Abstract
Autotrophic ammonia-oxidizing communities, which are responsible for the rate-limiting step of nitrification in most soils, have not been studied extensively in semiarid ecosystems. Abundances of soil archaeal and bacterial amoA were measured with real-time polymerase chain reaction along an elevation gradient in northern Arizona. Archaeal amoA was the predominant form of amoA at all sites; however, ratios of archaeal to bacterial amoA ranged from 17 to more than 1,600. Although size of ammonia-oxidizing bacteria populations was correlated with precipitation, temperature, percent sand, and soil C/N, there were no significant relationships between ammonia-oxidizing archaea populations and any of the environmental parameters evaluated in this study. Our results suggest that in these soils, archaea may be the primary ammonia oxidizers, and that ammonia-oxidizing archaea and ammonia-oxidizing bacteria occupy different niches.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Nitrification is the biological oxidation of ammonia (NH3) to nitrate (\({\text{NO}}_{\text{3}}^{\text{ - }} \)) and an essential step in the global nitrogen cycle. In soils, nitrification can result in nitrogen loss from an ecosystem. \({\text{NO}}_{\text{3}}^{\text{ - }} \) is leached much more readily than NH3. It also serves as an electron acceptor for denitrification, the reduction of \({\text{NO}}_{\text{3}}^{\text{ - }} \) to the gaseous forms NO, N2O, and N2. Nitrate leaching can result in groundwater pollution, and N2O is a potent greenhouse gas. The first and rate-limiting step of nitrification is the oxidation of NH3 to nitrite (\({\text{NO}}_{\text{2}}^{\text{ - }} \)). Extensive research has focused on ammonia-oxidizing microorganisms because their activity impacts agricultural yields, water quality, and global climate change [10].
Two groups of organisms, a monophyletic subset of the β-Proteobacteria and members of the non-extremophilic Crenarchaeota, are responsible for ammonia oxidation in most soils [11, 13]. While a robust body of research has demonstrated that autotrophic ammonia-oxidizing bacteria (AOB) contribute to soil ammonia oxidation, the role of ammonia-oxidizing archaea (AOA) remains unclear. Several lines of evidence suggest that AOA may be important to nitrification. First, metagenomic studies revealed that some crenarchaea contain genes with high similarity to the ammonia monooxygenase genes of AOB [17, 18]. Second, archaeal and bacterial versions of these genes are transcribed in the same soils [11]. Third, a chemoautotrophic crenarchaea that oxidizes NH3 to \({\text{NO}}_{\text{2}}^{\text{ - }} \) aerobically was isolated from an aquarium verifying that archaea capable of ammonia oxidation exist [9].
Ammonia monooxygenase, an enzyme essential for autotrophic ammonia oxidation, is encoded by the genes amoA, B, and C in AOB and AOA [13]. The amoA gene codes for the subunit containing the active site, and primer sets have been developed that detect either archaeal amoA or bacterial amoA in environmental samples [5, 16]. If the number of amoA copies per cell is similar in AOB and AOA, population densities of the two groups of ammonia oxidizers can be compared with real-time polymerase chain reaction (PCR) methods. Cultured AOB have two to three copies of the amoA gene [14]. Much less is known about the genome structure of AOA, but a study has suggested that marine AOA have one to three copies of amoA per cell [19].
A few studies have used amoA as a biomarker to compare the size of AOA and AOB populations. In a time series from the coastal North Sea, bacterial amoA was present in equal to or slightly greater numbers than archaeal amoA for most of the year, except from November until March when spikes in archaeal amoA abundance coincided with decreases in NH3 and increases in \({\text{NO}}_{\text{3}}^{\text{ - }} \). Bacterial amoA increased slightly during this period, but archaeal amoA abundance was 1–2 orders of magnitude greater than bacterial amoA [19]. In samples taken from a variety of European grassland and agricultural soils, archaeal amoA was always more abundant than bacterial amoA, although the ratio of archaeal amoA to bacterial amoA varied from 1.5 to more than 3,000. Increasing soil depth had an impact on this ratio due to declines in bacterial amoA; however, this result was not consistent across soil types [11]. To gain a clear picture of what factors regulate ammonia-oxidizing populations, AOA and AOB must be studied in a wide variety of habitats. Results from one system may be specific to the organisms found there.
Population sizes of AOB and AOA in semiarid soils have not yet been reported, and it remains unclear whether environmental factors will impact both ammonia-oxidizing populations in these systems similarly. The goal of this study was to compare the abundances of amoA genes, which served as a proxy for population size of AOB and AOA, along an elevation gradient in northern Arizona. Through regression analysis, the influence of environmental factors on the population densities of these two groups was evaluated.
Methods
Soils sampled in this study are from the C. Hart Merriam Elevation Gradient (http://www.mpcer.nau.edu/gradient) located on the San Francisco Peaks in northern Arizona (35°N latitude and 111°W longitude). The regional climate is semiarid, but sites along the 1,064 m gradient differ in temperature, precipitation, dominant vegetation, and soil type (Table 1). Five surface soil cores (4 cm diameter, 10 cm depth) were harvested from randomly chosen locations within 36 m2 sampling plots placed in open grassy areas at each site in May (post-snowmelt) and August (post-monsoon).
DNA was isolated from 0.5 g of frozen soil with the PowerSoil™ DNA Isolation Kit (MoBio Laboratories, Carlsbad, CA, USA) and further purified by ethanol precipitation. Cell lysis was achieved by bead beating for 15 s at setting 5.0 in a FastPrep® Instrument (Q-Bio Gene, Morgan Irvine, CA, USA), replacing step 5 of the MoBio protocol. Concentration and purity of DNA was determined with a BioPhotometer (Eppendorf, Hamburg, Germany), and all extractions were diluted to 5 ng DNA μl−1 in Tris-EDTA buffer.
Using the primers Arch-amoAF and Arch-amoAR [5], a 635 bp fragment of the archaeal amoA gene was amplified from samples harvested at the grassland site in August 2004. PCR reactions contained 0.2 μM primers, 1X PCR buffer, 0.2 mM dNTPs, 2 mM MgCl2, and 1 U of Platinum Taq DNA polymerase (Invitrogen, Carlsbad, CA, USA) in a final volume of 50 μl and were run in a PT-100 thermocycler (MJ research, Waltham, MA, USA). The cycling protocol was 2 min at 94°C followed by 30 cycles of 30 s at 94°C, 60 s at 55°C, 45 s at 72°C, and a final extension of 7 min at 72°C. Quality of PCR products was determined visually on a 0.5X TBE, 1.5% agarose gel. PCR products were cloned with the TOPO TA Cloning® Kit (Invitrogen), sequenced, and analyzed on a 3730xl DNA Analyzer (Applied Biosystems, Foster City, CA, USA). BLAST searches against the GenBank database verified that the PCR products were most closely related to other archaeal amoA sequences.
Archaeal amoA clones were pooled to create a standard curve of known archaeal amoA concentrations. Plasmids were extracted with the Perfect Prep Plasmid Mini Kit (Eppendorf, Hamburg, Germany) and quantified with a BioPhotometer. From the plasmid concentration and the length of the PCR®4-TOPO® vector (Invitrogen) with a 635 bp insert, amoA copy concentration was calculated. A series of 1:10 dilutions, ranging from 102 to 109 copies, was used as a standard curve for quantifying the abundance of archaeal amoA in soil DNA extractions. A 491-bp fragment of bacterial amoA was amplified from DNA extracted from the grassland site using primers amoA-1F and amoA-2R [16]. PCR conditions and the cycling protocol were identical to those described previously, except the extension step was decreased from 45 to 30 s. PCR products were cloned. Plasmids were extracted, quantified, and diluted to create a standard curve of known bacterial amoA concentration as described for archaeal amoA.
Real-time PCRs for both bacterial and archaeal amoA were run in an DNA Engine Opticon Real-Time PCR System (MJ Research) under the following conditions: 0.2 μM primers, 1X PCR buffer, 0.2 mM dNTPs, 4 mM MgCl2, 1.5 U of Platinum Taq DNA polymerase, and 0.25X sybr green (Invitrogen) in a final volume of 50 μl. Cycling protocols were 2 min at 94°C followed by 40 cycles of 30 s at 94°C, 60 s at 55°C, 45 s at 72°C for archaeal amoA or 30 s for bacterial amoA, and a final extension of 7 min at 72°C. Purity of PCR products was verified with melting curves. Threshold cycle (CT) was designated as the point when fluorescence intensity reached four times the standard deviation of baseline fluorescence.
Several soil characteristics predicted to affect the size of ammonia-oxidizing populations were measured. \({\text{NH}}_{\text{4}}^{\text{ + }} \) and \({\text{NO}}_{\text{3}}^{\text{ - }} \) concentrations of filtered 2 M KCl soil extractions were quantified by automated spectrophotometric analysis on a QuikChem® 8500 FIA System (Lachat Instruments, Loveland, CO, USA). The gravimetric water content of each soil sample was determined by calculating mass loss after drying 5 g of soil at 100°C for 24 h. Temperature and precipitation data were collected by weather stations located at each elevation gradient site. Soil pH was measured in water as described in Hendershot, Lalande, and Duquette (1993) [7]. Sieves, 2 and 0.05 mm, were used to determine soil texture. Soil carbon and nitrogen were measured with a Carlo Erba NC2100 Elemental Analyzer at the Colorado Plateau Stable Isotope Laboratory.
Results of the real-time PCRs were expressed in relative abundance, amoA copies per nanogram of DNA. These values were multiplied by the amount of DNA extracted from the soil sample to determine absolute abundance, amoA copies per gram of dry soil. To compare amoA copies per gram of soil among sites, it is necessary to assume DNA extraction efficiencies are similar for all soils. The benefit of absolute gene abundances is that they reflect differences among the sites in total microbial community size, which are substantial along the elevation gradient. This perspective is absent when relative abundances are compared. JMP 6.0 (SAS Institute, Cary, NC, USA) was used to run analyses of variance on amoA abundance by site, t tests to determine if there were differences in amoA abundances between sampling dates, and linear regressions of log transformed amoA abundances versus environmental factors.
Results
There were linear relationships between target gene copy number and CT values for bacterial amoA (y = −3.744 x +46.114; R 2 = 0.998) and archaeal amoA (y = −3.817 x +45.121; R 2 = 0.999) over a range of seven orders of magnitude with the real-time PCR protocols used in this study. The two primer sets had similar amplification efficiencies, 82.82% for archaeal amoA and 84.51% for bacterial amoA. Detection limits were ∼130 archaeal amoA copies and ∼200 bacterial amoA copies (Fig. 1).
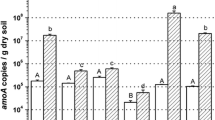
Archaeal amoA was more abundant than bacterial amoA at all sites along the C. Hart Merriam Elevation Gradient on both sampling dates (Fig. 2). The greatest differences in archaeal and bacterial amoA copies were found at the pinyon-juniper site in May and the grassland site in August where archaeal amoA was over three orders of magnitude more abundant than bacterial amoA. Mean archaeal amoA to bacterial amoA ratios were lowest at the ponderosa site, being 17 in August and 33 in May.
Abundance of bacterial amoA (open bars) and archaeal amoA (filled bars), expressed in copies per nanogram of DNA and copies per gram of soil, at the C. Hart Merriam Elevation Gradient sites in May and August 2004. Capital letters refer to significant differences determined by Tukey’s HSD (p < 0.05) in bacterial amoA abundance among sites at one sampling date, lowercase letters denote significant differences in archaeal amoA abundance. Site names are abbreviated: GB great basin, GL grassland, PJ pinyon-juniper, PP ponderosa pine, MC mixed conifer
There were significant differences in bacterial amoA abundance among elevation gradient sites, regardless of whether the data was expressed in copies per nanogram of DNA (May, F = 5.299, p < 0.01; August, F = 5.650, p < 0.01) or copies per gram of soil (May, F = 6.826, p < 0.01; August, F = 13.187, p < 0.001; Fig. 2). Archaeal amoA abundance also differed among elevation gradient sites when data was expressed in copies per nanogram of DNA (May, F = 6.713, p < 0.01; August, F = 5.443, p < 0.01) and copies per gram of soil (May, F = 10.505, p < 0.001; August, F = 22.971, p < 0.001; Fig. 2). Bacterial amoA abundance did not differ between the two sampling dates at any site except the pinyon-juniper where abundance was greater in August than in May (copies per nanogram of DNA, t = 3.446, p < 0.05; copies per gram of soil, t = 4.095, p < 0.01). Archaeal amoA abundance was greater in May than August at the great basin (copies per nanogram of DNA, t = 3.102, p < 0.05; copies per gram of soil, t = 4.409, p < 0.01) and pinyon-juniper sites (copies per nanogram of DNA, t = 6.216, p < 0.001; copies per gram of soil, t = 4.987, p < 0.001). At the ponderosa and mixed conifer sites, archaeal amoA abundance was greater in May than August when expressed in copies per nanogram of DNA (ponderosa, t = 2.638, p < 0.05; mixed conifer, t = 2.640, p < 0.05). However, seasonal differences in archaeal amoA abundance at the two highest elevation sites were not significant when abundances were expressed in copies per gram of soil. At the grassland site, archaeal amoA did not differ between sampling dates.
To determine what environmental factors influenced the abundance of archaeal and bacterial amoA in northern Arizona soils, a series of linear regressions were performed. Archaeal amoA abundance was not significantly related to any of the measured environmental variables, regardless of whether relative or absolute gene abundances were considered (Table 2). In contrast, regressions of precipitation, air temperature, and soil C/N against absolute bacterial amoA abundance (in copies per gram of soil) were all highly significant (Table 2, Fig. 3). In a multivariate analysis, these variables together accounted for nearly 80% of the variation found in bacterial amoA copies per gram of soil. There was a less significant relationship between bacterial amoA copies per gram of soil and percent sand by mass (Table 2). Soil C/N was the only measured parameter to be correlated with both relative and absolute bacterial amoA abundance (Table 2).
Discussion
Archaeal amoA was more abundant than bacterial amoA in all soils analyzed in this study, regardless of whether samples were taken during the post-snowmelt or post-monsoon seasons. Other studies have shown that in grassland and agricultural soils from Europe, as well as in the North Sea, archaeal amoA was more abundant than bacterial amoA [11, 19]. Our results from semiarid soils in northern Arizona add to the growing list of ecosystems where archaea appear to be the predominant ammonia oxidizer. This conclusion relies on the assumption that the number of amoA copies is similar in the genomes of AOA and AOB. An alternative explanation is that the genomes of AOA contain many more amoA copies than the genomes of AOB and differences in amoA copies do not reflect differences in cell densities. However it seems unlikely that AOA would have many more than the two to three amoA copies found in cultured AOB [14]. It is also possible that higher abundances of archaeal amoA were measured because AOA are more susceptible to lysis than AOB. AmoA abundance, expressed in copies per nanogram of DNA, can be interpreted as the relative abundance of AOB or AOA in a DNA extraction. While this is a useful measurement, it does not provide any information about the population densities of ammonia oxidizers in soil. By multiplying amoA copies per nanogram of DNA in an individual sample by the total amount of DNA extracted from that sample, amoA copies per gram of soil can be estimated. Recovery of DNA from soil may be influenced by soil type and microbial community composition [20]. Therefore, our estimates of amoA copies per gram of soil, that assume equal extraction efficiencies, could be biased toward sites with higher DNA extraction efficiencies. This assumption does not affect the comparison of archaeal versus bacterial amoA copies within a site.
Absolute bacterial amoA abundance, expressed in copies per gram of soil, was greatest at the sites with lower air temperatures and higher annual precipitation. While the individual effects of each of these climate variables cannot be determined from this study, there are several mechanisms by which climate could influence AOB populations. Temperature has been shown to directly impact AOB. Nitrifying activity in soil has an optimum temperature of 35°C, potential nitrification is highest when soils are incubated at moderate temperatures, and AOB community structure can be affected by incubation temperature [2, 3]. AOB growth may be limited in the lower elevation soils, which experience extremely high temperatures during the summer and are not shaded due to very sparse plant cover. These conditions will also result in decreased soil moisture, another factor known to be important in regulating AOB population size [3, 6, 8]. The role of soil moisture in determining bacterial amoA abundance in northern Arizona soils is supported by a positive relationship with precipitation. However, the regression of bacterial amoA abundance and soil moisture measured at the time of sampling was not significant. Soil moisture fluctuates dramatically in pulse-driven systems like those found in the southwestern USA [1]. It has been suggested that bacterial communities may be adapted to the moisture regime that they experience in the environment [4]. Populations of AOB, which have shown some resistance to desiccation, are likely to change on much slower time scales than soil moisture will fluctuate [3, 6]. The lack of significant relationships between archaeal amoA and temperature or precipitation suggests that AOA and AOB differ in their resistance to desiccation.
As the energy source for ammonia oxidizers, \({\text{NH}}_{\text{4}}^{\text{ + }} \) availability is likely to impact AOB and AOA populations. We did not observe significant relationships between bacterial or archaeal amoA abundance and \({\text{NH}}_{\text{4}}^{\text{ + }} \) concentration at the time of sampling. The frequent soil moisture fluctuations described previously result in pulses of nitrogen mineralization after precipitation events followed by periods of immobilization [1]. Studies utilizing molecular techniques in agricultural soils have suggested that AOB populations exhibit long-term, rather than short-term responses to changes in \({\text{NH}}_{\text{4}}^{\text{ + }} \). In soils that had been fertilized for more than 100 years there was no change in the abundance of AOB 3 days after fertilization and in a microcosm study abundance of AOB did not peak until 7 days after addition of ammonium [12, 15]. Due to slow-growth rates and the ability to maintain stable population density during periods of low \({\text{NH}}_{\text{4}}^{\text{ + }} \) availability, AOB abundance was greater in fertilized than control soils even eight and twelve months after \({\text{NH}}_4 ^{\text{ + }} \) treatment [12, 15]. Frequent changes in the nitrogen mineralization/immobilization status of semiarid soils, and long response times of AOB to both favorable and unfavorable conditions, may explain the absence of a relationship between \({\text{NH}}_{\text{4}}^{\text{ + }} \) availability and AOA or AOB abundance. \({\text{NO}}_{\text{3}}^{\text{ - }} \), the product of nitrification, was not significantly related to bacterial or archaeal amoA abundance in these soils. This is not surprising as both plants and microbes can utilize \({\text{NO}}_{\text{3}}^{\text{ - }} \) as a nitrogen source, and thus net changes in the \({\text{NO}}_{\text{3}}^{\text{ - }} \) pool do not solely reflect nitrifying activity.
The significant positive relationship between bacterial amoA abundance and soil C/N was an unexpected result. Under high C/N nitrogen demand by heterotrophs is high, and slow-growing AOB and presumably AOA face competition for available \({\text{NH}}_{\text{4}}^{\text{ + }} \) and oxygen. As a result, a negative relationship between AOB and soil C/N was expected. It is possible that a substantial portion of carbon may be in a recalcitrant form, making the C/N of bioavailable compounds lower than that of the total soil. Alternatively, soil C/N may covary with another environmental factor, such as net primary productivity, that impacts bacterial amoA abundance.
While our results, as well as those from other environments, suggest that AOA may be the predominant ammonia oxidizers, it is important to recognize that the contribution of AOA to ammonia oxidation rates has not yet been quantified. The lack of significant relationships between archaeal amoA abundance and environmental parameters indicates that there is much we do not yet understand about AOA. To gain a clear understanding of what factors regulate AOA and AOB populations, future studies should manipulate environmental parameters and use techniques that allow growth of ammonia-oxidizing organisms to be monitored, such as stable isotope probing.
References
Austin AT, Yahdjian L, Stark JM, Belnap J, Porporato A, Norton U, Ravetta DA, Schaeffer SM (2004) Water pulses and biogeochemical cycles in arid and semiarid ecosystems. Oecologia 141:221–235
Avrahami S, Liesack W, Conrad R (2003) Effects of temperature and fertilizer on activity and community structure of soil ammonia oxidizers. Environ Microbiol 5:691–705
Belser LW (1979) Population ecology of nitrifying bacteria. Annu Rev Microbiol 33:309–333
Fierer N, Schimel JP, Holden PA (2003) Influence of drying-rewetting frequency on soil bacterial community structure. Microb Ecol 45:63–71
Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB (2005) Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. P Natl Acad Sci U S A 102:14683–14688
Hastings RC, Butler C, Singleton I, Saunders JR, McCarthy AJ (2000) Analysis of ammonia-oxidizing bacteria populations in acid forest soil during conditions of moisture limitation. Lett Appl Microbiol 30:14–18
Hendershot WH, Lalande H, Duquette M (1993) Soil reaction and exchangeable acidity. In: Carter MR (ed) Soil sampling and methods of analysis. Lewis, Boca Raton, pp 141–145
Horz H-P, Barbrook A, Field CB, Bohannan BJM (2004) Ammonia-oxidizing bacteria respond to multifactorial climate change. P Natl Acad Sci U S A 101:15136–15141
Könneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA (2005) Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437:543–546
Kowalchuk GA, Stephen JR (2001) Ammonia-oxidizing bacteria: A model for molecular microbial ecology. Annu Rev Microbiol 55:485–529
Leininger S, Urich T, Schloter M, Schwark L, Qi J, Nicol GW, Prosser JI, Schuster SC, Schleper C (2006) Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442:806–809
Mendum TA, Sockett RE, Hirsch PR (1999) Use of molecular and isotopic techniques to monitor the response of autotrophic ammonia-oxidizing populations of the β subdivision of the class Proteobacteria in arable soils to nitrogen fertilizer. Appl Environ Microbiol 65:4155–4162
Nicol GW, Schleper C (2006) Ammonia-oxidising Crenarchaeota: important players in the nitrogen cycle? Trends Microbiol 14:207–212
Norton JM, Alzerreca JJ, Suwa Y, Klotz MG (2002) Diversity of ammonia monooxygenase operon in autotrophic ammonia-oxidizing bacteria. Arch Microbiol 177:139–149
Okano Y, Hristova KR, Leutenegger CM, Jackson LE, Denison RF, Gebreyesus B, Lebauer D, Scow KM (2004) Application of real-time PCR to study effects of ammonium on population size of ammonia-oxidizing bacteria in soil. Appl Environ Microbiol 70:1008–1016
Rotthauwe J-H, Witzel K-P, Liesack W (1997) The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol 63:4704–4712
Schleper C, Jurgens G, Jonuscheit M (2005) Genomic studies of uncultivated archaea. Nat Rev Microbiol 3:479–488
Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Dongying W, Paulsen I, Nelson KE, Nelson W, Fouts DE, Levy S, Knap AH, Lomas MW, Nealson K, White O, Peterson J, Hoffman J, Parson R, Baden-Tillson H, Pfannkoch C, Rogers Y-H, Smith HO (2004) Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66–74
Wuchter C, Abbas B, Coolen MJL, Herfort L, van Bleijswijk J, Timmers P, Strous M, Teira E, Herndl GJ, Middelburg JJ, Schouten S, Damsté JSS (2006) Archaeal nitrification in the ocean. P Natl Acad Sci U S A 103:12317–12322
Zhou J, Bruns MA, Tiedje JM (1996) DNA recovery from soils of diverse composition. Appl Environ Microbiol 62:316–322
Acknowledgments
We would acknowledge the following funding sources: Office of Science (BER), U.S. Department of Energy, Grant no. DE-FG02–04ER63883, the National Science Foundation, Grant No. DEB-0416223. K.A. was supported by an NSF IGERT fellowship (grant no. DGE-0549505). We would like to thank Paul Dijkstra, Joey Blankinship, and Ben Moan for their assistance with field work and site data.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Adair, K.L., Schwartz, E. Evidence that Ammonia-Oxidizing Archaea are More Abundant than Ammonia-Oxidizing Bacteria in Semiarid Soils of Northern Arizona, USA. Microb Ecol 56, 420–426 (2008). https://doi.org/10.1007/s00248-007-9360-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-007-9360-9