonia:ammonium ratio and described a highly specialized group of ammonia-oxidizing archaea (AOA) that occurs mainly in extremely acidic soils (pH 3.5–4.5), with consequent low availability of ammonia [7]. However, as the aim of the underlying study (UK Countryside Survey) was to characterize a large number of different soils with different land use types, other co-variables may have influenced AOA communities, and most of the acidic soils were from moorlands. Furthermore, the behavior of AOA in ultra-acidic soils, defined here as those with pH <3.5, where ammonia availability is even more reduced than in extreme acidic soils, is not clear. Therefore, in the present study, we focused on one land use type (forest) and included sites with soil pH values from 6.8 to 3.2 to (i) evaluate the role of pH as a driver for AOA in forest soil, (ii) evaluate the occurrence of the AOA clade in ultra-acidic forest soils that has been identified as a major contributor to ammonia oxidation in moorland soils with extreme acidic soil pH, and (iii) assess potential relationships between the ammonia oxidizer community structure and the potential nitrification rate.
Material and Methods
Topsoil was sampled from 27 experimental forest plots in Germany, which are part of the three sites of the Biodiversity Exploratories ( www.biodiversity-exploratories.de/): Schwäbische Alb (ALB, sites AEW1-9; Southern Germany), Hainich-Dün (HAI, sites HEW1-9, Central Germany), and Schorfheide-Chorin (SCH, sites SEW1-9, Northern Germany). Major study site characteristics are given in the Supplemental Material and Table S1. In brief, study sites of regions ALB and SCH were composed of age-class forests and natural forests; whereas in HAI besides age-class forests, single-tree selection systems were the predominant forest types. Spruce and beech tree species are prevalent in ALB and HAI; and at SCH sites, the vegetation is composed of beech and pine stands.
The soil pH was measured using 10 g sieved air-dried soil in 25 ml of 0.01 M CaCl2 solution. Based on these results, soils were grouped according to the following classification by the Agriculture Natural Resources Conservation Service (United States Department of Agriculture, USDA): ultra acidic (<3.5), extreme acidic (3.5–4.4), very strong acidic (4.5–5.0), strong acidic (5.1–5.5), moderate acidic (5.6–6.0), slight acidic (6.1–6.5), and neutral (6.6–7.3). Thus, the experimental design included 9 sites from each of the three exploratories, representing a pH gradient from ultra acidic (<3.5) to acido-neutral (>6.0). As the pH of the litter layer did not always follow the trend of the underlying mineral soil layers, and the pH of the litter layer is highly dynamic temporally, in response to the degradation status of the litter material, only the mineral soils were used for further analysis.
Ammonium and nitrate concentrations were determined colorimetrically [8]. The abundances of AOA and AOB were determined from DNA extracts based on the ammonia monooxygenase subunit A gene (amoA) using quantitative PCR (qPCR) [7, 8], and the diversity of AOA was assessed by amplicon-based pyrosequencing of amoA genes [7]; for details, see (Supplemental Material).
Results and Discussion
Archaeal amoA genes were detected in all DNA extracts derived from forest soils, even in those from ultra-acidic soils. The abundance of archaeal amoA genes ranged from 1 × 104 to 3.4 × 107 g−1 soil dry weight (Figure S1 A), with the highest abundance at the sites with a soil pH ranging from 3 to 4.5. The abundance of bacterial amoA genes was in the range of 2 × 104 to 4 × 106 g−1 soil dry weight (Figure S1 B), and AOA were more abundant than AOB in the majority of the sites, as assessed by the amoA gene abundance. This is consistent with the previous studies performed in some agricultural soils [8, 9], although AOB are often more abundant particularly in well-fertilized soils [10]. The highest AOA:AOB ratios were found at sites with the lowest pH values (SEW3,4,9; data not shown), confirming the previous data published by [11]. In the observed forest sites, archaeal abundance (determined via 16S rRNA gene abundance, data not shown) ranged from 106 to 107 g−1 dw soil in ALB and slightly higher values in HAI. The abundance was higher in SCH with values up to 2 × 108 g−1 dw soil. The ratios of archaeal 16S rRNA:archaeal amoA genes were similar in ALB and HAI sites (0.8–4.3 for ALB and 0.3–5.3 for HAI), with a few plots with ratios as high as 24. In the SCH region, the mean ratio was higher (mean value 24) with only two sites with ratios above 10 (data not shown). Bacterial 16S rRNA gene abundance was in the range of 108 for all sites. The ratios of 16S rRNA:bacterial amoA gene abundance were similar in range to HAI (mean value ca. 450) but varied strongly in ALB and SCH (80–4011 for ALB and 360–49,077 for SCH). A number of factors, in addition to pH, have been suggested as determining niche differentiation between AOA and AOB. These include ammonia concentration and mixotrophy, but both are based largely on the analysis of a very small number of laboratory cultures, with little evidence for either in soil [12]. A further factor with potential to influence relative abundance of AOA and AOB is the allelopathic inhibition of nitrification, and the allelopathic inhibition by monoterpenes or polyphenolic compounds on nitrification has been reported in coniferous forest soils [13]. This may explain the lower abundance for amoA genes in SCH sites with pine stands, but there is currently no evidence of the differential inhibition of AOA and AOB by such compounds. As pH influences the equilibrium of ammonium to ammonia, the latter being substrate for ammonia oxidation, an effect of different pH levels on PNR is expected. Interestingly, potential nitrification rate was weak, negatively correlated to soil pH (r = −0.518) as expected, but was influenced more by site-specific conditions, such as soil water content (r = −0.706), see table (S2). Thus, the highest potential nitrification rates were found in soils from the Schorfheide region, where soil water content was lowest (Table S1). Ammonium and nitrate concentrations at the time of sampling were in the range of 0.1–4.4 mg ammonium kg−1 dw soil and 0.5–7.7 mg nitrate kg−1 dw soil, and neither were correlated with AOA abundance (AOA—NO3 r = −0.063 and AOA—NH4 r = −0.221) or potential nitrification rate (PNR—NO3 r = −0.403 and PNR—NH4 r = −0.177), see table (S2). However, if AOB are favored by higher ammonia concentrations, the relatively high concentration of ammonia used in PNR assays will lead to greater rates in soils with high AOB:AOA ratios, while use of chlorate to inhibit nitrite oxidation may also favor AOB, which may be less sensitive to nitrite toxicity.
To investigate potential relationships between pH and AOA community structure, an amoA gene-targeted OTU-based approach was performed. To characterize the diversity of archaeal amoA gene-harboring communities for each site, indices for richness (Chao1) and evenness (Shannon) were calculated (Figure S3). AOA communities of sites with higher soil pH comprised remarkably higher OTU richness than sites with a more acidic soil pH. The distribution of OTUs was more even at sites with low pH, while a lower number of highly abundant OTUs dominated AOA communities at higher pH values. These observations indicate that pH indeed shapes the evenness and richness of AOA communities in forest soils as they seem to be more different under high pH levels, whereas fewer AOA seem to be adapted to low pH. This interpretation must, however, be treated with caution as saturation of rarefaction curves was not approached in the majority of the sites, indicating that full coverage had not been achieved (Figure S2), and comparisons between sites are complicated by analysis of different numbers of sequences. Two clusters dominated the archaeal amoA gene-harboring communities (Figure S4), on the basis of OTU-based dissimilarity, which clearly followed the pH values of the particular sites at all three study regions. The dominant clusters were separated at a pH of 4.6. The main clusters were confirmed by unifrac and parsimony tests (p <0.001). Interestingly, cluster A-included sites showed a pH <4.7 with generally reduced amoA gene diversity and more heterogeneous composition than cluster B.
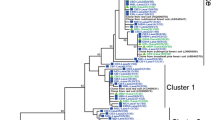
Overall, 10 major OTUs (representing >1 % in any sample) could be distinguished and were shared between the two clusters (Figure S5). OTUs 1 and 4 were highly abundant in ultra-acidic soils (<pH 3.5); whereas OTU 6 was highly abundant in one sample with pH 4.1, and OTU 2 was mainly representative of the sites with neutral-acidic soil pH (>pH 4.7). The phylogenetic analysis (Fig. 1) revealed OTU 4 to be closely related to the Nitrososphaera subcluster 7.2, confirming the data of [7] who also described a major group of AOA from agricultural soils with an ultra-acidic pH belonging to the same group. In contrast, OTU 1 showed large similarities to Nitrosotalea; this OTU has also been observed in an ultra-acidic upland soil in China [14].
Maximum likelihood phylogenetic analysis of partial archaeal amoA gene sequences detected in soil samples of grassland and forest (clustered at 97 % sequence identity, OTU1-99). Color code indicates the range of soil pH values where sequences were derived from (red, pH <3.5; pink, pH 3.6–4.6; blue, pH >4.7). Bar plots show relative abundance of each OTU in the different samples. Numbers in brackets show absolute abundance of the OTU. Number of OTUs derived from grassland: 45, forest; 10 and shared, 36. a OTUs related to the Nitrosopumilus/Nitrosotalea/Nitrososphaera sister cluster. b OTUs showing similarity to the species of the Nitrososphaera cluster. The grey bars indicate the most abundant sequences
The dataset of this experiment was compared with the sequences obtained in a previous study of agricultural soils [7], and the importance of soil pH for the AOA community structure was assessed using the same pipeline, including PCR and sequence data processing. The principal component analysis (PCA) (Fig. 2) indicated that agricultural and forest sites with a soil pH >4.5 showed little similarity in terms of shared AOA OTUs, whereas communities were similar in soils with pH < 3.5, regardless of vegetation type (agricultural vs. forest) or location and soil type. The PCA on grassland and forest sites was supplemented by further statistical analyses based on a PCA of the forest sites of the study (data not shown) to test the influence of selected environmental parameters, including soil moisture content, pH, ammonium, and nitrate, on the distribution of OTUs. Univariate regression of the first principal component revealed that pH value explained 43.8 % of variance, whereas soil moisture content accounted for only 19.6 % (data not shown). The percentages of explained variance for ammonium and nitrate concentration were negligibly low (0 % and 13.8 %, respectively). This supports our assumption that indeed, pH and not soil moisture content affects OTU composition in our forest sites.
Principal component analysis (PCA) of amoA gene-defined community structure in soils sampled from grassland (prefixed with GL) and forest (prefixed with F) with the influence of OTUs: length of the arrows show the strength of influence, the direction shows loading to samples investigated. R software was used for calculations. Datasets from the study of [7] and this study were compared to assess the relevance of pH as a general driver of AOA community structure
The importance of pH as driver of AOA composition in forest soils was further confirmed by a permutation test conducted for canonical correspondence analysis (p value <0.05, data not shown). This again clearly indicates that soil pH is a major driver of AOA community distribution in forest soils.
References
Boring L, Swank W, Waide J, Henderson G (1988) Sources fates and impacts of nitrogen inputs to terrestrial ecosystems: review and synthesis. Biogeochemistry 6:119–159
McGuirre D, Melillo J, Joyce L (2006) The role of nitrogen in the response of forest net primary production to elevated atmospheric carbon dioxide. Annu Rev Ecol Syst 26:473–503
Le Roux X, Poly F, Currey P, Commeaux C, Hai B, Nicol GW, Prosser JI, Schloter M, Attard E, Klumpp K (2008) Effects of aboveground grazing on coupling among nitrifier activity, abundance and community structure. ISME J 2:221–232
Long X, Chen C, Xu Z, Linder S, He J (2012) Abundance and community structure of ammonia-oxidizing bacteria and archaea in a Sweden boreal forest soil under 19-year fertilization and 12-year warming. J Soils Sediments 12:1124–1133
De Boer W, Kowalchuk GA (2001) Nitrification in acid soils: micro-organisms and mechanisms. Soil Biol Biochem 33:853–866
Pennington P, Ellis R (1993) Autotrophic and heterotrophic nitrification in acidic forest and native grassland soils. Soil Biol Biochem 25:1399–1408
Gubry-Rangin C, Hai B, Quince C, Engel M, Thomson BC, James P et al (2011) Niche specialization of terrestrial archaeal ammonia oxidizers. Proc Natl Acad Sci 108:21206–21211
Hai B, Diallo NH, Sall S, Haesler F, Schauss K, Bonzi M et al (2009) Quantification of key genes steering the microbial nitrogen cycle in the rhizosphere of sorghum cultivars in tropical agroecosystems. Appl Environ Microbiol 75:4993–5000
Adair K, Schwartz E (2008) Evidence that ammonia-oxidizing archaea are more abundant than ammonia-oxidizing bacteria in semiarid soils of northern Arizona, USA. Microb Ecol 56:420–426
Di HJ, Cameron KC, Shen JP, Winefield CS, O’Callaghan M, Bowatte S et al (2009) Nitrification driven by bacteria and not archaea in nitrogen-rich grassland soils. Nat Geosci 2:621–624
Nicol GW, Leininger S, Schleper C, Prosser JI (2008) The influence of soil pH on the diversity, abundance and transcriptional activity of ammonia-oxidizing archaea and bacteria. Environ Microbiol 10:2966–2978
Prosser JI, Nicol GW (2012) Archaeal and bacterial ammonia oxidisers in soil: the quest for niche specialisation. Trends Microbiol 20:523–531
Paavolainen L, Smolander A (1998) Nitrification and denitrification in soil from a clear-cut Norway spruce (Picea abies) stand. Soil Biol Biochem 30:775–781
He J, Shen J, Zhang L, Zhu Y, Zheng Y, Xu M et al (2007) Quantitative analyses of the abundance and composition of ammonia-oxidizing bacteria and ammonia-oxidizing archaea of a Chinese upland red soil under long-term fertilization practices. Environ Microbiol 9:2364–2374
Acknowledgments
We thank Theresa Klötzung for her excellent technical assistance with the laboratory work. We thank the managers of the three exploratories, Swen Renner, Sonja Gockel, Kerstin Wiesner, and Martin Gorke, for their work in maintaining the plot and project infrastructure; Simone Pfeiffer and Christiane Fischer for giving support through the central office; Michael Owonibi for managing the central data base; and Markus Fischer, Eduard Linsenmair, Dominik Hessenmöller, Jens Nieschulze, Daniel Prati, Ernst-Detlef Schulze, Wolfgang W. Weisser, and the late Elisabeth Kalko for their role in setting up the Biodiversity Exploratories project. The work has been (partly) funded by the DFG Priority Program 1374 “Infrastructure-Biodiversity-Exploratories”. Fieldwork permits were issued by the responsible state environmental offices of Baden-Württemberg, Thüringen, and Brandenburg (according to § 72 BbgNatSchG).
Author information
Authors and Affiliations
Corresponding author
Additional information
Barbara Stempfhuber, Marion Engel and Doreen Fischer have contributed equally to this work.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 4.08 mb)
Rights and permissions
About this article
Cite this article
Stempfhuber, B., Engel, M., Fischer, D. et al. pH as a Driver for Ammonia-Oxidizing Archaea in Forest Soils. Microb Ecol 69, 879–883 (2015). https://doi.org/10.1007/s00248-014-0548-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-014-0548-5