Abstract
Our goals were to evaluate the tolerance of mesorhizobia to acid and alkaline conditions as well as to investigate whether acid tolerance is related to the species or the origin site of the isolates. In addition, to investigate the molecular basis of acid tolerance, the expression of chaperone genes groEL and dnaKJ was analyzed using acid-tolerant and sensitive mesorhizobia. Tolerance to pH 5 and 9 was evaluated in liquid medium for 98 Portuguese chickpea mesorhizobia belonging to four species clusters. All isolates showed high sensitivity to pH 9. In contrast, mesorhizobia revealed high diversity in terms of tolerance to acid stress: 35 % of the isolates were acid sensitive and 45 % were highly tolerant to pH 5 or moderately acidophilic. An association between mesorhizobia tolerance to acid conditions and the origin soil pH was found. Furthermore, significant differences between species clusters regarding tolerance to acidity were obtained. Ten isolates were used to investigate the expression levels of the chaperone genes by northern hybridization. Interestingly, most acid-tolerant isolates displayed induction of the dnaK and groESL genes upon acid shock while the sensitive ones showed repression. This study suggests that acid tolerance in mesorhizobia is related to the pH of the origin soil and to the species cluster of the isolates. Additionally, the transcriptional analysis suggests a relationship between induction of major chaperone genes and higher tolerance to acid pH in mesorhizobia. This is the first report on transcriptional analysis of the major chaperones genes in mesorhizobia under acidity, contributing to a better understanding of the molecular mechanisms of rhizobia acidity tolerance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Agricultural practices and environmental changes increase the amount of land affected by acidity, limiting crop productivity worldwide. Most leguminous plants require a neutral or slightly acidic soil for growth, especially when they depend on symbiotic nitrogen fixation. Soil acidity affects the nodulation and nitrogen fixation processes undertaken by rhizobia since it reduces rhizobial persistence and survival in the soil as well as nodulation efficiency [20, 25].
In Portugal, most of the soils are acid mainly due to the agricultural practices and the mild and dry climate, which favors a fast mineralization of the organic matter [45]. This results in an important constraint to most agricultural crops, such as chickpea production. This legume has a great economic importance due to its use worldwide as food for both humans and animals. Although chickpea (Cicer arietinum L.) is a successful legume on alkaline soils [39], its symbiotic relationship with mesorhizobia is better adapted to acidity [24, 44].
To avoid losses in the productivity of leguminous crops in acidic soil conditions, the development of legume–rhizobia associations able to tolerate such stress [16] or the selection of rhizobia tolerant to low pH [42] are possible strategies. Chen et al. [13] described the pH range for mesorhizobia growth between 4 and 10, although the optimal pH range was between 6 and 8. Some exceptions have been identified as is the case of strains of M. loti that showed a high tolerance to acidity (pH 4) [14, 28] as well as chickpea mesorhizobia isolates that could grow at pH 3 [8].
Stress response in bacteria is essential for effective adaptation to changes in the environment. Bacteria have the ability to sense protein folding and other signals, leading to the activation of proteins such as molecular chaperones, proteases, and regulatory factors, which play an important role in promoting homeostasis under stress conditions, such as acidity [19, 21, 26]. Molecular chaperones recognize non-native states of other proteins and assist their folding and/or prevent their aggregation. The DnaK–DnaJ–GrpE and GroEL–GroES complexes are the best characterized molecular chaperone systems, especially in Escherichia coli [43].
Although several studies evaluated the tolerance to acid and alkaline pH of strains belonging to the Mesorhizobium genus [8, 31], little is known about the factors that determine acid tolerance in rhizobia. Furthermore, the molecular mechanisms enabling tolerant strains to endure low pH, and thus the molecular basis of rhizobia tolerance to acidity, remain mostly unknown.
The present study evaluates the tolerance to acid and alkaline conditions of a Portuguese chickpea rhizobia collection and investigates whether acid tolerance is related to the species cluster, origin soil pH, or geographical origin of the isolates. Additionally, it investigates changes in the expression levels of the major chaperone systems groESL and dnaKJ upon acidic shock, using tolerant and sensitive mesorhizobia isolates, belonging to several Mesorhizobium species.
Materials and Methods
Bacterial Isolates
A total of 98 isolates from a chickpea rhizobia collection, which covers almost all Portuguese territory including Madeira Island, were used in the present study (Table 1). These isolates were previously characterized regarding the 16S rRNA gene sequence, plasmid number, and symbiotic performance [1, 9]. The isolates used in this study were obtained from 26 sampling sites. All isolates were preserved in 30 % (v/v) glycerol at −80 °C and cultured in yeast extract mannitol (YEM) broth [46] at 28 °C for routine use.
pH Stress Tolerance
The pH stress tolerance of the bacterial isolates was screened by evaluating their growth based on optical density (OD) readings at 540 nm. The pH stresses and control conditions were performed according to Laranjo and Oliveira [31]. Briefly, the YEM medium was buffered with 25 mM homopiperazine-N, N′-bis-2-(ethanesulfonic acid) (Homopipes) for pH 5 and with 26 mM 2-amino-2-methyl-1,3-propanediol (AMPD) for pH 9. For control conditions, YEM was buffered with 20 mM 2-morpholinoethanesulfonic acid (MES) at pH 7. After overnight growth, bacterial cultures were standardized to an initial OD of 0.03 and grown for 48 h at 28 °C. Three replicas per isolate under each condition were used.
Statistical Analysis
In order to compare isolates tolerance, optical density values were converted into percentage values, considering growth at control conditions (pH 7) as 100 %. Average value and standard deviations of the three replicas were calculated. Statistical analyses were performed using SPSS 17.0 software (SPSS Inc., Chicago, USA). Both Kruskal–Wallis and the Welch tests were used when there is no homogeneity of variances in order to explore the relationship between stress tolerance (continuous dependent variable) and categorical independent variable, as for instance species group or province of origin. To identify categories that differ significantly from others, three different post hoc tests (Tamhane, Dunnett T3, and Games–Howell) were used. To detect structure in the relationships between categorical variables, the correspondence analysis (CA) was conducted as an exploratory data analysis technique [7]. In order to investigate whether distinct acid tolerance phenotypes were related to the pH value of the origin soil of the isolates, these were divided into four classes according to their growth at pH 5: sensitive (growth < 30 %), tolerant (growth between 30 % and 70 %), highly tolerant (growth between 70 % and 100 %), and moderately acidophilic (growth > 100 %).
Non-parametric correlations between percentages of growth at acid pH and symbiotic effectiveness as well as the soil pH value from the origin site of isolates were determined using Spearman’s rank order correlation coefficient.
RNA Extraction and Northern Hybridization
The transcription of groEL and dnaKJ genes was analyzed in ten isolates showing different phenotypes upon acid conditions in order to investigate the involvement of these genes in tolerance to acidity. The transcriptional levels of the major chaperone genes were evaluated by northern hybridization after an acid shock and compared to those of control conditions. Total RNA extraction was performed using cell cultures in exponential growth phase, submitted to an acidic shock in YEM (pH 3) for 1 h. Control RNA was extracted from cells grown in YEM (pH 7). Total RNA extraction was performed according to the protocol for rapid isolation of RNA from Gram-negative bacteria [6].
The non-radioactive DIG system (Roche Applied Science) was used for northern analysis. RNA samples were denatured in the loading buffer (50 % deionized formamide; 6.1 % formaldehyde; 1× MOPS) and separated by electrophoresis on a 1.5 % agarose gel containing 2 % formaldehyde in 1× MOPS (20 mM MOPS buffer, 5 mM sodium acetate, 2 mM EDTA, pH 7.0). After electrophoresis, capillary transfer into a positively charged nylon membrane (Roche Applied Science) was carried out in 20× SSC (3 M NaCl; 300 mM sodium citrate, pH 7.0). RNA was fixed by baking the membrane at 120 °C for 30 min. The groEL and dnaKJ RNA probes were obtained as previously described [3]. All RNA probes were obtained by in vitro transcription labeling, using DIG Northern Starter Kit (Roche Applied Science). The DNA probe for 16S rRNA was labeled using DIG High Prime DNA Labeling and Detection Starter Kit II (Roche Applied Science). The 16S rRNA gene PCR amplification was performed using DNA of Mesorhizobium mediterraneum Ca36T as previously described [3].
Hybridizations were carried out overnight at 68 °C after a pre-hybridization period of 30 min at the same temperature. For the 16S rRNA detection, the membranes were re-hybridized overnight at 50 °C with a DNA probe. After hybridization, stringency washes and immunological detection were performed according to the manufacturer’s instructions.
Hybridization signals were analyzed using ImageQuantTL™ v7.01 (GE Healthcare). The 16S rRNA signals were used as internal control of the amount of total RNA loaded. To determine the expression levels, the ratio between the transcript signals and the corresponding 16S rRNA signals was calculated and the number of folds was determined using the ratio of the previous value between control and the pH shock conditions minus 1.
Results
Evaluation of Mesorhizobia Tolerance to Acidic and Alkaline Stresses
Isolates from the chickpea mesorhizobia collection were tested for tolerance to pH 5 and 9 (Fig. 1). The screening revealed that chickpea mesorhizobia varies in terms of tolerance to acid conditions (pH 5). In contrast, all chickpea mesorhizobia were sensitive to alkaline conditions (pH 9). Interestingly, isolates from M. mediterraneum/M. temperatum and M. tianshanense clusters are highly sensitive to both tested pH stress conditions, whereas isolates belonging to the other two species clusters (M. ciceri/M. loti and M. huakuii/M. amorphae) showed high diversity in tolerance to pH 5. In total, only 35 % of the isolates tested were sensitive to acidic conditions. On the other hand, 45 % of the isolates were highly tolerant or prefer acidic pH. Moreover, isolates C-25, T-5, and Al-13 belonging to M. huakuii/M. amorphae cluster as well as isolates 27 and S-8 from the M. ciceri/M. loti cluster can be considered as moderately acidophilic (growth above 100 % in pH 5).
Statistical analysis indicated that there are significant differences between species clusters regarding their tolerance to acidic conditions (χ 2 = 125.822; df = 3; P < 0.01). For instance, M. ciceri/M. loti isolates showed the highest growth average at pH 5 while the M. tianshanense isolates showed the lowest growth average, and are significantly different from each other and from the other two species clusters. Actually, these results are reinforced by the post hoc tests, which indicate that the growth averages at pH 5 are significantly different among species clusters.
Similarly, the statistical analysis showed that provinces of origin are significantly different in terms of the isolates tolerance to acid pH (χ 2 = 102.260; df = 10; P < 0.01). The three provinces with the highest growth averages at pH 5 (Beira Alta, Trás-os-Montes e Alto Douro, Beira Litoral) were found to be significantly different from the provinces with the lowest growth average (Ribatejo, Algarve, Estremadura). In order to investigate whether the pH value of the sampling site is one of the explanations for the significant differences found between provinces of origin, the sampling soils were classified into three classes based on their pH values. Sampling soils with pH values below 6.5 were considered acid soils, whereas the neutral soils include soils with pH values ranging from 6.5 to 7.4 and the alkaline soils represented the soils with pH values above 7.4. The correspondence analysis biplot (Fig. 2) shows an association between the pH class of the origin soil and the isolate’s ability to tolerate pH 5. Additionally, a negative correlation was found between the isolates growth at acid pH and the pH value of the sampling soil (r = −0.358; P < 0.01). For instance, isolates from alkaline soils were more sensitive to pH 5 than the isolates from neutral or acidic soils. Sensitivity to acid conditions is clearly associated to alkaline soils whereas the acid tolerance is associated to neutral or acidic soils. Our results indicate an association (χ 2 = 156.863; df = 6; P < 0.01) between the origin soil pH of the isolates and species clusters. Moreover, a CA biplot also detects an association between species clusters and soil pH classes, where the M. ciceri/M. loti and M. huakuii/M. amorphae species clusters are associated to neutral soils and acidic soils, respectively, and the remaining species clusters are associated to the alkaline soils (Fig. 3).
Curiously, a positive correlation between tolerance to acidic conditions and symbiotic effectiveness was found (r = 0.139; P < 0.05). For example, the isolate G-55 showed a growth of 106 % at pH 5 and it is also highly efficient with a symbiotic effectiveness of 88 %.
Transcriptional Analysis of the Major Chaperones Genes upon Acidic Shock
In order to investigate the involvement of the major chaperone genes in tolerance to acidic conditions, the transcription of groEL and dnaKJ genes was analyzed. Using northern hybridization, the transcriptional levels of these chaperone genes upon an acidic shock were compared to those under control conditions. Based on the pH stress tolerance screening (Fig. 1), ten chickpea mesorhizobia isolates, from the four species clusters, with different phenotypes under acidic conditions, were selected. From the M. huakuii/M. amorphae species cluster, the following isolates were chosen: isolate PMI-6 as acid sensitive, V-15b and C-9 as acid tolerant, and AL-13 as moderately acidophilic. From the M. ciceri/M. loti cluster, the isolate PII-4 as acid sensitive, G-55 and G-10 as acid tolerant, and S-8 as moderate acidophilic were selected. The clusters M. tianshanense and M. mediterraneum/M. temperatum only include isolates sensitive to acidity, so only one isolate from each of these two clusters was chosen, namely ST-33 from the M. tianshanense and PM-14 from M. mediterraneum/M. temperatum.
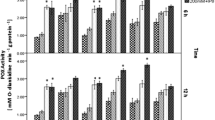
The northern blot analysis using the dnaKJ mRNA probe allows the detection of three different transcripts, with 1,917, 1,131, and 3,048 nucleotides in size, corresponding to the predicted mRNAs of dnaK, dnaJ, and also the bicistronic dnaKJ, respectively, according to the Mesorhizobium sp. MAFF303099 genome. A transcript with approximately 2 kb, which is consistent with the size of the dnaK gene transcript, was detected in all isolates under control conditions and upon acidic shock (Fig. 4a). The majority of the isolates showed an increase in the dnaK transcript levels (Fig. 5) after acidic shock.
Comparison of transcriptional analysis of the dnaK gene (a) and the groESL operon (b) between acid-tolerant and acid-sensitive isolates submitted to acidic shock. Northern blot hybridization of total RNA with probes specific for dnaKJ, groEL, and 16S rRNA under control conditions (C) and upon acidic shock (S)
The tolerant isolates belonging to M. ciceri/M. loti cluster showed an increase in the dnaK mRNA level after acidic shock when compared with the control. However, the sensitive isolate PII-4 showed a decrease in the expression of the dnaK gene after acidic shock. Similarly, in isolates from the M. huakuii/M. amorphae cluster, the level of dnaK gene transcription was positively related to the acid tolerance of the isolates since a slight repression of the dnaK gene was observed in the sensitive isolate (PMI-6) while induction was observed in the acid-tolerant ones (V-15b, C-9, and AL-13) (Fig. 5).
No significant changes in the transcriptional levels of dnaK gene were observed in the sensitive isolates neither from the M. tianshanense nor M. mediterraneum/M. temperatum clusters upon acidic shock.
Regarding the analysis of the groESL chaperone system, the groEL RNA probe allows the detection of two putative transcripts with 1,947 and 1,632 nucleotides, corresponding to the bicistronic groESL transcript and to the groEL transcript, respectively, according to the Mesorhizobium sp. MAFF303099 genome. In this study, the signal detected was approximately 2 kb, which corresponds to the bicistronic groESL mRNA (Fig. 4b). Most isolates showed an increase of the groESL mRNA transcript levels after the acidic shock.
Interestingly, isolates belonging to the M. ciceri/M. loti cluster presented the same pattern in the groESL chaperone gene transcription as obtained for the dnaK chaperone gene (Fig. 5): tolerant isolates (G-10, G-55, and S-8) showed an increase of the groESL mRNA levels while the sensitive isolate (PII-4) revealed a repression of this chaperone gene after acidic shock.
Concerning the cluster M. huakuii/M. amorphae, the tolerant isolates (V-15b and C-9) showed an increase in the groESL mRNA levels after acidic shock, whereas the sensitive isolate (PMI-6) showed repression of the groESL gene. Although the moderately acidophilic isolate (AL-13) showed a repression of the groESL gene, it shows one of the highest levels of dnaK gene induction upon acidic shock.
The acid-sensitive isolates belonging to the M. tianshanense and M. mediterraneum/M. temperatum cluster showed a slight induction of the groESL genes compared to the control.
For both dnaK and groESL transcription levels, the isolates from the M. ciceri/M. loti and M. huakuii/M. amorphae seem to exhibit a similar relationship between transcription levels and tolerance to acid pH, with the exception of the moderately acidophilic isolate AL-13 for the groESL analysis.
Discussion
In this study, evaluation of growth in acid and alkaline conditions was performed for 98 isolates from a collection of Portuguese native chickpea mesorhizobia previously characterized [1, 9]. All mesorhizobia showed sensitivity to alkaline stress, which is in agreement with previous studies [8, 31]. In contrast, the mesorhizobia isolates tested herein showed high diversity regarding tolerance to acid pH. Interestingly, almost half of the tested isolates were highly tolerant (growth > 70 %), including 11 isolates with a higher growth at pH 5 than at pH 7, indicative of moderately acidophilic isolates. High tolerance to low pH has been previously reported in Mesorhizobium species, namely M. huakuii [11], M. ciceri [38], M. loti [27], and M. amorphae [47], which are all able to grow at pH 5. On the other hand, M. mediterraneum and M. tianshanense cannot grow at pH 5 [12, 37]. Brígido et al. [8] have already identified moderately acidophilic chickpea mesorhizobia isolates. More recently, a high diversity in tolerance to acidic conditions within the Mesorhizobium genus was reported by Laranjo and Oliveira [31].
Amarger et al. [4] reported that tolerance to salinity, acidity, and alkalinity is more strain specific than species specific. However, other studies suggest that the tolerance to pH stress in rhizobia is species-related [8, 40]. Herein, the evaluation of tolerance to acidity of a large set of chickpea mesorhizobia suggests that acid tolerance phenotype is related to the species clusters. Significant differences between species clusters regarding tolerance to acidity were obtained. For instance, the majority of isolates from M. ciceri/M. loti cluster are acid tolerant whereas isolates belonging to M. tianshanense cluster are acid sensitive. Moreover, several studies in rhizobia have reported that stress tolerance seems to be species related namely temperature stress tolerance [3] and tolerance to copper [30] as well as antibiotic resistance [2].
An association between species clusters and origin soil pH of the isolates was found in Portuguese chickpea rhizobia [1] and recently in Chinese soybean rhizobia [34]. Our results are in agreement with these reports, suggesting that soil pH contributes to determine the species that prevail in the rhizobia population. This type of association was found in several studies addressing soil bacterial communities, indicating soil pH as the variable that better explains the population diversity and mainly the community composition [17]. On the other hand, in this study, the tolerance to acid pH of the chickpea mesorhizobia isolates was found to be associated to the origin soil pH, agreeing with some previous reports [8, 29, 41]. These results suggest that isolates collected from acidic or neutral soils may be more resistant to acidic environmental conditions than the ones from alkaline soils.
Interestingly, our results also reveal a positive correlation between the tolerance to acid pH and the symbiotic effectiveness of isolates, suggesting that the establishment of an effective symbiosis is related to tolerance to acidity. Bacterial persistence in soil is significantly dependent on soil pH, which may affect the symbiotic performance by reduction of the nodulation efficiency [10, 20, 25]. In addition, it is known that the pH in the rhizosphere of the leguminous host plant is lower due to the protons and organic acids excreted by the plants [35], suggesting that the rhizobial partner has to deal with this stressful condition to achieve effective symbiosis. Previous studies in our laboratory [8] indicated that the maximum symbiotic performance achieved by a specific strain is related to its optimal growth pH. This suggests that reaching a higher symbiotic performance of rhizobia inoculants in chickpea crops should require the selection of a strain whose preferred pH is similar to the pH of the soil to be cultivated. Similar results were found in peanut rhizobia by Angelini et al. [5].
Despite the fact that several studies have attempted to characterize the genes involved in tolerance to acidity, the molecular mechanisms to respond to acidity are still unknown in rhizobia. Molecular chaperones form a multi-protein network that prevent protein denaturation, and also help in the proper protein folding and refolding, transport, degradation, and regulation. The functioning of this network is particularly important under stress conditions, such as acidic shock [22]. Our results from the transcriptional analysis of ten mesorhizobia isolates show a relationship between higher levels of transcriptional induction of both dnaK and groESL genes upon acidic shock and a higher ability of mesorhizobia to tolerate acid pH. Upon acid shock, both dnaK and groESL genes were induced only in acid-tolerant isolates and not in sensitive isolates, with the exception of the moderately acidophilic isolate (AL-13), which showed a repression of the groESL gene but one of the highest levels of dnaK gene induction. These results suggest that increased expression of chaperone genes may contribute to a higher tolerance to acid stress in rhizobia. However, these results only refer to the isolates belonging to M. ciceri/M. loti and M. huakuii/M. amorphae species clusters since no comparison between tolerant and sensitive isolates was available for the remaining species. Foster [18] suggested a model to explain the acid response in Salmonella cells involving DnaK and GroEL due to their ability to refold acid denatured proteins. Studies in Streptococcus mutans revealed that dnaK and groEL are part of the general stress response being both induced during the acid shock response [32, 33, 36]. Other studies in E. coli showed that the expression of the chaperones DnaK, DnaJ, and GrpE was inducible under acid shock [49]. However, contrary to these results, dnaK and dnaJ chaperone genes were found to be down-regulated upon acidic stress in Streptococcus suis S2 [48].
In rhizobia, little is known about acid response. Recently, transcriptional analysis using Ensifer meliloti 1021 cells following an acidic upshift showed increased groEL5 transcript levels [15, 23]. Hellweg et al. [23] verified that the groEL5 gene was not immediately up-regulated after the pH shift, but slowly increased its expression level during the time course (1 h, pH 5.75).
Our previous studies in chickpea mesorhizobia suggest the existence of a relationship between higher levels of transcriptional induction of the dnaK and groESL chaperones genes and a higher ability of isolates to endure heat stress [3]. In contrast, no correlation between salt tolerance and expression levels of these chaperones genes in mesorhizobia was found [9]. Altogether, it seems that dnaK and groESL genes may be involved in acid and heat tolerance in chickpea mesorhizobia.
Here we evaluated the diversity of tolerance to acid and alkaline stress conditions of a collection of chickpea mesorhizobia isolates belonging to four species clusters and investigated possible relationships between acid tolerance phenotype of isolates and their species cluster, geographical origin, origin soil pH, and symbiotic effectiveness. Our findings suggest that tolerance to acid stress is related to the species clusters and to the origin soil pH. A clear relationship between induction of dnaK and groESL genes upon acidic shock and tolerance to acidic conditions was found, suggesting that induction of these chaperone genes is involved in the chickpea mesorhizobia tolerance to acid pH. However, further studies are required to clarify the role of these chaperone genes in acid stress tolerance of rhizobia.
References
Alexandre A, Brígido C, Laranjo M, Rodrigues S, Oliveira S (2009) A survey of chickpea rhizobia diversity in Portugal reveals the predominance of species distinct from Mesorhizobium ciceri and Mesorhizobium mediterraneum. Microb Ecol 58:930–941
Alexandre A, Laranjo M, Oliveira S (2006) Natural populations of chickpea rhizobia evaluated by antibiotic resistance profiles and molecular methods. Microb Ecol 51:128–136
Alexandre A, Oliveira S (2011) Most heat-tolerant rhizobia show high induction of major chaperone genes upon stress. FEMS Microbiol Ecol 75:28–36
Amarger N, Macheret V, Laguerre G (1997) Rhizobium gallicum sp. nov. and Rhizobium giardinii sp. nov., from Phaseolus vulgaris nodules. Int J Syst Bacteriol 47:996–1006
Angelini J, Taurian T, Morgante C, Ibanez F, Castro S, Fabra A (2005) Peanut nodulation kinetics in response to low pH. Plant Physiol Biochem 43:754–759
Ausubel FM, Brent R, Kingston RE, More DD, Seidman JG, Smith JA, Struhl K (1997) Current protocols in molecular biology. Wiley-Interscience, New York
Benzécri JP (1973) Analyse des données. Tome I: Analyse des correspondances. Tome II: La Classification. Dunod, Paris
Brígido C, Alexandre A, Laranjo M, Oliveira S (2007) Moderately acidophilic mesorhizobia isolated from chickpea. Lett Appl Microbiol 44:168–174
Brígido C, Alexandre A, Oliveira S Transcriptional analysis of major chaperone genes in salt-tolerant and salt-sensitive mesorhizobia. Microbiol Res. doi:10.1016/j.micres.2012.01.006
Brockwell J, Pilka A, Holliday RA (1991) Soil pH is a major determinant of the numbers of naturally occurring Rhizobium meliloti in noncultivated soils in central New South Wales. Aust J Exp Agr 31:211–219
Chen WX, Li GS, Qi YL, Wang ET, Yuan HL, Li JL (1991) Rhizobium huakuii sp. nov. isolated from the root-nodules of Astragalus sinicus. Int J Syst Bacteriol 41:275–280
Chen WX, Wang E, Wang SY, Li YB, Chen XQ (1995) Characteristics of Rhizobium tianshanense sp. nov., a moderately and slowly growing root-nodule bacterium isolated from an arid saline environment in Xinjiang, Peoples Republic of China. Int J Syst Bacteriol 45:153–159
Chen WX, Wang ET, Kuykendall D (2005) Genus VI. Mesorhizobium. In: Bergey’s manual of systematic bacteriology, vol. 2 (The Proteobacteria). Springer, New York, pp. 403–408
Correa OS, Barneix AJ (1997) Cellular mechanisms of pH tolerance in Rhizobium loti. World J Microbiol Biotechnol 13:153–157
de Lucena DKC, Puehler A, Weidner S (2010) The role of sigma factor RpoH1 in the pH stress response of Sinorhizobium meliloti. BMC Microbiol 10(265):17
Dilworth MJ, Howieson JG, Reeve WG, Tiwari RP, Glenn AR (2001) Acid tolerance in legume root nodule bacteria and selecting for it. Aust J Exp Agr 41:435–446
Fierer N, Jackson RB (2006) The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci USA 103:626–631
Foster JW (1991) Salmonella acid shock proteins are required for the adaptative acid tolerance response. J Bacteriol 173:6896–6902
Frydman J (2001) Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu Rev Biochem 70:603–647
Graham PH, Viteri SE, Mackie F, Vargas AT, Palacios A (1982) Variation in acid soil tolerance among strains of Rhizobium phaseoli. Field Crop Res 5(2):121–128
Hartl FU (1996) Molecular chaperones in cellular protein folding. Nature 381:571–579
Hartl FU, Hayer-Hartl M (2002) Protein folding—molecular chaperones in the cytosol: from nascent chain to folded protein. Science 295:1852–1858
Hellweg C, Puhler A, Weidner S (2009) The time course of the transcriptomic response of Sinorhizobium meliloti 1021 following a shift to acidic pH. BMC Microbiol 9:37
Howieson JG, O’Hara GW, Carr SJ (2000) Changing roles for legumes in Mediterranean agriculture: developments from an Australian perspective. Field Crop Res 65:107–122
Ibekwe AM, Angle JS, Chaney RL, vanBerkum P (1997) Enumeration and N-2 fixation potential of Rhizobium leguminosarum biovar trifolii grown in soil with varying pH values and heavy metal concentrations. Agric Ecosyst Environ 61:103–111
Jakob U, Gaestel M, Engel K, Buchner J (1993) Small heat shock proteins are molecular chaperones. J Biol Chem 268:1517–1520
Jarvis BDW, Pankhurst CE, Patel JJ (1982) Rhizobium loti, a new species of legume root nodule bacteria. Int J Syst Bacteriol 32:378–380
Jarvis BDW, van Berkum P, Chen WX, Nour SM, Fernandez MP, Cleyet-Marel JC et al (1997) Transfer of Rhizobium loti, Rhizobium huakuii, Rhizobium ciceri, Rhizobium mediterraneum, and Rhizobium tianshanense to Mesorhizobium gen. nov. Int J Syst Bacteriol 47:895–898
Kulkarni S, Nautiyal CS (1999) Characterization of high temperature-tolerant rhizobia isolated from Prosopis juliflora grown in alkaline soil. J Gen Appl Microbiol 45:213–220
Laguerre G, Courde L, Nouaim R, Lamy I, Revellin C, Breuil MC et al (2006) Response of rhizobial populations to moderate copper stress applied to an agricultural soil. Microb Ecol 52:426–435
Laranjo M, Oliveira S (2011) Tolerance of Mesorhizobium type strains to different environmental stresses. Anton Leeuw Int JG 99:651–662
Lemos JA, Luzardo Y, Burne RA (2007) Physiologic effects of forced down-regulation of dnaK and groEL expression in Streptococcus mutans. J Bacteriol 189:1582–1588
Lemos JAC, Chen YYM, Burne RA (2001) Genetic and physiologic analysis of the groE operon and role of the HrcA repressor in stress gene regulation and acid tolerance in Streptococcus mutans. J Bacteriol 183:6074–6084
Li QQ, Wang ET, Zhang YZ, Zhang YM, Tian CF, Sui XH et al (2011) Diversity and biogeography of rhizobia isolated from root nodules of Glycine max grown in Hebei province, China. Microb Ecol 61:917–931
Marschner H (2006) Mineral nutrition of higher plants. Academic, London
Matsui R, Cvitkovitch D (2010) Acid tolerance mechanisms utilized by Streptococcus mutans. Future Microbiol 5:403–417
Nour SM, Cleyet-Marel J-C, Normand P, Fernandez MP (1995) Genomic heterogeneity of strains nodulating chickpeas (Cicer arietinum L.) and description of Rhizobium mediterraneum sp. nov. Int J Syst Bacteriol 45:640–648
Nour SM, Fernandez MP, Normand P, Cleyet-Marel J-C (1994) Rhizobium ciceri sp. nov., consisting of strains that nodulate chickpeas (Cicer arietinum L.). Int J Syst Bacteriol 44:511–522
Rao DLN, Giller KE, Yeo AR, Flowers TJ (2002) The effects of salinity and sodicity upon nodulation and nitrogen fixation in chickpea (Cicer arietinum). Ann Bot 89:563–570
Reeve WG, Brau L, Castelli J, Garau G, Sohlenkamp C, Geiger O et al (2006) The Sinorhizobium medicae WSM419 IpiA gene is transcriptionally activated by FsrR and required to enhance survival in lethal acid conditions. Microbiology 152:3049–3059
Rodrigues C, Laranjo M, Oliveira S (2006) Effect of heat and pH stress in the growth of chickpea mesorhizobia. Curr Microbiol 53:1–7
Ruiz-Díez B, Fajardo S, Puertas-Mejia MA, Felipe MD, Fernandez-Pascual M (2009) Stress tolerance, genetic analysis and symbiotic properties of root-nodulating bacteria isolated from Mediterranean leguminous shrubs in Central Spain. Arch Microbiol 191:35–46
Sabate R, de Groot NS, Ventura S (2010) Protein folding and aggregation in bacteria. Cell Mol Life Sci 67:2695–2715
Siddique KHM, Loss SP, Regan KL, Jettner RL (1999) Adaptation and seed yield of cool season grain legumes in Mediterranean environments of south-western Australia. Aust J Agr Res 50:375–387
Torrent J, Barberis E, Gil-Sotres F (2007) Agriculture as a source of phosphorus for eutrophication in southern Europe. Soil Use Manage 23:25–35
Vincent JM (1970) A manual for the practical study of root-nodule bacteria, no. 15. IBP Handbook. Blackwell, Oxford
Wang ET, van Berkum P, Sui XH, Beyene D, Chen WX, Martinez-Romero E (1999) Diversity of Rhizobia associated with Amorpha fruticosa isolated from Chinese soils and description of Mesorhizobium amorphae sp nov. Int J Syst Bacteriol 49:51–65
Wei Y, Zeng X, Yuan Y, Jiang H, Zheng Y, Tan Y et al (2011) DNA microarray analysis of acid-responsive genes of Streptococcus suis serotype 2. Ann Microbiol 61:505–510
Zmijewski MA, Kwiatkowska JM, Lipinska B (2004) Complementation studies of the DnaK–DnaJ–GrpE chaperone machineries from Vibrio harveyi and Escherichia coli, both in vivo and in vitro. Arch Microbiol 182:436–449
Acknowledgments
This work has received funding from FCT (Fundação para a Ciência e a Tecnologia) and co-financed by EU-FEDER (PTDC/BIO/80932/2006). C. Brígido acknowledges a PhD fellowship (SFRH/BD/30680/2006) from FCT. We thank G. Mariano for technical assistance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Brígido, C., Oliveira, S. Most Acid-Tolerant Chickpea Mesorhizobia Show Induction of Major Chaperone Genes upon Acid Shock. Microb Ecol 65, 145–153 (2013). https://doi.org/10.1007/s00248-012-0098-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-012-0098-7