Abstract
Eastern Mediterranean sediments are characterized by the periodic occurrence of conspicuous, organic matter-rich sapropel layers. Phylogenetic analysis of a large culture collection isolated from these sediments revealed that about one third of the isolates belonged to the genus Photobacterium. In the present study, 22 of these strains were examined with respect to their phylogenetic and metabolic diversity. The strains belonged to two distinct Photobacterium populations (Mediterranean cluster I and II). Strains of cluster I were isolated almost exclusively from organic-rich sapropel layers and were closely affiliated with P. aplysiae (based on their 16S rRNA gene sequences). They possessed almost identical Enterobacterial Repetitive Intergenic Consensus (ERIC) and substrate utilization patterns, even among strains from different sampling sites or from layers differing up to 100,000 years in age. Strains of cluster II originated from sapropels and from the surface and carbon-lean intermediate layers. They were related to Photobacterium frigidiphilum but differed significantly in their fingerprint patterns and substrate spectra, even when these strains were obtained from the same sampling site and layer. Temperature range for growth (4 to 33°C), salinity tolerance (5 to 100‰), pH requirements (5.5–9.3), and the composition of polar membrane lipids were similar for both clusters. All strains grew by fermentation (glucose, organic acids) and all but five by anaerobic respiration (nitrate, dimethyl sulfoxide, anthraquinone disulfonate, or humic acids). These results indicate that the genus Photobacterium forms subsurface populations well adapted to life in the deep biosphere.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
In the recent years the marine deep subsurface received increasing attention. Not only geochemical analyses and modeling [13] but also radiotracer-based activity measurements [36, 37], direct microscopic, or viable counts [13, 44] revealed the presence of active microbial communities in up to 15-Ma-old marine sediments [61]. It was also shown that cell densities and metabolic activities in deeply buried sediments correlate with the availability of organic carbon [8, 37] and/or electron acceptors [11, 13, 14]. Extensive molecular analyses were performed and unraveled an unexpectedly large microbial diversity [24, 33], including some phylogenetic lineages appearing to be typical for deep subsurface habitats [8, 60]. However, because of the low fraction of sediment microbes that have been brought into pure culture [13, 55], there is a lack in knowledge of physiological adaptations of indigenous deep biosphere bacteria.
One aspect lowering the cultivation success might be the application of high substrate concentrations in standard microbiological media. Apparently, the use of media containing submillimolar substrate concentrations increased the cultivation efficiency and resulted in the subsequent isolation of a large culture collection from ancient Mediterranean sapropels [49]. These dark, periodically (approximately every 20,000 years) occurring sediment layers differ from other subsurface environments in their unusually high organic carbon contents (up to 30% of the dry weight [40]). It is assumed they were deposited during periods of intense precipitation with a high riverine influx of freshwater leading not only to increased productivity but also to haline stratification [27]. The latter prevented mixing of oxygen-rich surface and deep waters, eventually leading to anoxic conditions and enhanced preservation of organic material [40]. Sapropels are interspersed in-between carbonate-rich and extremely organic carbon poor hemipelagic sediments that were deposited under highly oligotrophic conditions like they prevail today. Although the organic material within the sapropels consists mainly of highly recalcitrant kerogen [22], sapropels were shown to be subsurface ‘hotspots’ with elevated microbial numbers and increased potential microbial activities [8, 9, 12].
The culture collection from sapropels of the Eastern Mediterranean included 98 strains covering 19 different phylotypes. Phylogenetic analysis revealed that about a third of the strains affiliated with the soil bacterium Rhizobium radiobacter [49]. The occurrence of this phylotype as a member of the deep biosphere was recently confirmed by molecular methods [50]. About another 30% of the strains belonged to the genus Photobacterium [49]. This genus was one of the first described bacterial taxa [6] and was originally considered to be generally associated with marine animals [41]. Although photobacteria were found to be widespread in marine sediments (e.g., Photobacterium profundum and P. frigidiphilum; [34, 46]), their prevalence in the culture collection obtained from the up to 120,000-year-old sapropels is unexpected and extends their ‘typical’ habitat range. Because Photobacterium sp. are among the dominant cultured subsurface bacterial groups, analysis of their physiology might help to deepen the knowledge of metabolic adaptations of sediment microbial communities.
Several recent studies have revealed a remarkable phylogenetic [1, 39, 53] or physiological [3, 20, 31] heterogeneity at the subspecies level within single bacterial taxa. The ecological significance of this phenomenon still remains elusive [16], but it was assumed that this microdiversity is, among others, a prerequisite for longevity of bacterial populations in changing environments [2, 10, 45]. In the present study, 22 Photobacterium strains originating from three different sampling sites in the Eastern Mediterranean Sea were examined with respect to their phylogenetic and metabolic diversity. The extent of the culture collection offered the opportunity to link variations in 16S rRNA genes and Enterobacterial Repetitive Intergenic Consensus (ERIC)-polymerase chain reaction (PCR) fingerprinting patterns to physiological differences and correlate these to spatial separation or to age and TOC-content of the sediment layers.
Materials and Methods
Sample Origin and Isolation of the Photobacterium Strains
All strains analyzed in this study were obtained from eastern Mediterranean sapropels and from hemipelagic carbon-lean intermediate layers sampled during R/V Meteor cruises M 40/4 and M 51/3 [9, 49]. Samples were taken from three different sites: Site 67 (34°48.83′N, 27°17.77′E, sampled in January 1998) and site 567 (34°48.79′N, 27°17.13′E, sampled in November 2001) were located approximately 200 m apart from each other and situated about 100 km east of the island of Crete (Greece). Site 575 (34°31.39′N, 31°46.40′E) was sampled in November 2001 and located about 65 km west of Cyprus. Each core was cut longitudinally, what left behind a potentially contaminated surface. After covering with cling film, rapidly freezing this surface with dry ice and lifting it off, 5 cm3 subsamples were retrieved aseptically from the undisturbed sediment underneath using cut-off sterile plastic syringes [8].
Most strains were isolated from highest positive dilutions of oxic and anoxic most-probable-number (MPN) series that were prepared onboard ship in a polyethylene chamber (AtmosBag, 280 l, Aldrich, Milwaukee, Wisconsin, USA) under a nitrogen atmosphere. Anoxic MPN plates were incubated in gas-tight plastic bags equipped with a gas generating and catalyst system (Anaerocult C mini, Merck, Darmstadt, Germany) [49]. MPN series were supplemented with different carbon sources and electron acceptors (Table 1). Strains 67TD and 67FSB were obtained from anoxic enrichments directly inoculated with sapropel material. For isolation under anoxic conditions, cultures were diluted in agar-solidified media in tubes (deep agar dilution series) under a N2 atmosphere, whereas aerobes were obtained by repeated streaking on agar plates [49].
Extraction of Nucleic Acids
Genomic DNA of the strains was extracted following a standard protocol with cell lysis by lysozyme, sodium dodecyl sulfate (SDS), and “freeze and thaw” cycles with subsequent purification as described by Süß et al. [50]. Purified nucleic acids were resuspended in Tris–EDTA (TE) buffer (10 mM Tris HCl, 1 mM EDTA, pH 8.0) and stored at −20°C.
Phylogenetic Analysis
The 16S rRNA genes of the strains were amplified and partial sequences determined as described elsewhere [49]. Sequences (892 to 1300 bp long) were compared to those available in the GenBank database using the BLASTN tool. Six partial sequences from Photobacterium sp. (strains J2, J4, J158, S1D, Z0F, Z1E) isolated from the same sites but lost before this study were included in the phylogenetic analysis. Phylogenetic trees were constructed using the ARB software package [29]. The maximum likelihood method was used for the construction of backbone-trees considering sequences of validly described species with sequence lengths of at least 1300 bp. Sequences of recently described species [46, 47, 52, 64] not yet available in the ARB database release were retrieved from GenBank. Sequences <1300 bp were added afterwards by parsimony interactive using a newly constructed specific Photobacterium filter. To verify the stability of the Mediterranean Photobacterium clusters, neighbor-joining and parsimony trees were calculated as described for the maximum likelihood method.
Enterobacterial Repetitive Intergenic Consensus-Polymerase Chain Reaction
To investigate subspecies diversity of closely related strains genomic fingerprinting using the primers ERIC1R and ERIC2 [58] was performed. The PCR reaction mix contained 1 U Red Taq DNA polymerase (Sigma, Munich, Germany) and the appropriate 10× buffer, diethylnitrophenyl thiophosphates (dNTPs; 200 μM each), MgCl2 (2.1 mM), bovine serum albumin (BSA; 0.2 ng μl−1), the primers ERIC1R and ERIC2 (5 pM each) and 4 ng μl−1 template DNA in a final volume of 50 μl. PCR was performed in a PerkinElmer thermocycler (PerkinElmer Gene Amp PCR System 9600, Wellesley, MA). The following protocol was used according to Versalovic et al. [58]: 4-min denaturation at 96°C, followed by 35 cycles with denaturation for 30 s at 94°C, annealing for 1 min at 52°C, and elongation for 8 min at 72°C. Post elongation was performed for 10 min at 72°C. Fragments were separated on 1% (w/v) agarose gels (90 V for 3 h). Gels were stained for 20 min with ethidium bromide and documented by means of a digital imaging system and the respective software (BioDoc Analyze Biometra, Göttingen, Germany). The resulting band patterns were analyzed using the software package GelCompar II version 2.5 (Applied Maths, St-Martens-Latem, Belgium). The computer-generated densitometric curves were compared using the Pearson coefficient. Dendrograms were generated by the unweighted-pair group method with arithmetic averages (UPGMA).
Growth Media
Artificial seawater reflecting the average element composition of the Mediterranean Sea was used as a basal medium. This medium contained (in g l−1): NaCl (24.3), MgCl2·6H2O (10), CaCl2·H2O (1.5), KCl (0.66), Na2SO4 (4), KBr (0.1), H3BO3 (0.025), SrCl2·6H2O (0.04), NH4Cl (0.021), KH2PO4 (0.0054), and NaF (0.003). The medium was supplemented with 1 mL L−1 trace element solution SL10 and 0.2 mL L−1 of a selenite and tungstate solution [62]. The oxic medium was buffered with HEPES (2.4 g L−1). The pH of the medium was adjusted to 7.2–7.4 with NaOH before autoclaving. After autoclaving, the medium was cooled under air and supplemented with vitamins [4] and sodium bicarbonate (final concentration, 0.2 g L−1). For routine growth, temperature, and pH tests, a dilute yeast extract-peptone medium [49] was used containing yeast extract (0.03 g·L−1), peptone (0.06 g·L−1), sodium lactate (5 mmol·L−1), glucose (1 mmol·L−1), sodium thiosulfate (1 mmol·L−1), vitamins, and sodium bicarbonate (0.2 g·L−1).
For anoxic incubations, a slightly different medium was used. It contained resazurin (0.25 mg L−1) as a redox indicator. After autoclaving, the anoxic medium was cooled under an atmosphere of N2/CO2 (80/20, v/v). Instead of HEPES, 30 mL L−1 of a 1 M sodium hydrogen carbonate solution was added as a buffer from a sterile stock. After addition of vitamins, the medium was carefully reduced by addition of sterile dithionite until the resazurin turned colorless. The pH of the reduced medium was set to 7.2–7.4 with sterile HCl or Na2CO3 if necessary.
Substrate Utilization Under Oxic Conditions
For tests on substrate utilization, 56 different carbon sources were chosen for growth tests under oxic conditions. Assays were set up in polystyrene microtiter plates (Costar 3795, Corning, New York, NY). Each well was filled with 180 μl oxic basal medium. The following carbon sources were provided (final concentrations are given in brackets): (1) complex substrates: peptone (0.05% w/v), casamino acids (0.05% w/v), and yeast extract (0.005% w/v); (2) polysaccharides: cellulose (0.05% w/v), starch (0.1% w/v), chitin (0.05% w/v), xylan (0.05% w/v), and laminarin (0.05% w/v); (3) mono- and disaccharides, sugar derivates: sucrose, cellobiose, maltose, trehalose, arabinose, xylose, fructose, glucose, mannose, rhammnose, mannitol, gluconate, and glucosamine (each at 5 mM); (4) organic acids: lactate and succinate (each at 10 mM), formate, acetate, malonate, fumarate, malate, 2-oxoglutarate, glycolate and pyruvate (each at 5 mM), butyrate (2.5 mM), tartrate (2 mM), citrate (2 mM), propionate (1 mM), capronate, caprylate, crotonate, and valerate (each at 0.5 mM); (5) alcohols: ethanol, n-propanol, n-butanol, ethylene glycol, glycerol (each at 5 mM), methanol (2 mM), and Tween80 (0.001% w/v); (6) l-amino acids: alanine, arginine, cysteine, glutamine, isoleucine and phenylalanine (each at 2 mM); and (7) miscellaneous compounds: betaine, benzoate, salicylate, and nicotinate (each at 2 mM). Cells were harvested from exponential phase liquid cultures and washed three times. Three replicates and two substrate free controls were inoculated for each strain and two inoculum free controls for each substrate. The plates were incubated at 20°C for 6 weeks. Growth was determined by fluorometry [30] and by phase contrast microscopy of selected wells.
Use of Electron Acceptors
The tests were performed in completely filled, screw cap glass tubes with anoxic artificial seawater as basal medium. Acetate (10 mM) was used as electron donor and carbon source. \( {\text{NO}}^{ - }_{3} \) (10 mM), Fe(OH)3 (40 mM), manganese oxides (20 mM), thiosulfate (10 mM), elemental sulfur (20 mM), dimethyl sulfoxide (DSMO, 10 mM), 9,10-anthraquinone-2,6-disulfonate (AQDS, 4 mM), and an iron-free humic acid suspension (1 mg mL−1, [7]) were chosen to test anaerobic growth with alternative electron acceptors. For each strain and electron acceptor combination, three replicates were inoculated and an inoculum-free control and a control without electron acceptor. The assays were incubated for 6 weeks at 20°C in the dark. Growth was monitored by phase contrast or epifluorescence microscopy after staining with 4′,6-diamidino-2-phenylindole (DAPI) if necessary. The conversion of nitrate to nitrite or ammonium was determined photometrically according to Grasshoff et al. [17]. Reduction of Fe(III) to Fe(II) was generally indicated by the disappearance of reddish ferric hydroxide and the formation of black precipitates. The utilization of Mn(IV) led to decolorization and finally disappearance of the brown manganese oxides and the precipitation of whitish manganese carbonates. The production of sulfide as a result of the reduction of thiosulfate, sulfite, or elemental sulfur was measured photometrically after addition of an acidic cupric solution at 436 nm in accordance to Widdel [Widdel F (1980) Ph.D. Dissertation University Göttingen]. Reduction of 9,10-anthraquinone-2, 6-disulfonic acid (AQDS) to the reduced anthrahydroquinone was measured photometrically at 450 nm [28].
Fermentative Growth
Fermentative growth was tested in completely filled screw cap glass tubes filled with anoxic artificial seawater supplied either with glucose (10 mM), a mixture of l-amino acids (alanine, threonine, lysine-monohydrochloride, arginine, asparagine, aspartate, leucine, isoleucine, glumatate, glutamine, and methionine, each 5 mM), organic acids (malate, fumarate, succinate, and lactate, each 15 mM), or a mixture of alcohols (methanol, ethanol, n-propanol, and n-butanol, each 10 mM) as substrates. For each substrate, three replicates and a substrate free control were incubated at 20°C for 6 weeks. Growth was monitored by phase contrast microscopy. Assays proven positive for glucose consumption were analyzed by reversed phase high-performance liquid chromatography (HPLC) using a Waters HPLC system (Waters, Milford, MA) equipped with a Syneri 4 μ Hydro-RP column (Phenomenex, Aschaffenburg, Germany) and using phosphate buffer (50 mM KH2PO4, pH 2.9) as eluent. As standards, 12 different carboxylic acids (oxalate, gluconate, formate, pyruvate, malate, 2-oxoglutarate, lactate, acetate, citrate, fumarate, succinate, and propionate) were used. Data analysis was done using the Millenium32 3.05.01. Software (Waters, Eschborn, Germany).
Similarity Analysis of Physiological Data
The results of all physiological tests were used for the construction of a matrix with a binary code for the presence or absence of each phenotypic trait. Similarity check was performed using the MVSP 3.1 Software (Kovach Computing Services, Pentreath, UK) and the Dice coefficient for calculation of the distance matrix.
Determination of Intact Phospholipids and Phospholipid Fatty Acids
Cells were grown in oxic media amended with lactate (10 mM), harvested at the end of the exponential growth phase by centrifugation, washed with phosphate buffer (130 mM NaCl, 5 mM NaH2PO4, 5 mM Na2HPO4, pH 7.4), freeze-dried, and stored at −20°C. Aliquots (30–100 mg) of the freeze-dried cells were extracted ultrasonically up to 10 times for 10 min each using a solvent mixture of methanol/dichloromethane/ammonium acetate buffer pH 7.6 (2:1:0.8 by volume) in centrifuge tubes (modified after Vancanneyt et al. [56]). After centrifugation at 2200×g at 15°C for 10 min, the supernatants were removed and collected in a separatory funnel. Dichloromethane and ammonium acetate buffer (pH 7.6) were added to the combined extracts to achieve a final ratio of methanol/dichloromethane/ammonium acetate buffer of 1:1:0.9 (by volume). After phase separation, the organic phase was removed, and the aqueous phase was re-extracted with dichloromethane five times. Combined extracts were dried over anhydrous sodium sulfate, evaporated to dryness and stored at −20°C.
Lipid extracts were dissolved in 1 mL dichloromethane/methanol 9:1 (by volume) and then chromatographically separated according to Zink and Mangelsdorf [65]. Two glass columns in sequence filled with pure silica (1 g silica 60, 63–200 μm, dried at 110°C for 16 h) and Florisil (1 g magnesium silica gel 150–250 μm, Merck, Darmstadt, Germany) were used to gain four fractions: (1) neutral lipids (eluted with 20 mL dichloromethane); (2) free fatty acids (50 mL methyl formate with 0.025% v/v pure acetic acid); (3) glycolipids (20 mL acetone), and (4) phospholipids. To obtain the phospholipid fraction, the Florisil column was removed, and only the silica column was eluted with 25 mL methanol. All fractions were evaporated to dryness and stored at −20°C.
Aliquots of the phospholipid fractions were transesterified with trimethylsulfonium-hydroxide as described by Müller et al. [32]. The methyl esters obtained were quantified by gas chromatography–flame ionization detector (GC-FID; Hewlett Packard HP 5890 Series II gas chromatograph, Hewlett Packard, Waldbronn, Germany) equipped with a DB-5HT capillary column (30 m 0.25 mm, 0.1 μm film thickness, J&W Folsom, CA) and identified by GC-MS using a Finnigan MAT SSQ 710B mass spectrometer (Finnigan-Thermoquest, San Jose, CA). The carrier gas was helium (constant pressure of 12 psi). The oven temperature was raised from 60°C (isothermal for 2 min) to 360°C at a rate of 3°C min−1 and held for 5 min. Mass spectra were collected in full scan mode (m/z 50–650, ionization energy 70 eV). Positions of double bonds were tentatively assigned by comparison with retention times of standards (Bacterial Acid Methyl Esters CP Mix; Supelco, Bellefonte, PA).
Results
Phylogenetic Affiliation of the Photobacterium Strains
Based on short 16S rRNA gene fragments analyzed previously, the isolates were originally found to be closely related to P. profundum [49]. However, several novel Photobacterium species have been described since then (P. aplysiae [47]; P. frigidiphilum [46]; P. lipolyticum [64], and P. rosenbergii [52]), whereas the former Hyphomicrobium indicum was transferred to this genus [63]. These sequences and almost complete 16S rRNA gene sequences (most of them 1100 to 1300 bp long) of the isolates from the Mediterranean Sea sediments were now included into the phylogenetic analysis.
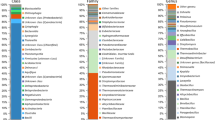
It turned out that the isolates shared between 96 and 100% sequence similarity of the 16S rRNA genes with each other. Their sequences affiliated with a phylogenetic branch containing Photobacterium aplysiae, P. frigidiphilum, P. indicum, P. lipolyticum, and P. profundum (Fig. 1). The strains were most closely related to either P. frigidiphilum or P. aplysiae (sequence similarities 97.1 to 99.5% and 96.8 to 99.5%, respectively), suggesting that the strains belong to two separate phylogenetic clusters (Mediterranean cluster I and II, Fig. 1). Although phylogenetic trees obtained by different methods showed some variation in the branching patterns (what is supported by the low bootstrap values on the maximum likelihood tree shown in Fig. 1), consistently, two separate clusters were obtained. Cluster I comprised strains obtained from all three sampling sites. Most of them were isolated from sapropels (strains S4, S10, S12, S14, S1E, J16, J34, 67FSB, and 67TD), whereas only two strains originated from hemipelagic surface (strain Z0F) and intermediate (strain Z1E) layers. In contrast, strains of cluster II were obtained exclusively from a single site (567) but appeared to be less confined to sapropels or hemipelagic sediments.
Maximum likelihood tree showing the phylogenetic positions of the two Photobacterium clusters and closely related taxa. Sequences of Shewanella (5) and Vibrio (5) were taken as outgroup and ingroup, respectively. 16S rRNA gene sequences of Mediterranean isolates were added to a backbone tree consisting of validly described species by maximum parsimony using the ARB software and a specific Photobacterium filter (Ludwig et al. [29]). Numbers at nodes are percentages bootstrap values, based on 1000 iterations
Phenotypic Characteristics
Microscopic inspection revealed that cells were rod-shaped, as considered to be typical for the genus Photobacterium. However, cells were often enlarged and irregularly shaped because of the accumulation of huge amounts of endogenous storage granula (Fig. 2). Sudan-black staining (0.05% w/v in ethanol abs) revealed that these consisted of poly-3-hydroxyalkanoate. The cells were motile by monopolar flagella. Luminescence was not observed.
According to their growth behavior on dilute yeast extract–peptone–glucose agar plates, the strains could be separated into two groups that reflected the two phylogenetic clusters. Cluster I strains were characterized by large cream-colored colonies that were formed normally within 2 days of incubation at 20°C. The remaining strains grew distinctly slower and needed several days to weeks of incubation at 20°C to form small whitish colonies (diameter generally less than 0.2 mm).
Molecular Characterization
For all strains tested, genomic fingerprinting based on ERIC-PCR yielded a clear and unique pattern of amplified DNA fragments of different lengths that could be used for cluster analysis (Fig. 3). Three groups of patterns were recognized. Strains of cluster I had very uniform ERIC band patterns clearly distinct from the other strains. Within this cluster, the strains grouped corresponding to the sampling site from which they were isolated.
Cluster analysis of ERIC-PCR fingerprinting band patterns of the Photobacterium strains. Strains of Mediterranean cluster II formed two subgroups (A and B). The dendrogram was calculated using Pearson correlation and UPGMA and is based on computer-generated densitometric curves obtained by analysis of negative images of ethidium bromide stained agarose gels
Strains belonging to cluster II yielded two separate subclusters based on ERIC band patterns (Mediterranean cluster II subgroup A and B, Fig. 3). Subgroup A comprised most of the strains (five of seven) originating from sapropel 1, and two strains (J43 and J156) isolated from the intermediate layer beneath (Z1). The second subgroup (B) exhibited the highest variability in band patterns and comprised strains originating from all four depths analyzed.
Denaturing gradient gel electrophoresis (DGGE) analysis revealed that all of the Photobacterium strains possessed up to five different rrn gene copies (differing in one to three bases in approximately 500 bp). Based on the presence or absence of certain bands in the DGGE gel, three subgroups were identified that confirmed the clusters defined on basis of the ERIC-PCR results (data not shown).
Aerobic Substrate Utilization
Under oxic conditions, the strains grew with a range of different substrates. The majority of strains utilized the complex substrates yeast extract, casamino acids, and peptone and poly- (starch, chitin) and monosccharides (glucose, fructose, and mannose), monocarboxylic acids (pyruvate, lactate), and tricarboxylic acid cycle intermediates (citrate, 2-oxoglutarate, succinate; Table 2). Only a very few strains (<4) grew with n-propanol, ethylene glycol, aromatic compounds, certain carboxylic, and amino acids (glycolate, tartrate, crotonate, isoleucine, and phenylalanine), or with C1 compounds (formate, and methanol).
With respect to their metabolic capacities significant differences between cluster I and II strains were observed. Strains of cluster I exhibited relatively uniform substrate utilization patterns (averaged pairwise similarities: 0.87 ± 0.05, Fig. 4) growing with 27 to 36 of the provided carbon sources. All of them grew with the amino acids alanine, arginine, asparagine, glutamine, proline, tryptophane, and on fumarate, malate, acetate, capronate, and glucosamine, whereas these substrates were utilized only by a very few strains belonging to cluster II.
Mediterranean cluster II strains showed irregular results (Fig. 4). Each strain exhibited its unique set of substrates that supported growth. When compared to the majority of the strains, a few isolates (J10, J33 and J15) appeared to be rather restricted in their metabolic capacities. However, despite aerobic growth on only 10 to 13 substrates, including yeast extract, glucose, and asparagine, these three strains nonetheless readily fermented carbohydrates, amino acids, and carboxylic acids (Table 1).
Anaerobic Metabolism
All strains were able to grow in the absence of oxygen by fermentation or by anaerobic respiration (Table 1). All isolates of cluster I reduced nitrate to nitrite, whereas five strains grew with DMSO and two with AQDS as electron acceptor. One of the isolates grew in the assay amended with humic acids. In contrast, only 8 of the 14 strains belonging to cluster II grew by anaerobic respiration. Seven of these strains used nitrate and five reduced DMSO. AQDS reduction or growth on humic acids was not observed for this cluster. It is interesting to note that in cluster II, nitrate reduction was found for most isolates from surface and the youngest sapropel S1 (six out of nine strains) but was less common in isolates originating from the deeper layers (one of five strains). None of the strains reduced manganese oxides, ferric hydroxide, sulfite, thiosulfate, or elemental sulfur.
All strains grew by fermentation of glucose. The mixture of dicarboxylic acids and lactate was fermented by all strains of cluster I and by 9 out of 14 strains of cluster II. Fermentative growth on amino acids was found for the majority of strains in both clusters. Two strains (67FSB and J43) grew even in the assay containing alcohols.
The fermentation assays with glucose as substrate were analyzed by HPLC. In the supernatant, four to six different acids were identified, with formate, 2-oxoglutarate, lactate, acetate, and citrate, being detected in almost all cultures and suggesting mixed acid fermentation (data not shown). With concentrations often exceeding 13 mmol·L−1, formate was found to be the major fermentation product, except for four strains of cluster II. These strains (J13, J18, J43, and J156), which were isolated from the deeper layers Z1 and S5, were remarkable as they also failed to reduce nitrate and to grow fermentatively with amino acids or carboxylic acids.
Temperature Range, Salinity Tolerance, and pH Requirements
With respect to their growth temperatures, no clear differences between the two clusters were observed. With exception of isolate J34 (cluster I, T min = 6°C), all strains grew at 4°C. Most of them revealed an upper temperature limit for growth at 33°C (Table 1), whereas four strains did not grow above 30°C and five still grew at 35°C. The strains were also quite uniform with respect to their salt and pH requirements. None of them grew at pH below 5.5, but they were tolerating rather alkaline conditions with pH values of up to 9.3. All strains required Na+ and salinities of at least 5‰ for growth. The upper salinity limit for growth was between 50 to 75‰. Strain J18 was an exception, growing at salinities of 100‰.
Phospolipid Content
Eight strains (four of each cluster) were analyzed for their phospholipids. Generally, all strains yielded very similar results with phosphatidyl glycerol (12-20%) and phosphatidyl ethanolamine (70–85%) as the dominant phosopholipid classes. Analysis of the polar lipid fatty acids revealed the dominance of n-16:0 (up to 32%), n-16:1 (up to 48%), and n-18:1 (up to 16.6%) fatty acids in both clusters (Table 2). Although all strains of cluster II contained n-20:5ω3 (0.3–2%), traces of this fatty acid type were found only in two of the four strains of cluster I. The latter strains, in turn, contained n-18:2 and saturated short-chain fatty acids (n-9:0 to n-13:0).
Discussion
In the present study, we have analyzed a large number Photobacterium strains isolated from subsurface sediments. The strains appear well adapted to their environment and form two distinct clusters that differ with respect to genomic diversity, their physiological capacities, lipid composition, and their habitat. Strains of cluster I were isolated almost exclusively from organic-rich sapropels, whereas for the presence of the second cluster, no correlation to certain sediment layers was found. Therefore, it can be concluded that the two clusters occupy different ecological niches.
Photobacterium sp. as Indigenous Subsurface Microbes
Although Photobacterium species were so far known from pelagic environments and surface sediments, this genus apparently forms subsurface populations that thrive in this inhospitable environment. It can be regarded as almost certain that the isolates are indigenous deep-biosphere representatives. Deep-well plates containing the MPN series from which the majority of strains were isolated had four additional rows that were not inoculated and served as control. None of the controls was proven positive, indicating that contamination during set-up of MPN series can be ruled out [49]. Furthermore, the isolates were repeatedly obtained during different sampling campaigns (1998 and 2001) and applying various cultivation conditions (Table 1). The high viable counts (up to 3.3% of the total count) [49] also indicate that Photobacterium sp. are numerically important and therefore most likely metabolically active members of the sediment communities. If they were inactive, it could be expected that their numbers strongly decreased with depth: This was not the case. In addition, dormant stages such as spores or cysts are not described for this genus [5].
If the Photobacterium strains represent indigenous subsurface microorganisms, they must be able to grow under in situ conditions. In fact, the isolates grew well under anoxic conditions and exhibited anaerobic pathways so far unknown for Photobacterium spp. [5] like fermentation of dicarboxylic or amino acids. The latter capacity was, however, recently inferred from genome sequencing data of the sediment-dwelling P. profundum [59].
The Eastern Mediterranean Sea is characterized by low primary production and sedimentation rates. Consequently, organic matter contents and microbial activities in the surface layers of the sediments are low (Table 3). Under these conditions, oxygen and nitrate may penetrate as deep as the top sapropel S1 [48, 57] and support growth of the Photobacterium strains. Profiles of Mn2+ and Fe2+ indicate that metal reduction is the dominant terminal oxidation process in this layer [57]. Although a clear drop in redox potential was found across sapropel 1, in the layers underneath, still oxidized conditions with positive redox potentials prevail [57]. This is in line with the low sulfate reduction rates observed in the younger sapropels S1 to S7 (Table 3) and might explain the recovery of only very few sulfate reducers from sapropels [49] and the presence of only negligible sulfate gradients even as deep as 100 m below surface [12]. Oxidized conditions, in turn, might favor growth of facultative anaerobes such as Photobacterium sp., rather than of strict anaerobes and might explain their predominance in the culture collection obtained from this habitat [49], but also other deep-sea sediments [13].
The finding that our isolates degrade a broad spectrum of monomeric and polymeric substrates classifies them as generalists, and this nutritional versatility might be a useful adaptation to their habitat. Although sapropels are characterized by the presence of highly recalcitrant polymeric material [22], easily degradable substrates like carbohydrates or peptides are still present at very low concentrations or adsorbed to the kerogen matrix of the sapropels [9]. However, the degradation capacities of our isolates might be even underestimated as all growth tests were performed under atmospheric pressure. Gene expression analyses of the deep-sea strain P. profundum SS9 indicated that certain metabolic pathways, such as fermentation of amino acids or the degradation of biopolymers, might be expressed only at elevated hydrostatic pressure [59].
Microdiversity in Subsurface Environments
The Photobacterium clusters investigated in this study differed with respect to their microdiversity. Mediterranean cluster I strains, which were predominantly isolated from sapropel layers, apparently maintained a highly stable genome organization over thousands of years while Mediterranean cluster II strains exhibited a higher level of microdiversity. However, this degree of genomic heterogeneity was much lower than that generally found in pelagic bacteria [20, 42] or those inhabiting surface sediments [3, 43]. At first sight, this might correlate with the highly constant environmental conditions in subsurface environments. Seasonal changes of physicochemical conditions and temperature [54], varying light conditions [31], or the increasing pressure from the sea surface to the deep-sea [26] were shown to stimulate the development of specifically adapted subpopulations within single species, and even protozoan grazing or virus infection [15] were supposed to increase microdiversity. These stimuli in combination with short generation times may stimulate genotypic and phenotypic diversification in bacteria from pelagic environments and surface sediments but cannot sufficiently explain existence of microdiversity in subsurface bacteria like our Photobacterium strains. In fact, genome stability can be interpreted as an indication for metabolic activity. Starving or resting cells were shown to enhance the generation of diversity [2], and ecologically successful populations do not necessarily need to develop a high genetic diversity [19]. This could explain the low level of diversity in Mediterranean cluster I. On the other hand, increased variability within a population, e.g., in Mediterranean cluster II, can be considered as complementary strategy for long-term survival. The development of multiple ecotypes allows an immediate response to a wide range of environmental conditions [18, 23, 31].
Although the concept of periodic selection predicts that two bacterial populations cannot occupy the same ecological niche and that one type would outcompete the other [35], Thompson et al. [53] reported on the consistent co-occurrence of a large diversity of Vibrio genomovars within a natural community. They concluded that most of the genotypic diversity they detected must be “ecologically neutral” and explained the co-occurrence with the erratic and unpredictable appearance of resources and grazing that could eliminate dominant subpopulations. These factors can be ruled out for subsurface bacteria. On the other hand, it might be that the low in situ growth rates do not allow one subpopulation overgrowing another and therefore enable co-existence. The highly complex and diverse structure of the sapropelic kerogen (e.g., polyphenylic and aliphatic residues, [22]) most likely requires a specialized bacterial community for degradation. Because Photobacterium cluster I exhibited low diversity and was detected mostly in the sapropels, it can be expected that it is involved in this process. Strains of cluster II in contrast were found in all layers and showed a higher diversity in terms of substrate utilization and genomic level. This might indicate a role as opportunistic bacterium, which quickly reacts to changing conditions, e.g., substrate availability. This indicates that the two Photobacterium clusters occupy distinct ecological niches.
References
Acinas SG, Klepac-Ceraj V, Hunt DE, Pharino C, Ceraj I, Distel DL, Polz MF (2004) Fine-scale phylogenetic architecture of a complex bacterial community. Nature 430:551–554
Arber W, Naas T, Blot M (1994) Generation of genetic diversity by DNA rearrangements in resting bacteria. FEMS Microbiol Ecol 15:5–14
Bagwell CE, Lovell CR (2000) Microdiversity of culturable diazotrophs from rhizoplanes of the salt marsh grasses Spartina alterniflora and Juncus roemerianus. Microb Ecol 39:128–136
Balch WE, Fox GE, Magrum LJ, Woese CR, Wolfe RS (1979) Methanogens: reevaluation of a unique biological group. Microbiol Rev 43:260–296
Baumann P, Baumann L (1984) Genus II Photobacterium Beijerinck 1889, 401 AL. In: Krieg, NR, Holt, JG (Eds.) Bergey’s Manual of Systematic Bacteriology. Williams and Wilkins, Baltimore, pp 539–545
Beijerinck MW (1889) Le Photobacterium luminosum. Bactérie lumineuse de la Mer Nord. Arch Néerl Sci 23:401–427
Benz M, Schink B, Brune A (1998) Humic acid reduction by Propionibacterium freudenreichii and other fermenting bacteria. Appl Environ Microbiol 64:4507–4512
Coolen MJL, Cypionka H, Sass AM, Sass H, Overmann J (2002) Ongoing modification of Mediterranean Pleistocene sapropels mediated by prokaryotes. Science 296:2407–2410
Coolen MJL, Overmann J (2000) Functional exoenzymes as indicators of metabolically active bacteria in 124,000-year-old sapropel layers of the eastern Mediterranean Sea. Appl Environ Microbiol 66:2589–2598
Cowan DA (2000) Microbial genomes-the untapped resource. Trends Biotechnol 18:14–16
Cowen JP, Giovannoni SJ, Kenig F, Johnson HP, Butterfield D, Rappe MS, Hutnak M, Lam P (2003) Fluids from aging ocean crust that support microbial life. Science 299:120–123
Cragg BA, Law KM, Cramp A, Parkes RJ (1998) The response of bacterial populations to sapropels in deep sediments of the Eastern Mediterranean (site 969). In: Robertson, AHF, Emeis, K-C, Richter, C, Camerlenghi, A (Eds.) Proc Ocean Drilling Program. Sci Results 160, College Station, Texas, pp 303–307
D’Hondt S, Jørgensen BB, Miller DJ, Batzke A, Blake R, Cragg BA, Cypionka H, Dickens GR, Ferdelman T, Hinrichs K-U, Holm NG, Mitterer R, Spivack A, Wang G, Bekins B, Engelen B, Ford K, Gettemy G, Rutherford SD, Sass H, Skilbeck CG, Aiello IW, Guerin G, House CH, Inagaki F, Meister P, Naehr T, Niitsuma S, Parkes RJ, Schippers A, Smith DC, Teske A, Wiegel J, Padilla CN, Acosta JLS (2004) Distributions of microbial activities in deep subseafloor sediments. Science 306:2216–2221
DeLong EF (2004) Microbial life breathes deep. Science 306:2198–2200
Feil EJ (2004) Small change: keeping pace with microevolution. Nat Rev Microbiol 2:483–495
Fuhrman JA, Campbell L (1998) Microbial microdiversity. Nature 393:410–411
Grasshoff K, Kremling K, Ehrhardt M (1999) Methods of Seawater Analysis, 3rd edn. Wiley-VCH, Weinheim
Gray ND, Howarth R, Rowan A, Pickup RW, Jones JG, Head IM (1999) Natural communities of Achromatium oxaliferum comprise genetically, morphologically, and ecologically distinct subpopulations. Appl Environ Microbiol 65:5089–5099
Hahn MW, Pockl M, Wu QL (2005) Low intraspecific diversity in a Polynucleobacter subcluster population numerically dominating bacterioplankton of a freshwater pond. Appl Environ Microbiol 71:4539–4547
Jaspers E, Overmann J (2004) Ecological significance of microdiversity: Identical 16S rRNA gene sequences can be found in bacteria with highly divergent genomes and ecophysiologies. Appl Environ Microbiol 70:4831–4839
Johnson RM, Weisrock WP (1969) Hyphomicrobium indicum sp. nov. Hyphomicrobiaceae Douglas. Int J Syst Bacteriol 19:295–307
Killops SD, Killops VJ (2004) An Introduction to Organic Geochemistry, 2nd edn. Blackwell, Oxford
Klepac-Ceraj V, Bahr M, Crump BC, Teske AP, Hobbie JE, Polz MF (2004) High overall diversity and dominance of microdiverse relationships in salt marsh sulphate-reducing bacteria. Environ Microbiol 6:686–698
Kormas KA, Smith DC, Edgcomb V, Teske A (2003) Molecular analysis of deep subsurface microbial communities in Nankai Trough sediments (ODP Leg 190, Site 1176). FEMS Microbiol Ecol 45:115–125
Kroon D, Alexander I, Little M, Lourens LJ, Matthewson A, Robertson AHF, Sakamoto T (1998) Oxygen isotope and sapropel stratigraphy in the Eastern Mediterranean during the last 3.2 million years. In: Robertson, AHF, Emeis, K-C, Richter, C, Camerlenghi, A (Eds.) Proc Ocean Drilling Program. Sci Results 160, College Station, Texas, pp 181–189
López-López A, Bartual SG, Stal L, Onyshchenko O, Rodríguez-Valera F (2005) Genetic analysis of housekeeping genes reveals a deep-sea ecotype of Alteromonas macleodii in the Mediterranean Sea. Environ Microbiol 7:649–659
Lourens LJ, Hilgen FJ, Gudjonsson L, Zachariasse WJ (1992) Late Pliocene to early Pleistocene astronomically forced sea surface productivity and temperature variations in the Mediterranean. Mar Micropaleontol 19:49–78
Lovley DR, Coates JD, Blunt-Harris EL, Phillips EJP, Woodward JC (1996) Humic substances as electron acceptors for microbial respiration. Nature 382:445–448
Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, Buchner A, Lai T, Steppi S, Jobb G, Forster W, Brettske I, Gerber S, Ginhart AW, Gross O, Grumann S, Hermann S, Jost R, Konig A, Liss T, Lussmann R, May M, Nonhoff B, Reichel B, Strehlow R, Stamatakis A, Stuckmann N, Vilbig A, Lenke M, Ludwig T, Bode A, Schleifer K-H (2004) ARB: A software environment for sequence data. Nucleic Acids Res 32:1363–1371
Martens-Habbena W, Sass H (2006) Sensitive determination of microbial growth by nucleic acid staining in aqueous suspension. Appl Environ Microbiol 72:87–95
Moore LR, Rocap G, Chisholm SW (1998) Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature 393:464–467
Müller KD, Schmid EN, Kroppenstedt RM (1998) Improved identification of mycobacteria by using the microbial identification system in combination with additional trimethylsulfonium hydroxide pyrolysis. J Clin Microbiol 36:2477–2480
Newberry CJ, Webster G, Cragg BA, Parkes RJ, Weightman AJ, Fry JC (2004) Diversity of prokaryotes and methanogenesis in deep subsurface sediments from the Nankai Trough, Ocean Drilling Program Leg 190. Environ Microbiol 6:274–287
Nogi Y, Masui N, Kato C (1998) Photobacterium profundum sp. nov., a new, moderately barophilic bacterial species isolated from a deep-sea sediment. Extremophiles 2:1–7
Palys T, Nakamura LK, Cohan FM (1997) Discovery and classification of ecological diversity in the bacterial world: the role of DNA sequence data. Int J Syst Bacteriol 47:1145–1156
Parkes RJ, Cragg BA, Bale SJ, Getliff JM, Goodman K, Rochelle PA, Fry JC, Weightman AJ, Harvey SM (1994) Deep bacterial biosphere in Pacific Ocean sediments. Nature 371:410–413
Parkes RJ, Webster G, Cragg BA, Weightman AJ, Newberry CJ, Ferdelman TG, Kallmeyer J, Jorgensen BB, Aiello IW, Fry JC (2005) Deep sub-seafloor prokaryotes stimulated at interfaces over geological time. Nature 436:390–394
Passier HF, Middelburg JJ, van Os BJH, De Lange GJ (1996) Diagenetic pyritisation under eastern Mediterranean sapropels caused by downward sulphide diffusion. Geochim Cosmochim Acta 60:751–763
Pukall R, Päuker O, Buntefuß D, Ulrichs G, Lebaron P, Bernard L, Guindulain T, Vives-Rego J, Stackebrandt E (1999) High sequence diversity of Alteromonas macleodii-related cloned and cellular 16S rDNAs from a Mediterranean seawater mesocosm experiment. FEMS Microbiol Ecol 28:335–344
Rohling EJ (1994) Review and new aspects concerning the formation of eastern Mediterranean sapropels. Mar Geol 122:1–28
Salle AM (1961) Fundamental Principles of Bacteriology, 5th edn. McGraw-Hill, New York, Toronto, London
Sass AM, Sass H, Coolen MJL, Cypionka H, Overmann J (2001) Microbial communities in the chemocline of a hypersaline deep-sea basin (Urania Basin, Mediterranean Sea). Appl Environ Microbiol 67:5392–5402
Sass H, Wieringa E, Cypionka H, Babenzien HD, Overmann J (1998) High genetic and physiological diversity of sulfate-reducing bacteria isolated from an oligotrophic lake sediment. Arch Microbiol 170:243–251
Schippers A, Neretin LN, Kallmeyer J, Ferdelman TG, Cragg BA, Parkes RJ, Jørgensen BB (2005) Prokaryotic cells of the deep sub-seafloor biosphere identified as living bacteria. Nature 433:861–864
Schloter M, Lebuhn M, Heulin T, Hartmann A (2000) Ecology and evolution of bacterial microdiversity. FEMS Microbiol Rev 24:647–660
Seo HJ, Bae SS, Lee J-H, Kim S-J (2005) Photobacterium frigidiphilum sp. nov., a psychrophilic, lipolytic bacterium isolated from deep-sea sediments of Edison Seamount. Int J Syst Evol Microbiol 55:1661–1666
Seo HJ, Bae SS, Yang SH, Lee JH, Kim S-J (2005) Photobacterium aplysiae sp. nov., a lipolytic marine bacterium isolated from eggs of the sea hare Aplysia kurodai. Int J Syst Evol Microbiol 55:2293–2296
Slomp CP, Thomson J, de Lange GJ (2002) Enhanced regeneration of phosphorus during formation of the most recent eastern Mediterranean sapropel (S1). Geochim Cosmochim Acta 66:1171–1184
Süß J, Engelen B, Cypionka H, Sass H (2004) Quantitative analysis of bacterial communities from Mediterranean sapropels based on cultivation-dependent methods. FEMS Microbiol Ecol 51:109–121
Süß J, Schubert K, Sass H, Cypionka C, Overmann J, Engelen B (2006) Widespread distribution and high abundance of Rhizobium radiobacter within Mediterranean subsurface sediments. Environ Microbiol 8:1753–1763
Ten Haven HL, De Lange GJ, McDuff RE (1987) Interstitial water studies of late Quarternary Eastern Mediterranean sediments with emphasis on early diagenetic reactions and evaporitic salt influences. Mar Geol 75:119–136
Thompson FL, Thompson CC, Naser S, Hoste B, Vandemeulebroecke K, Munn C, Bourne D, Swings J (2005) Photobacterium rosenbergii sp. nov. and Enterovibrio coralii sp. nov., vibrios associated with coral bleaching. Int J Syst Evol Microbiol 55:913–917
Thompson JR, Pacocha S, Pharino C, Klepac-Ceraj V, Hunt DE, Benoit J, Sarma-Rupavtarm R, Distel DL, Polz MF (2005) Genotypic diversity within a natural coastal bacterioplankton population. Science 307:1311–1313
Thompson JR, Randa MA, Marcelino LA, Tomita-Mitchell A, Lim E, Polz MF (2004) Diversity and dynamics of a North Atlantic coastal Vibrio community. Appl Environ Microbiol 70:4103–4110
Toffin L, Webster G, Weightman AJ, Fry JC, Prieur D (2004) Molecular monitoring of culturable bacteria from deep-sea sediment of the Nankai Trough, Leg 190 Ocean Drilling Program. FEMS Microbiol Ecol 48:357–367
Vancanneyt M, Witt S, Abraham WR, Kersters K, Frederickson HL (1996) Fatty acid content in whole-cell hydrolysates and phospholipid fractions of Pseudomonas: a taxonomic evaluation. Syst Appl Microbiol 19:528–540
Van Santvoort PJM, De Lange GJ, Thomson J, Cussen H, Wilson TRS, Krom MD, Ströhle K (1996) Active post-depositional oxidation of the most recent sapropel (S1) in sediments of the Eastern Mediterranean. Geochim Cosmochim Acta 60:4007–4024
Versalovic J, Koeuth T, Lupski JR (1991) Distribution of repetitive DNA-sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res 19:6823–6831
Vezzi A, Campanaro S, D’Angelo M, Simonato F, Vitulo N, Lauro FM, Cestaro A, Malacrida G, Simionati B, Cannata N, Romualdi C, Bartlett DH, Valle G (2005) Life at depth: Photobacterium profundum genome sequence and expression analysis. Science 307:1459–1461
Webster G, Parkes RJ, Fry JC, Weightman AJ (2004) Widespread occurrence of a novel division of bacteria identified by 16S rRNA gene sequences originally found in deep marine sediments. Appl Environ Microbiol 70:5708–5713
Wellsbury P, Mather I, Parkes RJ (2002) Geomicrobiology of deep, low organic carbon sediments in the Woodlark Basin, Pacific Ocean. FEMS Microbiol Ecol 42:59–70
Widdel F, Bak F (1992) Gram-negative mesophilic sulfate-reducing bacteria. In: Balows A, Trüper HG, Dworkin M, Harder W, Schleifer KH (Eds.) The Prokaryotes. A Handbook on the Biology of Bacteria: Ecophysiology, Isolation, Identification, Applications, 2nd edn. Springer, New York, pp 3352–3378
Xie C-H, Yokota A (2004) Transfer of Hyphomicrobium indicum to the genus Photobacterium as Photobacterium indicum comb. nov. Int J Syst Evol Microbiol 54:2113–2116
Yoon J-H, Lee J-K, Kim Y-O, Oh T-K (2005) Photobacterium lipolyticum sp. nov., a bacterium with lipolytic activity isolated from the Yellow Sea in Korea. Int J Syst Evol Microbiol 55:335–339
Zink KG, Mangelsdorf K (2004) Efficient and rapid method for extraction of intact phospholipids from sediments combined with molecular structure elucidation using LC-ESI-MS-MS. Anal Bioanal Chem 380:798–812
Acknowledgments
The support of the scientific party of RV Meteor cruises M40/4 and M51/3, with Christoph Hemleben as chief scientist is gratefully acknowledged. We thank two anonymous reviewers for their support and valuable discussion. Jürgen Rullkötter, Jürgen Köster, and Bernd Kopke are acknowledged for providing facilities for phospholipid analysis and for experimental help. This work was supported by a grant of the Deutsche Forschungsgemeinschaft.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Süß, J., Herrmann, K., Seidel, M. et al. Two Distinct Photobacterium Populations Thrive in Ancient Mediterranean Sapropels. Microb Ecol 55, 371–383 (2008). https://doi.org/10.1007/s00248-007-9282-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-007-9282-6