Abstract
Molluscan shells are composed of calcium carbonates, with small amounts of extracellular matrices secreted from mantle epithelial cells. Many types of shell matrix proteins (SMPs) have been identified from molluscan shells or mantle cells. The pen shell Atrina pectinata (Pinnidae) has two different shell microstructures, the nacreous and prismatic layers. Nacreous and prismatic layer-specific matrix proteins have been reported in Pteriidae bivalves, but remain unclear in Pinnidae. We performed transcriptome analysis using the mantle cells of A. pectinata to screen the candidate transcripts involved in its prismatic layer formation. We found Asprich and nine highly conserved prismatic layer-specific SMPs encoding transcript in P. fucata, P. margaritifera, and P. maxima (Tyrosinase, Chitinase, EGF-like proteins, Fibronectin, valine-rich proteins, and prismatic uncharacterized shell protein 2 [PUSP2]) using molecular phylogenetic analysis or multiple alignment. We confirmed these genes were expressed in the epithelial cells of the mantle edge (outer surface of the outer fold) and the mantle pallium. Phylogenetic character mapping of these SMPs was used to infer a possible evolutionary scenario of them in Pteriomorphia. EGF-like proteins, Fibronectin, and valine-rich proteins encoding genes each evolved in the linage leading to four Pteriomorphia (Mytilidae, Pinnidae, Ostreidae, and Pteriidae), PUSP2 evolved in the linage leading to three Pteriomorphia families (Pinnidae, Ostreidae, and Pteriidae), and chitinase was independently evolved as SMPs in Mytilidae and in other Pteriomorphia (Pinnidae, Ostreidae, and Pteriidae). Our results provide a new dataset for A. pectinata SMP annotation, and a basis for understanding the evolution of prismatic layer formation in bivalves.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Mollusks are among the most diverse phyla; they have evolved several ancient shell forms since the Cambrian. Mollusk shells are composed of calcium carbonates (aragonite or calcite) and extracellular matrix components such as polysaccharides, proteins, and glycoproteins (Levi-Kalisman et al. 2001). These extracellular matrix materials are incorporated into or form scaffolds around crystals, and are therefore involved in crystal morphology, polymorphism, nucleation, or growth orientation (Belcher et al. 1996). The recent development of high-throughput DNA sequencing and mass spectrometry techniques has facilitated the identification of many skeletal matrix proteins from calcified organisms such as corals, mollusks, brachiopods, and sea urchins (e.g., Ramos-Silva et al. 2013; Marie et al. 2010; Jackson et al. 2015; Mann et al. 2008), some of which have demonstrated rapid evolution (e.g., Jackson et al. 2006). Although their skeletons are composed of different crystal polymorphs and/or microstructures, some skeletal matrix proteins share the same functional domains (e.g., von Willebrand factor type A, chitin-binding domain, or carbonic anhydrase) across these metazoans (Jackson et al. 2010; Marie et al. 2012, 2013, 2017; Zhang et al. 2012; Luo et al. 2015; Shimizu et al. 2019; Zhao et al. 2018).
The pen shell Atrina pectinata (Pinnidae) is a large benthic bivalve (shell length ≤ 30 cm) that is widely distributed in tidal and sub-tidal environments from subtropical to tropical regions. Its large posterior adductor muscles or other soft parts such as the mantle and foot are commercially important as fisheries resources, especially in Asian countries. The shell of A. pectinata has two layers, composed of calcium carbonate polymorphs; the outer prismatic layer contains calcite, and the inner nacreous layer contains aragonite (Nudelman et al. 2007; Okumura et al. 2012). These layers are also present in pearl oysters; for example, Pinctada fucata is a model mollusk species for biomineralization during nacreous and prismatic layer formation. However the prismatic structure in A. pectinata is strictly different from that in P. fucata (Okumura et al. 2012) (Fig. 1). The prismatic layer in A. pectinata consists of a monolithic single crystal and the crystal defects are not observed. On the other hand, that in P. fucata has a large variance of lattice spacing and subgrain structure that is formed from the arrangements of organic macromolecules and contains the crystal defects (Okumura et al. 2012; Kintsu et al. 2017). Although these differences were probably caused by differences of their matrix proteins, it has not been reported their differences yet.
Many shell matrix proteins (SMPs) have been reported in P. fucata and other Pinctada species; previous studies have shown that SMPs differ between the nacreous and prismatic layers (Suzuki and Nagasawa 2013). For example, N16/Pearlin and Pif were first isolated from the nacreous layer of P. fucata (Samata et al. 1999; Suzuki et al. 2009; Suzuki 2020). These SMPs have been found in the nacreous layer in P. margaritifera and P. maxima (Kono et al. 2000; Montagnani et al. 2011; Suzuki et al. 2013, 2017), but not in the prismatic layer of either species (Marie et al. 2012). Other SMPs including chitinase have been found only in the prismatic layer in P. margaritifera, P. maxima, and P. fucata (Marie et al. 2012; Kintsu et al. 2017; Zhao et al. 2018). Interestingly, some nacreous or prismatic layer-specific SMPs have been identified in freshwater pearl oysters (Marie et al. 2017). These findings suggest that differences among shell structures and/or calcium carbonate polymorphs are produced by specific SMPs secreted by mantle epithelial cells in spatially different parts of the shell (Thompson et al. 2000; Marie et al. 2012). Although many SMPs were reported from the family Pteriidae including genus Pinctada, only three SMPs (Asprich, Caspartin, and Calprismin) were reported from the family Pinnidae including genus Atrina and Pinna (Gotliv et al. 2005; Marin et al. 2005). Asprich identified from the mantle tissue in A. rigida consists of many aspartic acid residues (Gotliv et al. 2005) like Aspein in P. fucata (Takeuchi et al. 2016b). The partial amino acid sequences of casprismin and caspartin were identified from the prismatic layer of Pinna nobilis (Marin et al. 2005). Casprismin is glycosylated protein and comprise of a pattern of four cysteine residues (CX6CX13CX6C). Caspartin consists of many aspartic acid residues at the first 75N-terminus and involves in the inhibition of the precipitation of calcium carbonate (Marin et al. 2005). The whole amino acid sequences of these proteins have not been revealed yet. It has not been conducted the transcriptome analysis using the mantle tissue to screen the candidate biomineralization genes in the family Pinnidae yet.
In this study, we focused on the prismatic layer of the pen shell A. pectinata and analyzed transcriptome sequences using the outer part of the mantle tissues corresponding to the prismatic layer. We first searched the prismatic layer-related transcripts previously reported in in the family Pinnidae (Asprich, Caspartin, and Calprismin). To understand the universality of the prismatic layer-related SMPs between the A. pectinata and the pearl oysters, we searched for the highly conserved prismatic layer-related transcripts previously reported in three kinds of pearl oysters (P. fucata, P. margaritifera, and P. maxima). We also performed multiple alignment analysis and/or molecular phylogenetic analysis and annotated 7 candidate transcripts that probably involve in the prismatic layer formation (Asprich, Tyrosinase, Calcification-related Chitinase [CR-CN], EGF-like proteins [EGF-like 1 and EGF-like 2], Calcification-related Fibronectin [CR-FN], prismatic uncharacterized shell protein 2 [PUSP2]) from A. pectinata. These conserved proteins in the prismatic layers of the pearl oysters may play important roles in the prismatic layer formation or calcite crystallization in A. pectinata, as reported in P. margaritifera, P. maxima, and P. fucata.
Materials and Methods
RNA Extraction and Transcriptome Analysis
We bought adults of the pen shell A. pectinata (shell length is over 25 cm) from Tsukiji market (Tokyo, Japan). To extract total RNA, 50 ~ 100 mg of mantle tissues (mixed mantle edge and pallium) was dissected from three individuals, and Sepasol®-RNA I (1 ml) was added to each sample. After homogenizing samples for 5 min, 0.2 ml of chloroform was added to precipitate the proteins in each sample. The sample was centrifuged at 19,000×g for 15 min at 4 °C, and the supernatant was mixed with 0.5 ml 2-propanol to precipitate total RNA. After centrifugation at 19,000×g for 10 min at 4 °C, the supernatant was collected and 75% ethanol (1 ml) was added to remove salt. The samples were centrifuged at 19,000×g for 5 min at 4 °C and the supernatant was removed, and then finally total RNA was dissolved to distilled water. The concentration of total RNA was assessed by the absorbance at 260 nm. Before the RNA library preparation, we checked the quality of total RNA sample by Agilent 2100 Bioanalyzer (Agilent Technology, California, USA), and the mRNA sample was enriched from total RNA using oligo (dT) beads. We then prepared 150 bp RNA libraries from 1 µg of mRNA sample using NEBNext UltraRNA Library Prep Kit (E7350L, Illumina, California, USA), and performed 150-base pair-end sequencing using NovaSeq 6000 (Illumina). Low-quality reads (base quality is less than 20) were removed from raw read data. Only high-quality paired end reads were assembled with Trinity v2.5.1 (Grabherr et al. 2011; Haas et al. 2013). Hierarchical clustering was performed based on multiple mapping events and expression pattern and removed redundance with CORSET (Davison and Oshlack 2014). We chose the longest transcripts of each cluster as unigenes. We mapped the transcripts for each of the reads back to the de novo assembled sequence by RSEM (Li and Dewey 2011) and calculated the number of fragments per kilobase of exon per million mapped reads (FPKM) of each transcript. Quality of the assembled sequences was calculated by the BUSCO v2/v3 (Simão et al. 2015) using metazoan dataset implemented in gVolante (ver. 1.2.1) web server (Nishimura et al. 2017, https://gvolante.riken.jp/index.html, last accessed November 14, 2020). Coding sequence (CDS) prediction was conducted two ways, and about 65% of unigenes (40,111/61,263) were translated from nucleotide sequences to amino acid sequences. CDS was extracted from unigene sequences and translated into peptide sequences based on the standard codon table when we found the similar sequences from the non-redundant protein sequence database (NR) of NCBI (http://blast.ncbi.nlm.nih.gov, last accessed November 14, 2020) and Swiss-Prot database (https://www.uniprot.org, last accessed November 14, 2020) by BLAST. Unigenes with no hit in BLAST were analyzed with ESTScan (3.0.3) (Iseli et al. 1999) to predict their CDS. These shot-gun raw read sequences and assembly sequences are available in the DNA Data Bank of Japan (Sequence Read Archive: DRR209159, BioProject: PRJDB9333, BioSample: SAMD00206368, TSA: ICPQ01000001-ICPQ01061263).
Gene Functional Annotation
To find similar sequences to unigenes and CDS, we performed Diamond (v0.8.22) search using non-redundant protein sequence databases (NR) of NCBI (http://blast.ncbi.nlm.nih.gov, last accessed February 14, 2020, the e-value threshold is 1.0e−5), or Swiss-Prot database (https://www.uniprot.org, last accessed February 14, 2020, the e-value threshold is 1.0e−5). Results from NR and Swiss-Prot searches were integrated to Blast2Go v2.5 (Götz et al. 2008). We then performed gene function and pathway classification using Ontology (GO) terms (e-value thresholds are 1.0e−6).
Prediction of Prismatic Layer-Related Transcripts in A. pectinata
We first searched the similar transcripts of A. pectinata to the previous known acidic protein Asprich in A. rigida (Gotliv et al. 2005) using BLASTP program. A multiple sequence alignment was performed with the online version of MAFFT (v7.310; http://mafft.cbrc.jp/alignment/server/index, last accessed February 14, 2020; Katoh et al. 2002) using whole part of amino acid sequences. We then searched the similar transcripts of A. pectinata to 25 prismatic layer-related SMPs (12 gene groups) in P. fucata that are highly conserved among other pearl oysters P. margaritifera and P. maxima using BLASTP program (e-value < 1.0e−10). We searched the signal peptide and the structural domains by SignalP (Petersen et al. 2011) and Pfam protein domain search (https://pfam.xfam.org, last accessed February 14, 2020 Finn et al. 2016,), respectively. Multiple sequence alignments were performed with the online version of MAFFT (v7.310; http://mafft.cbrc.jp/alignment/server/index, last accessed February 14, 2020; Katoh et al. 2002), and these sequences were trimmed by TrimAl (1.2rev59) (Capella-Gutiérrez et al. 2009). We chose the best-fit amino acid substitution model using MEGA (v5.1) (Tamura et al. 2011) and maximum likelihood trees with 100 bootstrap replications were constructed with the online version of RAxML (Stamatakis 2014).
cDNA Synthesis and Gene Cloning
Synthesis of cDNA was conducted from 1 µg of total RNA using Prime Script RT reagent kit (#RR037A, Takara, Tokyo, Japan) according the manufacturer’s protocol. Partial sequences of nine target genes of A. pectinata (ApeAsprich, ApeCR-CN1, ApeCR-CN2, ApeEGF-like 1, ApeEGF-like 2, ApeCR-FN, ApeMP10, ApeAlveolin, and ApePUSP2) were amplified with PCR using specific primers designed with reference to transcriptome data (Supplementary Table S1). These PCR amplicons were purified by the QIAquick PCR Purification Kit (#28104, Qiagen, Hilden, Germany) and were ligated into the pGEM-T easy vectors using a DNA ligation kit (#A1360, Promega). The vectors were then transformed to the competent Escherichia coli BL21 cells. Inserts of the vectors were sequenced by ABI3130 (Applied Biosystems, CA) with the standard protocols using T7 and SP6 primers, and nucleotide sequences of them (ApeAsprich, ApeCR-CN1, ApeCR-CN2, ApeEGF-like 1, ApeEGF-like 2, ApeCR-FN, ApeMP10, ApeAlveolin, and ApePUSP2) are available in the DNA Data Bank of Japan (LC574994-LC575002) (Supplementary Table S1).
Probe Synthesis and Section in situ Hybridization
Fragments amplified using T7 and SP6 primers were purified by the QIAquick PCR Purification Kit (#28104, Qiagen). Antisense probes were synthesized using DIG RNA labeling Mix (#11277073910, Roche), 10 mM Dithiothreitol (DTT), RNase ribonuclease inhibitor (#SIN201, Toyobo), T7 or SP6 RNA polymerase with 1X transcription buffer (#10881767001 or #10810274001, Roche), and purified PCR products (500 ng per reaction) according to manufacturer’s protocol. Probe synthesis reactions were performed at 37 °C for at least 3 h and then treated with DNase I (#M6101, Promega) at 37 °C for 1 h. Synthesized probes were purified using NucAway spin columns (#AM10070, Thermo Fisher Scientific) and stored at − 20 °C.
Adult mantle tissues of A. pectinata were fixed with fixation buffer (4% paraformaldehyde, 0.5 M NaCl, 0.1 M MOPS, and 2 mM EGTA) overnight at 4 °C. After washing with PBS, samples were dehydrated with a gradual series of ethanol/PBS (30/70, 50/50, 80/20) for 10 min each at 20 °C and stored in 80% ethanol at − 20 °C. Samples were rehydrated with PBS, followed by the replacement with a gradual series of sucrose/PBS (5/95, 10/90, 15/85) for 15 min each at 20 °C and with 30% sucrose in PBS for overnight at 4 °C. Removing excess 30% sucrose solution, samples were mounted in cryomolds with frozen section compound (FSC 22, Leica) and were immediately frozen in -80 °C freezer for overnight. Sections (thickness is 20 µm) were prepared with the cryosection (CM1900, Leica) and put on the MAS-coated microscope slides (#MAS02, Matsunami Glass IND. LTD., Osaka, Japan). Slides were dried at 20 °C for at least 1 h and stored in the sealed box at − 20 °C until staining.
The slides were dried at 20 °C for at least 1 h and washed twice with PBS for 5 min each. After treatment with Proteinase K (1 µg/ml in PBS) at 20 °C for 10 min, slides were quickly washed three times with PBS and immediately fixed with fixation buffer (4% PFA in PBS) for 30 min. Samples were treated with 0.1 M triethanolamine (TEA) (pH 8.0) for 5 min. Slides were acetylated twice with 0.25% (v/v) acetic anhydride in 0.1 M TEA for 5 min each and then washed twice with PBS for 5 min each. Pre-hybridization was conducted with pre-hybridization buffer (50% formamide, 5x SSC, 5 mM EDTA) at 55 °C for 1 h. Solution was replaced with hybridization buffer (50% formamide, 5x SSC, 5 mM EDTA, 100 μg /ml Torula RNA, 100 μg/ml Heparin, 0.1% Tween-20) containing the RNA probes (final concentration: 0.5–1 ng/μl) and it was preheated at 80 °C for 10 min and hybridized overnight at 55 °C.
Excess probes were removed by washing with pre-hybridization buffer at 55 °C for 10 min and then washed with a gradual series of wash buffer (2x SSC with 0.1% Tween-20, 0.2x SSC with 0.1% Tween-20, 0.05x SSC with 0.1% Tween-20) for 30 min each at 55 °C. Two washes in maleic acid buffer (MAB) pH 7.5 were performed at 20 °C for 5 min each. Blocking was performed with blocking buffer (2% blocking reagent Roche, 11096176001) and 0.1% sheep serum for 1 h, and then slides were treated with antibody solution (1:1500 dilution of anti-Digoxigenin-AP Fab fragments in blocking buffer; Roche, 11093274910) overnight at 4 °C.
Excess antibody was removed by washing three times with MAB at 20 °C for 20 min each and replaced solution with alkaline phosphatase buffer (0.1 M Tris pH 9.5, 0.1 M NaCl, 0.1% Tween-20) for 10 min. Color development with NBT/BCIP was conducted with NBT/BCIP detection buffer (0.1 M Tris pH 9.5, 0.1 M NaCl, 50 mM MgCl2, 10% N,N-dimethylformamide [DMF], 2% NBT/BCIP stock solution; Roche, 11681451001) in the dark. Color development was stopped by washing with PBT, and samples were fixed with 4% PFA in PBS for 1 h at 20 °C. Three washes in PBS were performed for 5 min each, and slides were mounted with 80% glycerol.
Results and Discussion
Gene Complement and Annotation
After removing low-quality reads, we obtained 81,287,282 reads and 111,987 transcripts by de novo transcriptome assembly using the Trinity software. After filtering and clustering using the Corset software (Davison and Oshlack 2014), we obtained 62,275 contigs and 61,263 unigenes. The final assembly unigenes ranged from 201 to 13,685 bases, with an average length of 1,107 bases. Among the clean reads, 81.33% were successfully mapped back to the filtered de novo transcriptome, and 92.33% of core (universal single-copy orthologous) metazoan genes were completely or partially detected from 61,263 unigenes (Supplementary Table S2). Thus, the quality of the filtered de novo transcriptome was sufficient for transcriptome data analysis. BLASTX search results indicated that 23,263 (37.97%) and 17,742 (28.96%) unigenes were similar to known proteins in the NCBI nr protein database and Swiss-Prot database, respectively, using a cutoff e-value of 1e-5 (Supplementary Tables S3 and S4). The gene functions of 21,035 unigenes (90.4% and 34.34% of annotated and total unigenes, respectively) were predicted using Gene Ontology (GO) terms, which were classified into three groups: biological process (BP), cellular component (CC), and molecular function (MF) (Supplementary Fig. S1, Table S5). The most common major BPs were cellular, metabolic, and single-organism processes (Supplementary Fig. S1), and the most common MF was binding (GO:0005488) and/or catalytic activity (Supplementary Fig. S1).
Identification of the Candidate Prismatic Layer SMPs Encoding Transcripts in A. pectinata
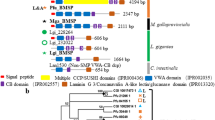
We first identified the previous known three SMPs identified from Pinnidae (Asprich, Caspartin, and Calprismin) and only found Asprich from A. pectinata. We then seek to find highly conserved prismatic layer SMPs from three kinds of pearl oysters P. fucata, P. margaritifera, and P. maxima using BLASTP program. We found at least 25 prismatic layer SMPs were conserved among them (Supplementary Table S6). We then searched similar sequence of these conserved SMP through the transcriptome data of A. pectinata; 17 of 25 prismatic layer SMPs were identified (e-values < 1.0e−10) (Fig. 2, Supplementary Table S6). These proteins were classified into eight groups: tyrosinase, chitinase, EGF-like proteins (EGF-like), fibronectin, V-rich proteins, and prism uncharacterized protein 2 (PUSP2) (Fig. 2). We further performed molecular phylogenetic analysis and gene expression analysis, focusing on these conserved prismatic layer SMPs.
Prediction of the prismatic layers-related SMPs encoding transcripts in A. pectinata. Comparison of the highly conserved prismatic SMPs in three kinds of pearl oysters (P. fucata, P. margaritifera, and P. maxima) with the transcriptome of A. pectinata (this study), the genome and the proteome of C. gigas (Zhang et al. 2012), and the proteome of M. galloprovincialis (Gao et al. 2015) by BLASTP program (e-value < 1.0e–10, details shown in Supplementary Table S6). The circle colors blue, black, and orange represent data source from proteome, genome, and transcriptome. G, genome; P, proteome; T, transcriptome (Color figure online)
Asprich
Some acidic shell matrix proteins (MSP1, Aspein, Asprich, Caspartin, Pif and MPP1) that are composed of many acidic residues (aspartic acid and glutamic acid) have been identified from mollusks shells (Sarashina and Endo 2001; Tsukamoto et al. 2004; Gotliv et al. 2005; Marin et al. 2005; Samata et al. 2008; Suzuki et al. 2009). For example, Aspein was first identified as an acidic SMP in P. fucata (Tsukamoto et al. 2004). This acidic protein is found in many aspartic acids (60.41% of a full-length PfuAspein, except for a signal peptide region) and is among the most acidic SMPs in mollusks (Tsukamoto et al. 2004). Homologs of aspein have been isolated from other Pteriidae species including P. maxima, P. penguin, and Isognomon perna (Isowa et al. 2012). Expression analysis and in vivo crystallization experiments have shown that aspein plays important roles in calcite formation in the prismatic layer (Takeuchi et al. 2008). Although Pteriidae species are closely related, their amino acid sequences are diverse (Isowa et al. 2012). The acidic protein Asprich is similar to aspein, found in A. rigida (AriAsprich), where it is present only in the prismatic layer (Gotliv et al. 2005). We found the transcript Asprich (Ape_Cluster-22716.14618) from A. pectinata (ApeAsprich), which is similar to AriAsprich (A. rigida), in the transcriptome of a closely related species (A. pectinata). This result demonstrates the good quality of our transcriptome de novo assembly for feature analysis, since Asprich contains many repeat sequences. However it is difficult to compare ApeAsprich with the C-terminus of AriAsprich, because Ape_Cluster-22716.14618 is a partial sequence (Fig. 3). We thus amplified the full length of Aspein sequence by PCR with specific primers and compared it with AriAspein. ApeAspein contained signal peptides and the polyaspartic acid domain (D-domain), like AriAsprich (Gotliv et al. 2005) or aspein (Tsukamoto et al. 2004; Isowa et al. 2012); however, we did not find DEAD repeats, which may have Mg-binding capabilities (Fig. 3). The sequences around the D-domain of ApeAsprich showed wide variation compared with those of the closely related species A. rigida, although their signal peptides were completely conserved. Some SMPs contain partially similar sequences, including some specific domains that interact with proteins or other organic substances such as chitin among mollusks (Marie et al. 2011). However, some acidic amino acid-rich sequences that interact with calcium carbonate crystals can show greater diversity than specific domains, such as C-terminal amino acid sequences of Pif in P. fucata (Suzuki et al. 2013; Suzuki and Nagasawa 2013). Our results support this hypothesis, suggesting that the molecular evolution of acidic proteins like Asprich is faster than that of other proteins.
Alignment of Asprich. Alignment of the amino acid sequences of ApeAsprich (A. pectinata) and seven AriAsprichs (A. rigida). Blue characters indicate aspartic acid (D) residues. Red and orange boxes indicate signal peptide and DEAD repeat region, respectively. ‘*’Indicates positions which have a single, fully conserved residue. ‘:’ and ‘.’ indicate that ‘strong’ and ‘weak’ similar groups are fully conserved, respectively (Color figure online)
Tyrosinase
Tyrosinase is a copper-containing enzyme that catalyzes hydroxylation of monophenols and oxidation of o-diphenols. Many tyrosinases have been reported as SMPs in mollusk species (Nagai et al. 2007; Zhang et al. 2012; Liao et al. 2015). Tyrosinase is a well-conserved domain among mollusk SMPs (Shimizu et al. 2019). Tyrosinases are also involved in pigmentation, similar to melanisation (Naraoka et al. 2003; Liao et al. 2015), and periostracum formation (Sánchez-Ferrer et al. 1995; Zhang et al. 2006). Interestingly, the tyrosinase gene family has expanded in mollusks (Aguilera et al. 2014) and members of this family have independently evolved as SMPs in several taxa (Aguilera et al. 2014; Shimizu et al. 2019). We identified 15 transcripts containing tyrosinase domains (PF00264) from the A. pectinata transcriptome by BLASTP program (e-values < 1.0e−10) (Supplementary Fig. S3). Phylogenetic analysis using whole part of amino acid sequences showed that tyrosinases were diverse in A. pectinata and expressed in mantle tissues, but did not form a clade with shell-related tyrosinases of the bivalves C. gigas, P. fucata, P. margaritifera, and P. maxima with high bootstrap support (≥ 50%) (Fig. 4) (Nagai et al. 2007; Marie et al. 2012; Zhang et al. 2012; Zhao et al. 2018). These results demonstrate the difficulty of examining biomineralization-related tyrosinases in A. pectinata without proteome analysis.
Phylogeny of Tyrosinase in mollusks. The maximum likelihood tree was inferred from 115 tyrosinase amino acid sequences under the LG + Г model (170 positions, 100 bootstrap replicates). Bootstrap support values < 50% are not shown. Black circles on nodes indicate high bootstrap values (≥ 70%) and white circles indicate medium bootstrap values (50 ~ 70%). Branch lengths are proportional to the expected number of substitutions per site, as indicated by the scale bar. Red, blue, and green circles indicate proteins that have been identified as SMPs from nacreous, prismatic layers, and whole shells of C. gigas, respectively. Ape, Atrina pectinata; Cgi, Crassostrea gigas; Hsa, Homo sapiens; Lgi, Lottia gigantea; Mmu: Mus musculus; Nve, Nematostella vectensis; Pfu, Pinctada fucata. Pmarg, P. margaritifera; Pmax, P. maxima (Color figure online)
Chitinase
Many mollusk shells are composed of chitin, which is an extracellular matrix component that plays important roles in organic framework formation (Schonitzer and Weiss 2007; Suzuki et al. 2007). Chitinase catalyzes chitin degradation. Like other SMPs, Chitinase containing the glycoside hydrolase family 18 (GH18) domain (PF00704) has been identified in mollusk shells (Marie et al. 2012; Yonezawa et al. 2016; Kintsu et al. 2017; Zhao et al. 2018). Functional analyses using allosamidin, which is a chemical inhibitor of GH18 chitinase, showed that chitinolytic enzymes control the thickness of organic layers on shell surfaces in the pond snail Lymnaea stagnalis (Yonezawa et al. 2016) or the fine control of prismatic layer microstructure construction in P. fucata (Kintsu et al. 2017). We found 15 transcripts with sequences similar to Chitinase (PfuClp1, PfuClp3, PfuPU2, and PfuPNU3) containing the GH18 domain in the A. pectinata transcriptome by BLASTP program (Supplementary Fig. S3). Molecular phylogenetic analysis using whole part of amino acid sequences showed that two transcripts (Ape_Cluster-22716.11152 and Ape_Cluster-22716.15449) formed well-supported clusters with calcification-related chitinase protein 1 and 2 (CR-CN1 and CR-CN2) identified from Pinctada prismatic shell layers (Marie et al. 2012; Kintsu et al. 2017; Zhao et al. 2018) (Fig. 5). These results suggest that Ape_Cluster-22716.11152 (ApeCR-CN1) and Ape_Cluster-22716.15449 (ApeCR-CN2) correlate with prismatic layer microstructure formation as in P. fucata (Kintsu et al. 2017).
Phylogeny of Chitinase in mollusks. The maximum likelihood tree was inferred from 95 chitinase amino acid sequences under the LG + Г model (336 positions, 100 bootstrap replicates). Bootstrap support values < 50% are not shown. Black circles on nodes indicate high bootstrap values (≥ 70%) and white circles indicate medium bootstrap values (50 ~ 70%). Branch lengths are proportional to the expected number of substitutions per site, as indicated by the scale bar. Red, blue, and green circles indicate proteins that have been identified as SMPs from nacreous, prismatic layers, and whole shells of C. gigas respectively. Ape, Atrina pectinata; Cgi, Crassostrea gigas; Hsa, Homo sapiens; Lgi, Lottia gigantea; Mga: M. galloprovincialis; Mmu: Mus musculus; Pfu, Pinctada fucata; Pmarg, P. margaritifera; Pmax, P. maxima (Color figure online)
EGF-like Proteins
EGF-like domain-containing proteins have been reported as SMP in bivalves (Marie et al. 2011, 2012; Liu et al. 2015; Gao et al. 2015; Zhao et al. 2018; Iwamoto et al. 2020). All of them were identified from calcite structures including prismatic layer, fibrous prismatic layer, and chalky layer. We found three transcripts (Ape_Cluster-22716.12182, Ape_Cluster-22716.16837, Ape_Cluster-22716.12019) with sequences similar to EGF-like protein (PfuPU12) in the A. pectinata transcriptome by BLASTP program (e-values < 1.0e−10) (Fig. 6a). All of them have two EGF-like domains in the N-terminal region. We then conducted phylogenetic analysis using EGF-like proteins from three kinds of pearl oysters, P. fucata, P. margaritifera, and P. maxima, the pacific oyster, C. gigas, and the limpet, L. gigantea. The result of the molecular phylogenetic analysis using whole part of amino acid sequences showed that EGF-like proteins separated two groups; Ape_Cluster-22716.16837 (ApeEGF-like-1) formed a monophyletic group with other bivalves EGF-like-1 proteins (Fig. 6b), and Ape_Cluster-22716.12182 (ApeEGF-like-2) formed a sister clade with other bivalves EGF-like-2 proteins including PfuPU12 (Fig. 6b). These results suggest that ApeEGF-like-1 (Ape_Cluster-22716.16837) and ApeEGF-like-2 (Ape_Cluster-22716.12182) probably involve in prismatic layer formation or calcite aggregation as in C. gigas (Iwamoto et al. 2020).
Schematic representation and phylogeny of EGF-like proteins in mollusks. a The domain structures of EGF-like proteins in A. pectinata. b The maximum likelihood tree was inferred from 18 EGF-like amino acid sequences under the LG + Г model (339 positions, 100 bootstrap replicates). Bootstrap support values < 50% are not shown. Black circles on nodes indicate high bootstrap values (≥ 70%) and white circles indicate medium bootstrap values (50 ~ 70%). Branch lengths are proportional to the expected number of substitutions per site, as indicated by the scale bar. Red, blue, and green circles indicate proteins that have been identified as SMPs from nacreous, prismatic layers, and whole shells of C. gigas, respectively. Ape, Atrina pectinata; Cgi, Crassostrea gigas; Lgi, Lottia gigantea; Mga: M. galloprovincialis; Pfu, Pinctada fucata. Pmarg, P. margaritifera; Pmax, P. maxima (Color figure online)
Fibronectin-like Proteins
Fibronectin proteins containing some fibronectin type 3 (FN3) domains have been reported as SMPs in bivalves (Marie et al. 2011, 2012; Liu et al. 2015; Gao et al. 2015; Zhao et al. 2018). In human, it has been known that FN proteins have calcium-binding ability (Amphlett and Hrinda 1983) and involves in calcium oxalate crystal growth inhibition and aggregation (Khamchun et al. 2019). Although three or more Calcification-related Fibronectin-like proteins (CR-FN) were identified as SMPs from each species including three kinds of pearl oysters, P. fucata, P. margaritifera, and P. maxima and the pacific oyster, C. gigas, their evolutionary processes were unknown. We found two transcripts (Ape_Cluster-22716.3775 and Ape_Cluster-11531.0) with sequences similar to CR-FN proteins (PfuPU3, PU5, PU15, and PU16) in the A. pectinata transcriptome by BLASTP program (e-values < 1.0e−10) (Fig. 7a). Ape_Cluster-22716.3775 contains four FN3 domains, and Ape_Cluster-11531.0 contains three FN3 domains and two immunoglobulin I-set domains (Fig. 7a). The result of phylogenetic analysis using whole part of amino acid sequences showed that Ape_Cluster-22716.3775 (ApeCR-FN) formed a monophyletic group with other bivalves CR-FN proteins except for PfuPU16 and PmaxFN1 (Fig. 7b). This result suggests that the duplications of CR-FN proteins were independently occurred several in the pacific oyster C. gigas and the common ancestor of three kinds of pearl oysters (P. fucata, P. margaritifera, and P. maxima). These results suggest that Ape_Cluster-22716.3775 (ApeCR-CN) probably interacts with calcium ion and involves in the prismatic layer formation.
Schematic representation and phylogeny of Fibronectin in mollusks. a The domain structures of Fibronectin proteins in A. pectinata. b The maximum likelihood tree was inferred from 34 Fibronectin amino acid sequences under the LG + Г model (213 positions, 100 bootstrap replicates). Bootstrap support values < 50% are not shown. Black circles on nodes indicate high bootstrap values (≥ 70%) and white circles indicate medium bootstrap values (50 ~ 70%). Branch lengths are proportional to the expected number of substitutions per site, as indicated by the scale bar. Red, blue, and green circles indicate proteins that have been identified as SMPs from nacreous, prismatic layers, and whole shells of C. gigas, respectively. Ape, Atrina pectinata; Cgi, Crassostrea gigas; Lgi, Lottia gigantea; Mga: M. galloprovincialis; Pfu, Pinctada fucata. Pmarg, P. margaritifera; Pmax, P. maxima (Color figure online)
Valine-Rich Proteins (MP10 and Alveolin-like Protein)
Valine-rich protein encoding gene called PFMP10 was first isolated from the mantle of the pearl oyster P. fucata (Liu et al. 2007), and later MP10 was identified as SMP from the prismatic layers of P. fucata, P. margaritifera, and P. maxima (Marie et al. 2012; Liu et al. 2007; Zhao et al. 2018). Marie et al. (2012) also found other valine-rich protein called Alveolin-like protein from the prismatic layers of P. margaritifera and P. maxima. MP10 and Alveolin-like proteins were the second and the third abundant prismatic layer SMPs in P. margaritifera (Marie et al. 2012). These two valine-rich proteins MP10 and Alveolin-like encoding genes were tandemly arranged in the same scaffold of P. fucata (Takeuchi et al. 2016a). We found three MP10-like transcripts (ApeMP10) and three Alveolin-like (ApeAlveolin-like) transcripts from the A. pectinata transcriptome (Fig. 8a). The result of the molecular phylogenetic analysis using whole part of amino acid sequences showed that these transcripts formed a monophyletic group with MP10 proteins or with Alveolin-like proteins (Fig. 8b).
Schematic representation and phylogeny of valine-rich proteins (MP10 and Alveolin-like) in bivalves. a The domain structures of valine-rich proteins (MP10 and Alveolin-like) in A. pectinata. b The maximum likelihood tree was inferred from 18 valine-rich amino acid sequences (MP10 and Alveolin-like) under the WAG + Г model (213 positions, 100 bootstrap replicates). Bootstrap support values < 50% are not shown. Black circles on nodes indicate high bootstrap values (≥ 70%) and white circles indicate medium bootstrap values (50 ~ 70%). Branch lengths are proportional to the expected number of substitutions per site, as indicated by the scale bar. Blue circles indicate proteins that have been identified as SMPs from prismatic layer. Ape, Atrina pectinata; Cgi, Crassostrea gigas; Mga: M. galloprovincialis; Pfu, Pinctada fucata. Pmarg, P. margaritifera; Pmax, P. maxima (Color figure online)
Prism Uncharacterized Shell Protein 2 (PUSP2)
Fifty prism SMPs were found from P. margaritifera (Marie et al. 2012), and 19 of them were classified as prism uncharacterized proteins (PUSPs). Four kinds of PUSPs (PUSP1, 2, 4, 5) were well conserved in three kinds of pearl oysters P. fucata, P. margaritifera, and P. maxima (Supplementary Table S6) and only one PUSP2-like transcript (Ape_Cluster-22716.11342) was found from the transcriptome of A. pectinata (ApePUSP2). PUSP2 proteins have signal peptide and some low complexity regions but not contain any specific domains (Fig. 9a). ApePUSP2 (Ape_Cluster-22716.11342) has signal peptide and one low complexity region in the C-terminus (Fig. 9a). We also found PUSP2-like protein from the pacific oyster C. gigas, and this protein identified from the shell of C. gigas (Zhang et al. 2012; Feng et al. 2017). We multiple aligned the amino acid sequences of PUSP2 and found some conserved residues (Fig. 9b). The C-terminus of PUSP2 in pearl oysters has conserved aspartic acid (D)-rich site (Fig. 9b). However, we could not find D-rich site from the PUSP2-like proteins of A. pectinata and C. gigas (Fig. 9b). Instead of D-rich site, glutamic acid (E) and serine (S) rich site were found form the C-terminus of ApePUSP2 (Fig. 9b). This ES-rich site possibly has similar function to D-rich site of PUSP2 in the pearl oyster, because glutamic acid and aspartic acid are negative charged amino acid and interact with calcium ion.
Schematic representation and alignment of PUSP2 in bivalves. a The domain structure of prismatic uncharacterized shell protein 2 (PUSP2) in A. pectinata. b Alignment of the amino acid sequences of PUSP2 in Atrina pectinata, Crassostrea gigas, Pinctada fucata, P. margaritifera, and P. maxima. Blue and red characters indicate acidic amino acids aspartic acid (D) and glutamic acid (E), respectively. Red, blue, and orange boxes indicate signal peptide, E-rich region, and D-rich region, respectively. ‘*’Indicates positions which have a single, fully conserved residue. ‘:’ and ‘.’ indicate that ‘strong’ and ‘weak’ similar groups are fully conserved, respectively. Ape, Atrina pectinata; CGI, Crassostrea gigas; Pfu, Pinctada fucata. Pmarg, P. margaritifera; Pmax, P. maxima (Color figure online)
Expression of the Candidate Prismatic Layer SMPs Encoding Transcripts in the Mantle Edge
The results of molecular phylogenetic analysis or multiple alignment analysis showed that 7 transcripts (ApeAsprich, ApeCR-CN1, ApeCR-CN2, ApeEGF-like 1, ApeEGF-like 2, ApeCR-FN, ApeMP10, ApeAlveolin-like, and ApePUSP2) probably involve in the prismatic layer formation. We then confirmed their expression regions in the mantle edge that is related to the prismatic layer formation using in situ hybridization. All of these transcripts were expressed in the outer surface of the outer fold and outer pallium as expected (Fig. 10), and only ApeMP10 and ApeAlveolin-like were expressed both in outer and inner surfaces of the outer fold (Fig. 10). No signals were detected at the dorsal region of the mantle and the middle and the inner folds in all transcripts (Fig. 10).
Gene expressions of nine transcripts in the mantle edge. Expression of Asprich (a), CR-CN1 (b), CR-CN2 (c), EGF-like 1 (d), EGF-like 2 (e), CR-FN (f), Alveolin-like (g), MP10 (h), and PUSP2 (i) in the mantle edge of A. pectinata. j Schematic of nine genes expression (a–i). Blue and red colors indicate the common expression region of nine genes (outer surface of the outer fold and pallium) and the specific expression region of Alveolin-like and MP10 (inner surface of the outer fold), respectively. IF, inner fold; MF, middle fold; P, pallium; OF, outer fold (Color figure online)
Molecular Evolution of the Prismatic Layer-Related Genes in Pteriomorphia
Prismatic layers consist of calcite crystals were observed in the shells of Pteriomorphia except for some family. Numerous kinds of SMPs were identified from the shells of three families of Pteriomorphia using transcriptome and proteome analyses (Pteriidae, Marie et al. 2012, Liu et al. 2015; Zhao et al. 2018; Ostreidae, Zhang et al. 2012, Feng et al. 2017; Mytilidae, Gao et al. 2015; Liao et al. 2015). However, the SMPs in Pinnidae remain unclear. We focused on the conserved prismatic layer-related SMPs in in three kinds of pearl oysters (P. fucata, P. margaritifera, and P. maxima). In his study, we should consider the limitations of transcriptome analysis; the de novo assembly cannot completely identify all transcripts and may contain fragmented transcripts, partial transcripts (lack of 3′ or 5′ terminus), and assembly errors. However, we found some candidate homologues of these SMPs (ApeCR-CN1, ApeCR-CN2, ApeEGF-like 1, ApeEGF-like 2, ApeCR-FN, ApeMP10, ApeAlveolin-like, and ApePUSP2) from the transcriptome data of A. pectinata (Pinnidae) (Figs. 2, 3, 4, 5, 6, 7, 8, 9 and 10). To understand the evolutionary scenario of these prismatic layer-related SMPs, we compared five groups of them (CR-CN, EGF-like proteins, CR-FN, V-rich proteins, and PUSP2) among four Pteriomorphia species (P. fucata, C. gigas, A. pectinata, and M. galloprovincialis) and found different evolutionary patterns among five groups (Fig. 11).
Molecular evolutions of five groups of prismatic layer-specific SMPs in Pteriomorphia. Reconstructions of five prismatic layer-specific SMPs in Atrina pectinata, Crassostrea gigas, Pinctada fucata, and Mytilus galloprovincialis. Boxes and star marks on the phylogeny indicate possible gene duplication event and possible acquisition event, respectively. CR-CN, calcification-related chitinase; CR-FN, calcification-related fibronectin; MP10, mantle protein 10; PUSP2, Prismatic uncharacterized shell protein 2
CR-CNs were found from four Pteriomorphia species (Figs. 2 and 5). The result of molecular phylogeny of CR-CNs showed that CR-CNs in M. galloprovincialis are different clades with CR-CN1 and CR-CN2 in P. fucata, C. gigas, and A. pectinata (Fig. 5b). This result suggests that CR-CNs were independently evolved as SMPs in Mytilidae (CR-MCN) and in the derived Pteriomorphia (A. pectinata, C. gigas, and P. fucata) (Fig. 11). CR-CN was duplicated at least in the common ancestor of Pinnidae, Ostreidae, and Pteriidae, because all of them have two kinds of CR-CNs (CR-CN1 and CR-CN2) (Figs. 5 and 11). EGF-like proteins were found from four Pteriomorphia species (Figs. 2 and 6). Duplication of EGF-like protein encoding gene was probably occurred in the common ancestor of Pinnidae, Ostreidae, and Pteriidae, because only one EGF-like protein (EGF-like 1/2) was found from M. galloprovincialis (Figs. 6 and 11). Three paralogs of CR-FN were found from three kinds of pearl oysters P. fucata, P. margaritifera, and P. maxima (Figs. 2 and 7) but only one CR-FN was found from M. galloprovincialis, A. pectinata (Figs. 2 and 7), and other CR-FN paralogs were found from C. gigas (Figs. 2 and 7). This result suggests that the gene duplication was independently occurred twice in the common ancestor of Pteriidae and in that of Ostreidae (Fig. 11). Two kinds of V-rich proteins called MP10 and Alveolin-like proteins were found from A. pectinata, C. gigas, P. fucata, P. margaritifera, and P. maxima (Figs. 2 and 8), and only Alveolin-like proteins were found from M. galloprovincialis (Figs. 2 and 8). The genes of MP10 and Alveolin-like were tandemly arranged in the same scaffold of P. fucata (Takeuchi et al. 2016a). Thus the origin of MP10 and Alveolin-like encoding genes were probably same and the tandem duplication was occurred in the common ancestor of Pinnidae, Ostreidae, and Pteriidae (Fig. 11). We found PUSP2 from A. pectinata, C. gigas, P. fucata, P. margaritifera, and P. maxima but not from M. galloprovincialis (Figs. 2 and 9). It is relatively difficult to find PUSP2-like proteins than other characterized SMPs such as CR-CN, EGF-like protein, and CR-FN, because PUSP2 does not contain some specific domains except for signal peptide (Fig. 9a). However, we found PUSP2-like protein from A. pectinata but not from M. galloprovincialis (Fig. 9). Thus PUSP2 was possibly evolved as SMPs at least in the common ancestor of Pinnidae, Ostreidae, and Pteriidae (Fig. 11).
Conclusion
We performed transcriptome analysis of mantle cells of the pen shell A. pectinata and obtained a total 61,263 unigenes, which included core (universal single-copy orthologous) genes of metazoans and will be useful for future studies. We focused on 17 highly conserved prismatic layer-related SMPs from the pearl oysters P. fucata, P. margaritifera, and P. maxima. We performed molecular phylogenetic analyses and identified 10 candidate transcripts that possibly related with prismatic layer formation from A. pectinata (Asprich, Tyrosinase, CR-CN1, CR-CN2, EGF-like 1, EGF-like 2, CR-FN, MP10, Alveolin-like, and PUSP2), with evolutionary processes that differ among genes. We identified the unique acidic protein Asprich encoding transcript in A. pectinata. Although A. pectinata is closely related to A. rigida, Asprich contains many mutations such as insertion/deletion and transition/transversion following nonsynonymous substitution. The tyrosinase gene family is among the most conserved SMPs in mollusks; however, members of this family have expanded in mollusks and evolved independently as SMPs in several species. The SMPs EGF-like 1, EGF-like 2, CR-FN, and Alveolin-like encoding genes each evolved once in the linage leading to four Pteriomorphia families (Mytilidae, Pinnidae, Ostreidae, and Pteriidae), and PUSP2 also evolved in the linage leading to three Pteriomorphia families (Pinnidae, Ostreidae, and Pteriidae). CR-CN was independently evolved as SMPs in Mytilidae (CR-MCN) and in other Pteriomorphia (Pinnidae, Ostreidae, and Pteriidae) (CR-CN1 and CR-CN2). This is the first report to apply transcriptome analysis to confirm predicted biomineralization candidate genes in Pinnidae. Our results provide a new dataset for understanding the molecular mechanisms and evolution of genes related to the prismatic layer formation in bivalves.
Data Availability
Transcriptome data are available in the DNA Data Bank of Japan (DDBJ) (Sequence Read Archive: DRR209159, BioProject: PRJDB9333, BioSample: SAMD00206368, TSA: ICPQ01000001-ICPQ01061263). Other sequence data (ApeAsprich, ApeCR-CN1, ApeCR-CN2, ApeEGF-like 1, ApeEGF-like 2, ApeCR-FN, ApeMP10, ApeAlveolin, and ApePUSP2) are available in the DDBJ under accession numbers LC574994-LC575002.
References
Aguilera F, McDougall C, Degnan BM (2014) Evolution of the tyrosinase gene family in bivalve molluscs: independent expansion of the mantle gene repertoire. Acta Biomater 10(9):3855–3865. https://doi.org/10.1016/j.actbio.2014.03.031
Amphlett GW, Hrinda ME (1983) The binding of calcium to human fibronectin. Biochem Biophys Res Commun 111:1045–1053. https://doi.org/10.1016/0006-291x(83)91405-5
Belcher A, Wu X, Christensen R et al (1996) Control of crystal phase switching and orientation by soluble mollusc-shell proteins. Nature 381:56–58. https://doi.org/10.1038/381056a0
Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T (2009) trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics (Oxford, England) 25:1972–1973. https://doi.org/10.1093/bioinformatics/btp348
Davison NM, Oshlack A (2014) Corset: enabling differential gene expression analysis for de novoassembled transcriptomes. Genome Biol 15:410. https://doi.org/10.1186/s13059-014-0410-6
Feng D, Li Q, Yu H, Kong L, Du S (2017) Identification of conserved proteins from diverse shell matrix proteome in Crassostrea gigas: characterization of genetic bases regulating shell formation. Sci Rep 7:45754. https://doi.org/10.1038/srep45754
Finn RD, Coggill P, Eberhardt RY et al (2016) The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res 44:D279–D285. https://doi.org/10.1093/nar/gkv1344
Gao P, Liao Z, Wang XX, Bao LF, Fan MH, et al (2015) Layer-by-layer proteomic analysis of mytilus galloprovincialis shell. PLoS One 10(7):e0133913. https://doi.org/10.1371/journal.pone
Gotliv B, Kessler N, Sumerel JL et al (2005) Asprich: a novel aspartic acid-rich protein family from the prismatic shell matrix of the bibalve Atrina rigida. Chem Bio Chem 6:304–314. https://doi.org/10.1002/cbic.200400221
Götz S, García-Gómez JM, Terol J, Williams TD, Nagaraj SH, et al (2008) High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res 36(10):3420–35. https://doi.org/10.1093/nar/gkn176
Grabherr MG, Haas BJ, Yassour M et al (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:644–652. https://doi.org/10.1038/nbt.1883
Haas BJ, Papanicolaou A, Yassour M et al (2013) De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc 8:1494–1512. https://doi.org/10.1038/nprot.2013.084
Iseli C, Jongeneel CV, Bucher P (1999) ESTScan: a program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. Proc Int Conf Intell Syst Mol Biol 138–48.
Isowa Y, Sarashina I, Setiamarga DHE, Endo K (2012) A comparative study of the shell matrix protein aspein in pterioid bivalves. J Mol Evol 75:11–18. https://doi.org/10.1007/s00239-012-9514-3
Iwamoto S, Shimizu K, Negishi L, Suzuki N, Nagata K, Suzuki M (2020) Characterization of the chalky layer-derived EGF-like domain-containing protein (CgELC) in the pacific oyster, Crassostrea gigas. J Struct Biol 212(1):107594. https://doi.org/10.1016/j.jsb.2020.107594
Jackson DJ, McDougall C, Green K et al (2006) A rapidly evolving secretome builds and patterns a sea shell. BMC Biol 4:40. https://doi.org/10.1186/1741-7007-4-40
Jackson DJ, McDougall C, Woodcroft B et al (2010) Parallel evolution of nacre building gene sets in molluscs. Mol Biol Evol 27:591–608. https://doi.org/10.1093/molbev/msp278
Jackson DJ, Mann K, Häussermann V et al (2015) The Magellania venosa biomineralizing proteome: a window into brachiopod shell evolution. Gen Biol Evol 7:1349–1362. https://doi.org/10.1093/gbe/evv074
Liu C, Li S, Kong J, Liu Y, Wang T, Xie L, Zhang R (2015) In-depth proteomic analysis of shell matrix proteins of Pinctada fucata. Sci Rep 5:17269. https://doi.org/10.1093/gbe/evv074
Katoh K, Misawa K, Ki K, Miyata T (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30:3059–3066. https://doi.org/10.1093/nar/gkf436
Khamchun S, Sueksakit K, Chaiyarit S, Thongboonkerd V (2019) Modulatory effects of fibronectin on calcium oxalate crystallization, growth, aggregation, adhesion on renal tubular cells, and invasion through extracellular matrix. J Biol Inorg Chem 24:235–246. https://doi.org/10.1007/s00775-019-01641-w
Kintsu H, Okumura T, Negishi L et al (2017) Crystal defects induced by chitin and chitinolytic enzymes in the prismatic layer of Pinctada fucata. Biochem Biophys Res Commun 489:89–95. https://doi.org/10.1016/j.bbrc.2017.05.088
Kono M, Hayashi N, Samata T (2000) Molecular mechanism of the nacreous layer formation in Pinctada maxima. Biochem Biophys Res Commun 269:213–218. https://doi.org/10.1006/bbrc.2000.2274
Levi-Kalisman Y, Falini G, Addadi L, Weiner S (2001) Structure of the nacreous organic matrix of a bivalve mollusk shell examined in the hydrated state using cryo-TEM. J Struct Biol 135:8–17. https://doi.org/10.1006/jsbi.2001.4372
Li B, Dewey CN (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform 12:323. https://doi.org/10.1186/1471-2105-12-323
Liao Z, Bao L, Fan M et al (2015) In-depth proteomic analysis of nacre, prism, and myostracum of Mytilus shell. J Proteomics 122:26–40. https://doi.org/10.1016/j.jprot.2015.03.027
Liu HL, Liu SF, Ge YJ et al (2007) Identification and characterization of a biomineralization related gene PFMG1 highly expressed in the mantle of Pinctada fucata. Biochemistry 46:844–851. https://doi.org/10.1021/bi061881a
Luo Y-J, Takeuchi T, Koyanagi R et al (2015) The Lingula genome provides insights into brachiopod evolution and the origin of phosphate biomineralization. Nat Commun 6:8301. https://doi.org/10.1038/ncomms9301
Mann K, Poustka AJ, Mann M (2008) The sea urchin (Strongylocentrotus purpuratus) test and spine proteomes. Proteome Sci 6:22. https://doi.org/10.1186/1477-5956-6-22
Marie B, Marie A, Jackson DJ et al (2010) Proteomic analysis of the organic matrix of the abalone Haliotis asinina calcified shell. Proteome Sci 8:54. https://doi.org/10.1186/1477-5956-8-54
Marie B, Trinkler N, Zanella-Cléon I et al (2011) Proteomic identification of novel proteins from the calcifying shell matrix of the Manila clam Venerupis philippinarum. Mar Biotechnol 13:955–962. https://doi.org/10.1007/s10126-010-9357-0 (PMID:21221694)
Marie B, Joubert C, Tayalé A et al (2012) Different secretory repertoires control the biomineralization processes of prism and nacre deposition of the pearl oyster shell. Proc Natl Acad Scie USA 109:20986–20991. https://doi.org/10.1073/pnas.1210552109
Marie B, Jackson DJ, Ramos-Silva P et al (2013) The shell-forming proteome of Lottia gigantea reveals both deep conservations and lineage-specific novelties. FEBS J 280:214–232. https://doi.org/10.1111/febs.12062
Marie B, Arivalagan J, Mathéron L et al (2017) Deep conservation of bivalve nacre proteins highlighted by shell matrix proteomics of the Unionoida Elliptio complanata and Villosa lienosa. J R Soc Interface. https://doi.org/10.1098/rsif.2016.0846
Marin F, Amous R, Guichard N et al (2005) Caspartin and Calprismin, two proteins of the shell calcitic prisms of the Mediterranean fan mussel Pinna nobilis. J Biol Chem 275:20667–20675. https://doi.org/10.1074/jbc.M506526200
Montagnani C, Marie B, Marin F et al (2011) Pmarg-pearlin is a matrix protein involved in nacre framework formation in the pearl oyster Pinctada margaritifera. Chem Bio Chem 12:2033–2043. https://doi.org/10.1002/cbic.201100216
Nagai K, Yano M, Morimoto K, Miyamoto H (2007) Tyrosinase localization in mollusc shells. Comp Biochem Physiol B 146:207–214. https://doi.org/10.1016/j.cbpb.2006.10.105
Naraoka T, Uchisawa H, Mori H et al (2003) Purication, characterization and molecular cloning of tyrosinase from the cephalopod mollusk, Illex argentinus. Eur J Biochem 270:4026–4038. https://doi.org/10.1046/j.1432-1033.2003.03795.x
Nishimura O, Hara Y, Kuraku S (2017) gVolante for standardizing completeness assessment of genome and transcriptome assemblies. Bioinformatics 15(33):3635–3637
Nudelman F, Chen HH, Goldberg HA, Weiner S, Addadi L (2007) Lessons from biomineralization: comparing the growth strategies of mollusk shell prismatic and nacreous layers in Atrina rigida. Faraday Discuss 136:9–25. https://doi.org/10.1039/b704418f
Okumura T, Suzuki M, Nagasawa H, Kogure T (2012) Microstructural variation of biogenic calcite with intracrystalline organic macromolecules. Cryst Growth Des 12:224–230. https://doi.org/10.1021/cg200947c
Petersen TN, Brunak S, von Heijne G, Nielsen H (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. https://doi.org/10.1038/nmeth.1701
Ramos-Silva P, Kaandorp J, Huisman L et al (2013) The skeletal proteome of the coral Acropora millepora: the evolution of calcification by co-option and domain shuffling. Mol Biol Evol 30:2099–2112. https://doi.org/10.1093/molbev/mst109
Samata T, Hayashi N, Kono M et al (1999) A new matrix protein family related to the nacreous layer formation of Pinctada fucata. FEBS Lett 462:225–229. https://doi.org/10.1016/s0014-5793(99)01387-3
Samata T, Ikeda D, Kazikawa A et al (2008) A novel phosphorylated glycoprotein in the shell matrix of the oyster Crassostrea nippona. FEBS J 275:2977–2989. https://doi.org/10.1111/j.1742-4658.2008.06453.x
Sánchez-Ferrer A, Rodríguez-López JN, García-Cánovas F, García-Carmona F (1995) Tyrosinase: a comprehensive review of its mechanism. Biochim Biophys Acta 1247(1):1–11. https://doi.org/10.1016/0167-4838(94)00204-t
Sarashina I, Endo K (2001) The complete primary structure of molluscan shell protein 1 (MSP-1), an acidic glycoprotein in the shell matrix of the scallop Patinopecten yessoensis. Mar Biotechnol 3:362–369. https://doi.org/10.1007/s10126-001-0013-6
Schonitzer V, Weiss I (2007) The structure of mollusc larval shells formed in the presence of the chitin synthase inhibitor Nikkomycin Z. BMC Struct Biol 7:71. https://doi.org/10.1186/1472-6807-7-71
Shimizu K, Kimura K, Isowa Y et al (2019) Insights into the evolution of shells and love darts of land snails revealed from their matrix proteins. Genome Biol Evol 11:380–397. https://doi.org/10.1093/gbe/evy242
Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM (2015) BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31:3210–3212. https://doi.org/10.1093/bioinformatics/btv351
Stamatakis A (2014) RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics (Oxford, England). https://doi.org/10.1093/bioinformatics/btu033
Suzuki M (2020) Structural and functional analyses of organic molecules regulating biomineralization. Biosci Biotechnol Biochem 84(8):1529–1540. https://doi.org/10.1080/09168451.2020.1762068
Suzuki M, Nagasawa H (2013) Mollusk shell structures and their formation mechanism. Can J Zool 91:349–366. https://doi.org/10.1139/cjz-2012-0333
Suzuki M, Sakuda S, Nagasawa H (2007) Identification of chitin in the prismatic layer of the shell and a chitin synthase gene from the Japanese pearl oyster Pinctada fucata. Biosci Biotechnol Biochem 71(7):1735–1744. https://doi.org/10.1271/bbb.70140
Suzuki M, Saruwatari K, Kogure T et al (2009) An acidic matrix protein, Pif, is a key macromolecule for nacre formation. Science 11:1388–1390. https://doi.org/10.1126/science.1173793
Suzuki M, Iwashima A, Kimura M, Kogure T, Nagasawa H (2013) The molecular evolution of the Pif family proteins in various species of mollusks. Mar Biotechnol 15:145–158. https://doi.org/10.1007/s10126-012-9471-2
Suzuki M, Kogure T, Nagasawa H (2017) Studies on the chemical structures of organic matrices and their functions in the biomineralization processes of molluscan shells. AGri-Biosci Monogr 7:25–39. https://doi.org/10.5047/agbm.2017.00702.0025
Takeuchi T, Sarashina I, Iijima M, Endo K (2008) In vitro regulation of CaCO3 crystal polymorphism by the highly acidic molluscan shell protein Aspein. FEBS Lett 582:591–596. https://doi.org/10.1016/j.febslet.2008.01.026
Takeuchi T, Koyanagi R, Gyoja F et al (2016a) Bivalve-specific gene expansion in the pearl oyster genome: implications of adaptation to a sessile lifestyle. Zool Lett 2:3. https://doi.org/10.1186/s40851-016-0039-2
Takeuchi T, Yamada L, Shinzato C, Sawada H, Satoh N (2016b) Stepwise Evolution of Coral Biomineralization Revealed with Genome-Wide Proteomics and Transcriptomics. PLoS One 11(6):e0156424. https://doi.org/10.1371/journal.pone.0156424
Tamura K, Peterson D, Peterson N et al (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Boil Evol 28:2731–2739. https://doi.org/10.1093/molbev/msr121
Thompson JB, Paloczi GT, Kindt JH et al (2000) Direct observation of the transition from calcite to aragonite growth as induced by abalone shell proteins. Biophys J 79:3307–3312. https://doi.org/10.1016/S0006-3495(00)76562-3
Tsukamoto D, Sarashina I, Endo K (2004) Structure and expression of an unusually acidic matrix protein of pearl oyster shells. Biochem Biophys Res Commun 320:1175–1180. https://doi.org/10.1016/j.bbrc.2004.06.072
Yonezawa M, Sakuda S, Yoshimura E, Suzuki M (2016) Molecular cloning and functional analysis of chitinases in the fresh water snail, Lymnaea stagnalis. J Struct Biol 196:107–118. https://doi.org/10.1016/j.jsb.2016.02.021
Zhang C, Xie LP, Huang J, Chen L, Zhang RQ (2006) A novel putative tyrosinase involved in periostracum formation from the pearl oyster (Pinctada fucata). Biochem Biophys Res Commun 342:632–639. https://doi.org/10.1016/j.bbrc.2006.01.182
Zhang G, Fang X, Guo X et al (2012) The oyster genome reveals stress adaptation and complexity of shell formation. Nature 490:49–54. https://doi.org/10.1038/nature11413
Zhao R, Takeuchi T, Luo YJ et al (2018) Dual gene repertoires for larval and adult shells reveal molecules essential for Molluscan shell formation. Mol Biol Evol 35:2751–2761. https://doi.org/10.1093/molbev/msy172
Acknowledgements
We thank Dr Takeshi Takeuchi (Okinawa Institute of Science and Technology Graduate University) for help in annotation of shell matrix proteins and molecular phylogenetic analysis. We also thank Dr Yosuke Ono, Dr Aya Takesono, and Dr Testu Kudoh (University of Exeter) for advice with section in situ hybridization. We also thank Dr Masaei Kanematsu (National Research Institute of Fisheries and Environment of Inland Sea) for the gift of adult specimens of A. pectinata. We thank two professional editors who are both native speakers of English for editing English in this manuscript.
Funding
This research was partially funded by Grant-in-Aid for Scientific Research B (JP19H03045) and Grant-in-Aid for Scientific Research on Innovative Areas IBmS: JSPS KAKENHI Grant Number JP19H05771.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics Approval
All animal collection and experiments followed the Collecting and Experimental Ethics Policy of the University of Tokyo.
Additional information
Handling editor: John Bracht.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Shimizu, K., Kintsu, H., Awaji, M. et al. Evolution of Biomineralization Genes in the Prismatic Layer of the Pen Shell Atrina pectinata. J Mol Evol 88, 742–758 (2020). https://doi.org/10.1007/s00239-020-09977-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-020-09977-7