Abstract
Various novel proteins have been identified from many kinds of mollusk shells. Although such matrix proteins are believed to play important roles in the calcium carbonate crystal formation of shells, no common proteins that interact with calcium carbonate or that are involved in the molecular mechanisms behind shell formation have been identified. Pif consists of two proteins, Pif 80 and Pif 97, which are encoded by a single mRNA. Pif 80 was identified as a key acidic protein that regulates the formation of the nacreous layer in Pinctada fucata, while Pif 97 has von Willebrand factor type A (VWA) and chitin-binding domains. In this study, we identified Pif homologues from Pinctada margaritifera, Pinctada maxima, Pteria penguin, Mytilus galloprovincialis, and in the genome database of Lottia gigantea in order to compare their primary protein sequences. The VWA and chitin-binding domains are conserved in all Pif 97 homologues, whereas the amino acid sequences of the Pif 80 regions differ markedly among the species. Sequence alignment revealed the presence of a novel significantly conserved sequence between the chitin-binding domain and the C-terminus of Pif 97. Further examination of the Pif 80 regions suggested that they share a sequence that is similar to the laminin G domain. These results indicate that all Pif molecules in bivalves and gastropods may be derived from a common ancestral gene. These comparisons may shed light on the correlation between molecular evolution and morphology in mollusk shell microstructure.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The mollusk shell is a hard tissue consisting of calcium carbonate and organic matrices. Among the various components of shell microstructures, the nacreous structure, or “nacre”, has been investigated most intensively because of its fantastically regulated structure and importance to the pearl agriculture industry. The nacreous layer is composed of aragonite tablets surrounded by an organic framework (chitin and matrix proteins) that form the compartment microstructure (Gregoire 1957; Wada 1961). Chitin acts as a scaffold for the organic framework between the aragonite tablets. Each mineral tablet is a few micrometers in diameter and its thickness is in the order of submicrometers. Although each tablet diffracts as a single crystal, recent studies have revealed a composite nanostructure on a variety of different scales (Takahashi et al. 2004). It is believed that the small amount of organic matrix contained in each layer plays important roles in the formation of the nacreous microstructure (Weiner and Hood 1975; Falini et al. 1996; Belcher et al. 1996). The assembly of mineral and organic components is guided partly by enzymatic and partly by self-assembly mechanisms, which are currently not well understood.
There are many reports about the identification of matrix proteins from the nacreous layer. Nacrein (Miyamoto et al. 1996), MSI60 (Sudo et al. 1997), lustrin A (Shen et al. 1997), N16/pearlin (Samata et al. 1999; Miyashita et al. 2000), perlucin, perlustrin (Weiss et al. 2000), N14, N66 (Kono et al. 2000), mucoperlin (Marin et al. 2000), AP7, AP24 (Michenfelder et al. 2003), perlwapin (Treccani et al. 2006), perlinhibin (Mann et al. 2007), N19 (Yano et al. 2007), and Pif (Suzuki et al. 2009) were identified from the nacreous layer and their full sequences were determined. Naturally, the proteins that play particular physiological roles, such as enzymes, hormones, extracellular matrix components, and antibodies, are highly conserved across all mollusk species. However, the matrix proteins in the nacreous layer of bivalves and gastropods do not have common domains, and there are almost no sequence similarities between them. Nonetheless, there are some homologous matrix proteins related to the nacreous layer in species belonging to the same genus, such as N16 and N14 (Kono et al. 2000), because of the limited time since their divergence. The N-terminal region of perlustrin is similar to part of the sequence of lustrin A (Weiss et al. 2000). These conserved sequences may be derived from the insulin-like growth factor-binding protein. As the molecular weight of these two proteins is completely different, the similarity of these two proteins may be caused by convergent evolution. In addition, homologues of nacrein, which has carbonic anhydrase activity, were identified from some mollusk species. Although the GXN or GN regions related to calcium carbonate interaction are conserved among nacrein and its homologues, they contain a repeated domain that differs in length, which means that the total length of these proteins varies to some extent (Kono et al. 2000; Miyamoto et al. 2003). These GXN or GN repeat sequences may be derived from other genes by transposon mechanism or gene duplication. Recently, the whole genome sequence of Lottia gigantea, a gastropod species, was analyzed and almost all genes in its genome can now be searched in a database (Lottia genome, 2008). However, homologues of matrix proteins from bivalve shells were not identified in the genome database of L. gigantea. This may indicate that the sequences of proteins that interact with calcium carbonate have mutated much more quickly than other proteins, making it difficult to identify homologues in phylogenetically distant species.
Pif is a matrix protein that consists of two proteins, Pif 97 and Pif 80, encoded by a single mRNA. After initial production as a single translated product, these two proteins are separated at the dibasic cleavage site (RMKR) located at the end of the Pif 97 sequence by Kex2-like protease (Burgess and Kelly 1987). Pif 97 has a von Willebrand factor type A (VWA) domain associated with protein–protein interaction and a chitin-binding domain. In contrast, Pif 80 has aragonite binding activity. Functional analyses revealed that Pif plays important roles in the aragonite formation of the nacreous layer in Pinctada fucata (Suzuki et al. 2009). This suggests that Pif is probably conserved in mollusk species that have the nacreous layer. Even if the sequence that interacts with calcium carbonate (part of Pif 80) varies markedly among species as described above, there may still be conserved domains like VWA and chitin-binding domains in Pif 97 that interact with other organic matrices. Because it has been shown that domains involved in the interactions between organic compounds, such as a ligand and its receptor, or a matrix and its enzyme, are conserved in many species, it is expected that the chitin-binding and VWA domains of Pif 97 are conserved well among distant species. To investigate the evolution of sequences related to calcium carbonate interaction, we searched for Pif homologues from various mollusk species using various methods and compared them. First, we identified Pif homologues by homology cloning from Pinctada margaritifera and Pinctada maxima, which belong to the same genus as P. fucata. Using the same method, we could also identify a Pif homologue from Pteria penguin, which belongs to a different genus. Pif homologues of P. margaritifera were also discovered by other groups using an expressed sequence tag (EST) database (Joubert et al. 2010; Berland et al. 2011). Subsequently, we tried to amplify the PCR products from Mytilus galloprovincialis, which belongs to Mytiloida, Mytilidae that is phylogenetically distant from P. fucata within the Bivalvia (Temkin 2010) using primers designed on the basis of the sequence of the chitin-binding domain in Pif 97. However, no homologue of Pif in M. galloprovincialis could be amplified, suggesting that the primary sequence of the Pif homologue in M. galloprovincialis is not similar to that in P. fucata. Thus, we used the calcium carbonate-binding assay to identify a Pif homologue (Blue Mussel Shell Protein, BMSP) from the nacreous layer of M. galloprovincialis. The detailed data were reported previously (Suzuki et al. 2011). Finally, we identified four Pif homologues from the genome database (Lottia gigantea v1.0, http://genome.jgi-psf.org/Lotgi1/Lotgi1.home.html, accessed on 17 May, 2012) of L. gigantea, which is a gastropod. Images of the shells of P. fucata, M. galloprovincialis, and L. gigantea are shown in Fig. 1e–g, respectively.
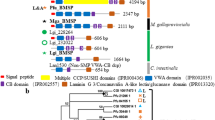
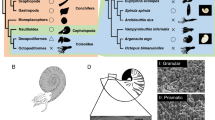
Pif molecules in various mollusk species. a The schematic representation of each Pif molecule from various species. White box, signal peptide; slashed box, VWA domain; vertical-lined box, chitin-binding domain; horizontal-lined box, chitin-binding domain-like sequence; black box, aragonite-binding protein; high-density vertical-lined box, aragonite and calcite-binding protein; gray box, the homologous sequence with aragonite-binding protein; cross-lined box, LG domain; asterisk, dibasic cleavage site. The frames at the bottom indicate the figures showing the alignment in each region. b–d The SEM picture of each shell microstructure. b The cross-section of the nacreous layer in P. fucata. c The cross-section of the nacreous layer in M. galloprovincialis. d The cross-section of prismatic structure with minor irregular spherulitic prismatic structure (M + 2 layer) in Lottia dorsuosa closely related to L. gigantea phylogenetically. e The shell of P. fucata. f The shell of M. galloprovincialis. g The shell of L. gigantea. Pif, Pif from P. fucata; pmPif, Pif homologue from P. margaritifera; pmxPif, Pif homologue from P. maxima. ppPif, Pif homologue from P. penguin. BMSP (Blue Mussel Shell Protein), Pif homologue from M. galloprovincialis. LG228264, LG232022, LG233460, and LG236719, Pif homologue from the genome database of L. gigantea. Arrows indicate the nacreous layer (e, f) and M + 2 layer (g)
Materials and Methods
Samples
Live P. margaritifera, P. maxima, and P. penguin were kind gifts from Tasaki Shinju Co. Ltd. The live mantle tissues are crashed and powdered in liquid nitrogen. The frozen powder was put into 1 ml ISOGENE (Wako, Japan) for 5 min. Then, 0.2 ml chloroform was added to this solution. After centrifugation, the supernatant was collected and RNA was purified by ethanol precipitation. The extracted RNA solution was stored at −80 °C.
Nucleotide sequence analysis
The sequences and positions of primers used in this experiment are shown in Fig. S1a–c and Table S1. First-strand cDNA was synthesized with 1 μg of total RNA using a SMARTTM RACE cDNA Amplification Kit (ClonTech, USA) according to the manufacturer’s instructions. For the first reverse transcription-polymerase chain reaction (RT-PCR), degenerate oligonucleotide primers were designed based on the amino acid sequences of the chitin-binding domain in Pif 97 of P. fucata. A cDNA fragment of each Pif homologue was amplified by two rounds of PCR. In the first round of PCR, the first-strand cDNA was used as a template, and the amplification was primed by a set of two primers. In the second PCR, the first PCR product was used as a template, and amplification was primed by a set of nested primers. The following program was used for PCR amplification: 30 cycles of 30 s at 94 °C (3 min 30 s for the first cycle only), 30 s at 55 °C, and 1 min at 72 °C (3 min for the last cycle only).
5′- and 3′-Rapid Amplification of cDNA Ends
The sequences and positions of primers used for 5′- and 3′-rapid amplification of cDNA ends (RACE) are shown in Fig. S1a–c and Table S1. The specific primers for each Pif homologue were prepared based on the nucleotide sequence of the cDNA fragments amplified by RT-PCR. cDNA fragments encoding the 5′-region were amplified by two rounds of PCR. In the first round of PCR, the first-strand cDNA was used as a template, and amplification was primed by a set of two primers. In the second round of PCR, the first PCR product was used as a template, and the amplification was primed by another set of primers, but because the 5′-RACE adaptor binds to various places in mRNA, we obtained various reverse transcript products with different lengths. Hence, we used many primer sets to amplify the 5′-RACE sequence. The following program was used for PCR amplification: 5 cycles of 5 s at 94 °C and 3 min at 72 °C; 5 cycles of 5 s at 94 °C, 10 s at 70 °C, and 3 min at 72 °C; and 25 cycles of 5 s at 94 °C, 10 s at 68 °C, and 3 min at 72 °C.
The specific primers of each Pif homologue were prepared based on the nucleotide sequence of the cDNA fragments amplified by RT-PCR. A cDNA fragment encoding the 3′-region of Pif homologue genes was amplified by two rounds of PCR. In the first round of PCR, the first-strand cDNA was used as a template, and amplification was primed by a set of primers. In the second round of PCR, the first PCR product was used as a template, and amplification was primed by a set of primers. Since the 3′-RACE adaptor binds to the adenine-rich region (lysine-rich site) of the dibasic cleavage site, various reverse transcription products with different lengths were obtained. Therefore, we used various primers to amplify the 3′-RACE sequence. The following program was used for PCR amplification: 30 cycles of 30 s at 94 °C (3 min 30 s for the first cycle only), 30 s at 55 °C, and 1 min at 72 °C (3 min for the last cycle only).
Nucleotide Sequence Analysis
All PCR products were ligated into a pGEM-T (Easy) vector (Promega, USA) according to the manufacturer’s instructions. Both strands of the inserted DNA were sequenced on an ABI PRISM 310 Genetic Analyzer (Applied Biosystems, USA) using a BigDye Terminator Ver. 3.1 (Applied Biosystems).
Results
Pif of P. fucata
We reported that the Pif protein from the nacreous layer of P. fucata plays important roles in the formation of this layer. Pif consists of two proteins, Pif 97 and Pif 80, encoded by a single mRNA. Pif 97 has VWA and chitin-binding (peritrophin A type) domains, while Pif 80 has aragonite binding function. These two proteins are separated at a dibasic cleavage site at the end of the sequence of Pif 97. The detailed sequence and functional analyses of Pif have previously been reported (Suzuki et al. 2009).
The calculated molecular mass and pI of Pif 97 before post-translational modifications are 58.8 kDa and 4.65, respectively (Table 1). In contrast to Pif 97, Pif 80 is rich in Asp (28.5 %), Glu (4.1 %), Lys (18.7 %), and Arg (10.9 %) residues. Its calculated molecular mass and pI before post-translational modifications are 54.2 kDa and 4.99, respectively (Table 2).
Pif of P. margaritifera
To identify a Pif homologue from P. margaritifera, primers were designed on the basis of the conserved sequence of the chitin-binding domain sequence of Pif. The entire cDNA sequence of the Pif homologue from P. margaritifera (pmPif) was determined by a combination of RT-PCR and 5′- and 3′-RACE (Fig. S2a). Sequence analysis of the 3,373-bp cDNA of pmPif revealed an open reading frame encoding a protein consisting of 1,011 amino acids, with the translation initiation codon (ATG) at nucleotide position 37. The first 24 amino acids apparently comprise a signal peptide because of their high hydrophobicity. An in-frame stop codon is located at nucleotide position 3,070, and a putative polyadenylation signal (AATTAA) is located 12 nucleotides upstream from the poly(A) tail. It is thus very likely that this cDNA represents a copy of the full length of the pmPif mRNA (Fig. S2a).
On the basis of the predicted amino acid sequence, pmPif may also be divided into two proteins, pmPif 97 and 80, at the sequence of RIKR located at amino acid position 555 located at the C-terminal end of pmPif 97. pmPif 97 contains VWA and chitin-binding domains. The calculated mass and pI of pmPif 97 before any post-translational modifications are 60.5 kDa and 4.60, respectively (Table 1). In contrast to pmPif 97, pmPif 80 is rich in Asp (25.6 %), Glu (5.1 %), Lys (16.3 %), and Arg (11.3 %) residues. The calculated mass and pI of pmPif 80 before any post-translational modifications are 53.3 kDa and 5.01, respectively (Table 2). These features of pmPif are very similar to those of Pif.
The Pif homologues from P. margaritifera (Pmarg-Pif) were reported from a study using EST analysis (Joubert et al. 2010). Four repeated sequences (LVKEIERRKSDDKKSFDD) were found in pmPif 80. These repeated sequences were also found in the pmPif 80 analyzed in this study, although there are some amino acid differences between the proteins in this previous study and the present work.
The last six amino acids of the fourth repeat in our sequence of pmPif 80 are replaced by GNGRRT, a difference which may be associated with the different habitat from which the sample was obtained.
Pif of P. maxima
A Pif homologue from P. maxima (pmxPif) was identified by the same method as that used for P. margaritifera. The sequence length and amino acid composition of pmxPif are almost the same as those of pmPif (see Tables 1 and 2; see Fig. S2b for detailed information).
Pif of P. penguin
A Pif homologue from P. penguin (ppPif) was identified by the same method as that used for P. margaritifera. Although the sequence length and amino acid composition of ppPif 97 are similar to those of Pif 97, pmxPif 97, and pmPif 97 (Table 1), the molecular mass and amino acid composition of ppPif 80 are different from those of other Pif 80 molecules in Pinctada species (Table 2; Fig. S2c). The proportion of Asp residues in ppPif 80 was found to be about half of those of other Pif molecules in Pinctada species. Furthermore, the proportions of Lys and Arg in ppPif 80 are much lower than those of other Pif molecules in Pinctada species. The sequence shows that there is no DDRK repeat in ppPif 80, indicating that point mutations or sequence deletion/insertion must have occurred in ppPif 80 during the period since the common ancestor of these species.
Pif of M. galloprovincialis (BMSP)
BMSP was identified from the nacreous layer of M. galloprovincialis using the calcium carbonate-binding assay. BMSP also consists of two proteins, BMSP 120 and BMSP 100, encoded by a single mRNA. BMSP 120 has four VWA domains and one chitin-binding domain. In contrast, BMSP 100 has calcium carbonate (both aragonite and calcite) binding ability. Since the distribution of the domains in BMSP 120 is similar to that of Pif 97 and the chemical property of BMSP 100 with regard to calcium carbonate binding is similar to that of Pif 80, BMSP could be confidently classified as a Pif homologue in M. galloprovincialis. The detailed sequence and functional analyses of BMSP have previously been reported (Suzuki et al. 2011). The molecular weight and amino acid composition of BMSP are completely different from those of the Pif homologues of P. fucata, P. margaritifera, P. maxima, and P. penguin. BMSP has a basic pI value and contains a lot of Trp residues.
Pif of L. gigantea
Recently, the whole genome sequences of L. gigantea were revealed (Lottia gigantea v1.0, http://genome.jgi-psf.org/Lotgi1/Lotgi1.home.html). This is the first achievement of its kind in mollusks. We identified four Pif homologues (protein ID LG228264, LG232022, LG233460, and LG236719) from the database. LG228264, LG232022, and LG233460 have one VWA domain and one chitin-binding domain, which are arranged in the same order and at similar positions to those in the Pif molecules of bivalves. In contrast, LG236719 has three VWA domains and one chitin-binding domain. Although all of these proteins have an N-terminal section corresponding to Pif 97, they do not have a C-terminal section corresponding to Pif 80 (Fig. 1a), suggesting that these gene models are incomplete partial sequences.
Alignment of the Partial Sequences of the Pif Homologues
A schematic representation of alignment of the Pif homologue sequences from various species is shown in Fig. 1a. Although Pinctada species and P. penguin have one VWA domain in one Pif molecule, BMSP from M. galloprovincialis and LG236719 from L. gigantea have multiple VWA domains in one Pif molecule (Fig. 2). Most of these VWA domains contain the conserved sequence of the metal ion-dependent adhesion site (MIDAS DXSXSXXXXTXXXXD) (Lee et al., 1995). In contrast, BMSP and the Pif homologues of L. gigantea have only a DXSXS motif. As the T and D residues in the adhesion site are not always needed for binding to metal ions (Whittaker and Hynes 2002), this short conserved MIDAS motif may serve as a sequence to bind metal ions. However, the DXSXS sequence was found to be changed to DXSXD in the Pif homologues of Pinctada species and P. penguin. These results suggest that the MIDAS motif is not always necessary for the function of the VWA domain in the shell.
Amino acid sequence alignment of the VWA domains. BMSP has four VWA domains, namely, BMSP-1, BMSP-2, BMSP-3, and BMSP-4, from the N-terminal side. LG236719 has three VWA domains, namely, LG236719-1, LG236719-2, and LG236719-3, from the N-terminal side. Asterisks indicate the MIDAS motif, DXSXS (abbreviations and sources: see the legend of Fig. 1)
The peritrophin A-type chitin-binding domain is conserved in all Pif homologues (Fig. 3). Although the similarity of the primary protein sequences is very low, six cysteine residues that form three disulfide bonds and contribute to the molecular motional restraint of the β-sheet structure that is required for binding to chitin are completely conserved.
Amino acid sequence alignment of the chitin-binding domains. Asterisks indicate the completely conserved six cysteine residues among all chitin-binding domains. The disulfide bonds are illustrated at the top. For the invertebrate chitin-binding domains, the disulfide bonds are predicted according to the disulfide bonds determined in tachycitin (Kawabata et al. 1996) (abbreviations and sources: see the legend of Fig. 1)
Downstream of the typical chitin-binding domain, the BLAST conserved domain search showed that there is a similar type of chitin-binding domain (chitin-binding domain-like sequence) with a high e-value (around 0.5). This high value means that the domain has low homology with the chitin-binding domain and its real function is obscure. In this region, almost all cysteine residues are also conserved (Fig. 4).
Amino acid sequence alignment from the downstream of the chitin-binding domain. Amino acid sequence alignment from the downstream after the chitin-binding domain to the dibasic cleavage sites (P. fucata, P. margaritifera, P. maxima, and P. penguin), to the C-terminal end of BMSP 120 (M. galloprovincialis), and to the end of sequence (L. gigantea). The box indicates the LG domain in LG233460. The single asterisk frame indicates the incomplete chitin-binding domain (chitin-binding domain-like sequence). The double asterisks frame indicates the N-terminal side of the sequences of the LG domain that are conserved in all Pif homologues. The triple asterisks frame indicates the C-terminal side of the sequences of the LG domain. The underlines indicate the dibasic cleavage sequence (white triangles indicate the position). Plus means the sequence position that has the possibility to bind calcium ion in LG domain (abbreviations and sources: see the legend of Fig. 1)
The amino acid sequences in the region between the chitin-binding domain and the dibasic cleavage site are highly conserved in all Pif homologues (Fig. 4), suggesting that all Pif homologues are derived from a common ancestral gene. This conserved region may have some important roles in the function of Pif, although the exact nature of this role is unclear.
Although the sequences of Pif 80, which has aragonite binding ability in Pinctada species, are highly conserved within this genus, the sequence found in P. penguin shows many deletion and amino acid changes compared with them (Fig. 5). The proportions of Asp, Glu, Lys, and Arg in ppPif 80 are about half of those in Pif 80 of Pinctada species. In addition, the primary protein sequence, amino acid composition, and pI of BMSP 100 are completely different from those of Pif 80 in Pinctada species and P. penguin, although BMSP 100 also has the ability to bind to calcium carbonate (both aragonite and calcite). These results indicate that the tertiary structure of the protein and the distribution of charge in the surface residues may be more important than the primary protein sequence for the binding of calcium and carbonate ions on the surface of crystals. Therefore, highly frequent mutations might have occurred in Pif molecules. The C-terminal sequences of the Pif homologues in L. gigantea are too short to compare with the other Pif 80 homologues.
Amino acid sequence alignment of Pif 80 in Pinctada species and P. penguin. The triple asterisks frame indicates the C-terminal side of the sequences of the LG domain. The line indicates the poly(Ala) region (abbreviations and sources: see the legend of Fig. 1)
Phylogenic Analyses of Pif
To investigate the evolutionary relationships among the Pif homologues, a phylogenetic tree was constructed with the Clustal W program (Thompson et al. 1994) using the neighbor joining algorithm (Saitou and Nei 1987) (Fig. 6). Since the VWA domain sequences are conserved in all Pif homologues, they were used for phylogenetic analyses. Pif, pmPif, and pmxPif derived from the Pinctada genus were classified as a monophyletic group. ppPif derived from the Pteria genus was identified as being closely related to the Pinctada genus but was placed outside the Pif derived from Pinctada. The four VWA domains in BMSP were defined as a monophyletic group and placed more distantly from the Pinctada group than ppPif. The three VWA domains in LG236719 were also identified to be a monophyletic group. These results indicate that the phylogenetic tree of VWA domains is consistent with the phylogeny identified using rRNA in mollusks. The four VWA domains in BMSP constitute a single clade as the three VWA domains in LG236719 do, indicating that gene duplication probably occurred after the divergence of each family, but not before the divergence of each class.
Phylogenetic tree showing the evolutionary relationship of Pif molecules using VWA domain sequences. The N–J tree was generated by using the method N–J of program ClustalW. Numbers represent bootstrap values (‰). The bar represents 0.1 substitutions per site (abbreviations are the same as those in the legend to Figs. 1 and 2)
Discussion
This study demonstrated that Pif homologues are present in both bivalves and gastropods, suggesting that Pif homologues act via the same basic mechanism in shell calcification.
The VWA domain is involved in protein–protein interaction. The discovery of multiple VWA domains suggests that this feature might be beneficial to mollusks in terms of facilitating shell formation, which is essential for their survival. A previous work revealed that the large matrix protein complex in the shell binds calcium carbonate crystal and regulates crystal nucleation, polymorph, and morphology (Matsushiro et al. 2003). Since the VWA domain is found in various extracellular matrix proteins and integrin receptors, the protein–protein interaction of the VWA domain in Pif 97 may be a key mechanism by which a complex of shell matrix proteins is formed for the production of an organic sheet. Homologues of dermatopontin, which was first identified from the shell of Biomphalaria glabrata (Marxen et al. 2003), were discovered in two species of Pulmonata (Sarashina et al. 2006). Almost all species in Pulmonata have multiple dermatopontin genes, and this multiplication might have occurred independently in the two species. Similarly to the multiple duplication of dermatopontin, multiple VWA domains in BMSP or LG236719 were each classified into a single cluster in phylogenetic analyses (Fig. 6), indicating that the duplication of the VWA domains in Pif occurred after speciation and the ancestral molecule of Pif might have benefited from having more VWA domains to make the shell. On the other hand, all Pif homologues have one chitin-binding domain, and the existence of an incomplete chitin-binding domain encoded downstream of the typical chitin-binding domain suggests that one chitin-binding domain is sufficient to bind to chitin in the shell.
Since the VWA and chitin-binding domains are useful for interaction with organic matrices, they are present in various kinds of molecules. The VWA domains are found in various extracellular matrix proteins that function in scaffolds and for cell adhesion as well as with integrin receptors (Tuckwell 1999; Whittaker and Hynes 2002). The chitin-binding domains are found in chitinase and peritrophic matrix proteins (Elvin et al. 1996; Shen and Jacobs-Lorena 1999; Suetake et al. 2000). The genes encoding these domains can move to other positions in the genome via transposon mechanism or gene duplication. However, with the exception of Pif, no molecules that contain both a VWA domain and a chitin-binding domain have been identified. To investigate the correlation between the mechanism of formation of shell microstructure and the matrix proteins in the shell, the origin of Pif molecules should be investigated in Monoplacophora, Polyplacophora, and Aplacophora, which are the common ancestors of both bivalves and gastropods, in the future.
While the aragonite-binding proteins (Pif 80) of Pif and its homologues are highly conserved within the Pinctada genus, ppPif 80 has some differences in terms of its sequence from Pif 80 and its homologues of Pinctada species; furthermore, the primary sequence of BMSP 100 is completely different from those of the Pif 80 molecules of Pinctada species and P. penguin. ppPif 80 has about half the number of charged amino acid residues of Pif of Pinctada species, and poly(Ala) residues are present instead of the Asp-, Lys-, and Arg-rich region (Table 2; Fig. 5). A similar poly(Ala) region was also identified in MSI60, which was identified from the nacreous layer of P. fucata (Sudo et al. 1997). This result suggests two possibilities regarding molecular evolution: (1) convergent evolution might have caused this change in ppPif 80 of P. penguin and (2) the DDRK site might have arisen only in the Pinctada genus. On the other hand, BMSP 100 does not contain many charged amino acid residues but contains many Trp residues (Table 2). Trp and aromatic amino acid residues are found in some extracellular matrix proteins like keratin (Marshall and Gillespie 1982). However, the function of Trp residues in BMSP is unclear. The homologues of aspein, the products of which are extremely rich in Asp and are related to the formation of the prismatic layer in P. fucata (Tsukamoto et al. 2004), have been identified as asprich from Atrina rigida (Gotliv et al. 2005). Although both aspein and asprich contain many Asp residues, there are no typical common motifs. A. rigida also expresses variously sized production of asprich-related genes in the mantle. These findings suggest that amino acid sequences related to interactions with calcium carbonate can still retain their function after diverging to some extent.
The amino acid sequences of the region between the chitin-binding domain of Pif 97 and the N-terminal residue of the protein of Pif 80 are highly conserved among all Pif homologues (Fig. 4). Although the function of this region is unknown, it may have some important functions for the whole Pif molecule. The C-terminal end of this region contains a dibasic cleavage site only in the Pif homologues of the Pinctada species and P. penguin. LG233460 has a laminin G (LG) domain from 496 to 637 amino acid residues at the C-terminus (Fig. 4). The LG domain is a calcium ion-mediated receptor for steroids, integrins, heparin, and α-dystroglycan and is involved in interaction with extracellular matrix proteins (Tryggvason 1993; Yurchenco et al. 1993; Yu and Talts 2003). Two Asp residues in the LG domain are involved in binding calcium ions (Hohenester et al. 1999). The N-terminal sequences of the LG domain are conserved in all Pif homologues. The C-terminal sequences of the LG domain are conserved only in L. gigantea. However, the C-terminal part in each of the Pinctada species and P. penguin is replaced by another sequence containing a dibasic cleavage site at the C-terminus (Fig. 4). The C-terminal parts of the LG domains in the Pinctada genus and P. penguin are located at the center of the Pif 80 sequences (Fig. 5). Figure 7 shows the amino acid sequence alignment of the central part of Pif 80 molecules of the Pinctada species and P. penguin (the region indicated by triple asterisks in Fig. 5) and the C-terminal part in LG233460 (the region indicated by triple asterisks in Fig. 4). As the basic amino acid residues play important roles in binding to ligands, there are many basic regions in LG domains. Previous studies also revealed that the dibasic site (RRKRR), a cleavage site, is present in the central part of the sequence in LG3 modules of laminin α2 chain (Talts et al. 1998; Talts and Timpl 1999; Smirnov et al. 2002). Cleavage at this site produces two peptide fragments and is required for complex formation in order to bind to other extracellular matrix proteins. These findings indicate that the sequence between the N- and C-terminal regions of the LG domain containing a dibasic cleavage site in the Pinctada genus and P. penguin exists to facilitate complex formation in the organic framework. As a result, Asp residues that are related to calcium ion binding and basic residues (Arg and Lys) that are related to ligand binding probably obtained the function of binding to aragonite crystal. As such, mutation and natural selection may have resulted in Pif 80 containing many Asp, Arg, and Lys residues. The findings in this study suggest that the Asp-rich sequence of Pif 80 may be derived from the calcium binding ability of the LG domain and that the dibasic cleavage site has evolved via mutation of the LG domain. The laminin receptor has a transmembrane domain and is located on the cell surface for cell adhesion to the basement membrane and signal transduction following binding events. However, the binding ability of the LG domain in Pif family proteins against the laminin receptor is unknown. In contrast, BMSP 100 does not contain any sequences related to the LG domain. The N-terminal amino acid of BMSP 100 is Met, but there are also many Trp residues, as in the case of keratin, suggesting that BMSP 100 is derived from another gene that encodes one of the extracellular matrix proteins. The putative molecular evolution of Pif is summarized in Fig. 8.
Amino acid sequence alignment of the C-terminal side of the sequence in the LG domain. a The schematic representation of amino acid sequence of Pif from Pinctada species, P. penguin, and LG233460. The vertical- and horizontal-lined box indicates the position of the sequence aligned in b. The meanings of other boxes and sign are the same as those in the legend to Fig. 1. b The triple asterisks frame indicates the C-terminal side of the sequences of the LG domain. The sequences of Pinctada species and P. penguin are from the middle part of the sequence in Pif 80 molecules (the region of the triple asterisks in Fig. 5). The sequence of LG233460 is from the last part of the LG domain (the region of the triple asterisks in Fig. 4) (abbreviations and sources: see the legend of Fig. 1)
The schematic representation of molecular evolution in Pif. Pif of the common ancestor of both bivalves and gastropods had the VWA, chitin-binding, and LG domains. This original Pif has evolved to Pifs in bivalves and gastropods. Pif in Gastropoda, Patellogastropoda, Lottiidae, L. gigantea has the VWA domains, chitin-binding domain, and LG domain. LG domain is conserved well. Pif in Bivalvia, Mytiloida, Mytilidae, M. galloprovincialis has the VWA domains and chitin-binding domain. The N-terminal side of the sequence of the LG domain is conserved in the end of BMSP 120. However, the C-terminal side of the sequence of the LG domain is replaced by another gene, BMSP 100. Pif in Bivalvia, Pterioida, Pteriidae species have the single VWA domain and chitin-binding domain. The dibasic cleavage site was introduced in the middle of LG domain and Asp-, Arg-, and Lys-rich sequences were increased to interact with aragonite crystals (black box). White box, signal peptide; slashed box, VWA domain; vertical-lined box, chitin-binding domain; horizontal-lined box, chitin-binding domain-like sequence; black box, Asp-, Arg-, and Lys-rich aragonite-binding sequence; vertical- and horizontal-lined box, aragonite and calcite-binding protein (BMSP 100); cross-lined box, LG domain; asterisk, dibasic cleavage site
The nacreous layer of the Pinctada species and P. penguin has a much more finely controlled compartmental structure than that of M. galloprovincialis (Fig. 1b, c). L. gigantea does not have a nacreous layer but rather a prismatic structure with a minor irregular spherulitic prismatic structure with aragonite crystals surrounded by an organic framework (Fuchigami and Sasaki 2005; Suzuki et al. 2010) (Fig. 1d). The Pif molecules are probably factors that universally function to make organic frameworks with chitin. The crystal morphology surrounded by an organic framework in the shell may depend on the structure of Pif homologues.
Matrix proteins in the nacreous layer vary among species, even closely related ones. Recent studies have revealed that perlucin and perlwapin are found in bivalves and gastropods (Marie et al. 2011). However, the similarities of these molecules between the bivalves and gastropods are very low, and the function of the conserved residues is unknown. These low levels of similarity inhibit our ability to understand the molecular evolution of calcification in mollusk shell. To clarify the correlation between various matrix proteins and shell microstructure, comparisons of all the proteins in the shells of various species are required.
References
Belcher AM, Wu XH, Christensen RJ, Hansma PK, Stucky GD, Morse DE (1996) Control of crystal phase switching and orientation by soluble mollusc-shell proteins. Nature 381:56–58
Berland S, Marie A, Duplat D, Milet C, Sire JY, Bedouet L (2011) Coupling proteomics and transcriptomics for the identification of novel and variant forms of mollusk shell proteins: a study with P. margaritifera. Chembiochem 12:950–961
Burgess TL, Kelly RB (1987) Constitutive and regulated secretion of proteins. Annu Rev Cell Biol 3:243–293
Elvin CM, Vuocolo T, Pearson RD, East IJ, Riding GA, Eisemann CH, Tellam RL (1996) Characterization of a major peritrophic membrane protein, peritrophin-44, from the larvae of Lucilia cuprina. cDNA and deduced amino acid sequences. J Biol Chem 271:8925–8935
Falini G, Albeck S, Weiner S, Addadi L (1996) Control of aragonite or calcite polymorphism by mollusk shell macromolecules. Science 271:67–69
Fuchigami T, Sasaki T (2005) The shell structure of the recent Patellogastropoda (Mollusca: Gastropoda). Paleontol Res 9:143–168
Gotliv BA, Kessler N, Sumerel JL, Morse DE, Tuross N, Addadi L, Weiner S (2005) Asprich: a novel aspartic acid-rich protein family from the prismatic shell matrix of the bivalve Atrina rigida. Chembiochem 6:304–314
Gregoire C (1957) Topography of the organic components in mother-of pearl. J Biophys Biochem Cytol 3:797–808
Hohenester E, Tisi D, Talts JF, Timpl R (1999) The crystal structure of a laminin G-like module reveals the molecular basis of alpha-dystroglycan binding to laminins, perlecan, and agrin. Mol Cell 4:783–792
Joubert C, Piquemal D, Marie B, Manchon L, Pierrat F, Zanella-Cleon I, Cochennec-Laureau N, Gueguen Y, Montagnani C (2010) Transcriptome and proteome analysis of Pinctada margaritifera calcifying mantle and shell: focus on biomineralization. BMC Genomics 11:613–625
Kawabata S, Nagayama R, Hirata M, Shigenaga T, Agarwala KL, Saito T, Cho J, Nakajima H, Takagi T, Iwanaga S (1996) Tachycitin, a small granular component in horseshoe crab hemocytes, is an antimicrobial protein with chitin-binding activity. J Biochem 120:1253–1260
Kono M, Hayashi N, Samata T (2000) Molecular mechanism of the nacreous layer formation in Pinctada maxima. Biochem Biophys Res Commun 269:213–218
Lee JO, Rieu P, Arnaout MA, Liddington R (1995) Crystal structure of the A domain from the alpha subunit of integrin CR3 (CD11b/CD18). Cell 80:631–638
Mann K, Siedler F, Treccani L, Heinemann F, Fritz M (2007) Perlinhibin, a cysteine-, histidine-, and arginine-rich miniprotein from abalone (Haliotis laevigata) nacre, inhibits in vitro calcium carbonate crystallization. Biophys J 93:1246–1254
Marie B, Le Roy N, Zanella-Cleon I, Becchi M, Marin F (2011) Molecular evolution of mollusc shell proteins: insights from proteomic analysis of the edible mussel Mytilus. J Mol Evol 72:531–546
Marin F, Corstjens P, de Gaulejac B, de Vrind-de JE, Westbroek P (2000) Mucins and molluscan calcification. Molecular characterization of mucoperlin, a novel mucin-like protein from the nacreous shell layer of the fan mussel Pinna nobilis (Bivalvia, Pteriomorphia). J Biol Chem 275:20667–20675
Marshall RC, Gillespie JM (1982) The tryptophan-rich keratin protein fraction of claws of the lizard Varanus gouldii. Comp Biochem Physiol B 71:623–628
Marxen JC, Nimtz M, Becker W, Mann K (2003) The major soluble 19.6 kDa protein of the organic shell matrix of the freshwater snail Biomphalaria glabrata is an N-glycosylated dermatopontin. Biochim Biophys Acta 1650:92–98
Matsushiro A, Miyashita T, Miyamoto H, Morimoto K, Tonomura B, Tanaka A, Sato K (2003) Presence of protein complex is prerequisite for aragonite crystallization in the nacreous layer. Mar Biotechnol (N Y) 5:37–44
Michenfelder M, Fu G, Lawrence C, Weaver JC, Wustman BA, Taranto L, Evans JS, Morse DE (2003) Characterization of two molluscan crystal-modulating biomineralization proteins and identification of putative mineral binding domains. Biopolymers 70:522–533
Miyamoto H, Miyashita T, Okushima M, Nakano S, Morita T, Matsushiro A (1996) A carbonic anhydrase from the nacreous layer in oyster pearls. Proc Natl Acad Sci USA 93:9657–9660
Miyamoto H, Yano M, Miyashita T (2003) Similarities in the structure of nacrein, the shell-matrix protein, in a bivalve and a gastropod. J Moll Stud 69:87–89
Miyashita T, Takagi R, Okushima M, Nakano S, Miyamoto H, Nishikawa E, Matsushiro A (2000) Complementary DNA cloning and characterization of pearlin, a new class of matrix protein in the nacreous layer of oyster pearls. Mar Biotechnol (N Y) 2:409–418
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–25
Samata T, Hayashi N, Kono M, Hasegawa K, Horita C, Akera S (1999) A new matrix protein family related to the nacreous layer formation of Pinctada fucata. FEBS Lett 462:225–229
Sarashina I, Yamaguchi H, Haga T, Iijima M, Chiba S, Endo K (2006) Molecular evolution and functionally important structures of molluscan dermatopontin: implications for the origins of molluscan shell matrix proteins. J Mol Evol 62:307–318
Shen Z, Jacobs-Lorena M (1999) Evolution of chitin-binding proteins in invertebrates. J Mol Evol 48:341–347
Shen X, Belcher AM, Hansma PK, Stucky GD, Morse DE (1997) Molecular cloning and characterization of lustrin A, a matrix protein from shell and pearl nacre of Haliotis rufescens. J Biol Chem 272:32472–32481
Smirnov SP, McDearmon EL, Li S, Ervasti JM, Tryggvason K, Yurchenco PD (2002) Contributions of the LG modules and furin processing to laminin-2 functions. J Biol Chem 277:18928–18937
Sudo S, Fujikawa T, Nagakura T, Ohkubo T, Sakaguchi K, Tanaka M, Nakashima K, Takahashi T (1997) Structures of mollusc shell framework proteins. Nature 387:563–564
Suetake T, Tsuda S, Kawabata S, Miura K, Iwanaga S, Hikichi K, Nitta K, Kawano K (2000) Chitin-binding proteins in invertebrates and plants comprise a common chitin-binding structural motif. J Biol Chem 275:17929–17932
Suzuki M, Saruwatari K, Kogure T, Yamamoto Y, Nishimura T, Kato T, Nagasawa H (2009) An acidic matrix protein, Pif, is a key macromolecule for nacre formation. Science 325:1388–1390
Suzuki M, Kameda J, Sasaki T, Saruwatari K, Nagasawa H, Kogure T (2010) Characterization of the multilayered shell of a limpet, Lottia kogamogai (Mollusca: Patellogastropoda), using SEM-EBSD and FIB-TEM techniques. J Struct Biol 171:223–230
Suzuki M, Iwashima A, Tsutsui N, Ohira T, Kogure T, Nagasawa H (2011) Identification and characterisation of a calcium carbonate-binding protein, blue mussel shell protein (BMSP), from the nacreous layer. Chembiochem 12:2478–2487
Takahashi K, Yamamoto H, Onoda A, Doi M, Inaba T, Chiba M, Kobayashi A, Taguchi T, Okamura T, Ueyama N (2004) Highly oriented aragonite nanocrystal–biopolymer composites in an aragonite brick of the nacreous layer of Pinctada fucata. Chemm Comm 8:996–997
Talts JF, Timpl R (1999) Mutation of a basic sequence in the laminin alpha2LG3 module leads to a lack of proteolytic processing and has different effects on beta1 integrin-mediated cell adhesion and alpha-dystroglycan binding. FEBS Lett 458:319–323
Talts JF, Mann K, Yamada Y, Timpl R (1998) Structural analysis and proteolytic processing of recombinant G domain of mouse laminin alpha2 chain. FEBS Lett 426:71–76
Temkin I (2010) Molecular phylogeny of pearl oysters and their relatives (Mollusca, Bivalvia, Pterioidea). BMC Evol Biol 10:342–369
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–80
Treccani L, Mann K, Heinemann F, Fritz M (2006) Perlwapin, an abalone nacre protein with three four-disulfide core (whey acidic protein) domains, inhibits the growth of calcium carbonate crystals. Biophys J 91:2601–2608
Tryggvason K (1993) The laminin family. Curr Opin Cell Biol 5:877–882
Tsukamoto D, Sarashina I, Endo K (2004) Structure and expression of an unusually acidic matrix protein of pearl oyster shells. Biochem Biophys Res Commun 320:1175–1180
Tuckwell D (1999) Evolution of von Willebrand factor A (VWA) domains. Biochem Soc Trans 27:835–840
Wada K (1961) Crystal growth of molluscan shells. Bull Nat Pearl Res Lab 7:703–785
Weiner S, Hood L (1975) Soluble protein of the organic matrix of mollusk shells: a potential template for shell formation. Science 190:987–9
Weiss IM, Kaufmann S, Mann K, Fritz M (2000) Purification and characterization of perlucin and perlustrin, two new proteins from the shell of the mollusc Haliotis laevigata. Biochem Biophys Res Commun 267:17–21
Whittaker CA, Hynes RO (2002) Distribution and evolution of von Willebrand/integrin A domains: widely dispersed domains with roles in cell adhesion and elsewhere. Mol Biol Cell 13:3369–3387
Yano M, Nagai K, Morimoto K, Miyamoto H (2007) A novel nacre protein N19 in the pearl oyster Pinctada fucata. Biochem Biophys Res Commun 362:158–163
Yu H, Talts JF (2003) Beta1 integrin and alpha-dystroglycan binding sites are localized to different laminin-G-domain-like (LG) modules within the laminin alpha5 chain G domain. Biochem J 371:289–299
Yurchenco PD, Sung U, Ward MD, Yamada Y, O'Rear JJ (1993) Recombinant laminin G domain mediates myoblast adhesion and heparin binding. J Biol Chem 268:8356–8365
Acknowledgments
We are grateful to Tasaki Shinju Co. Ltd in Hyogo Prefecture, Japan, for providing us with the shells and live oysters of P. margaritifera, P. maxima, and P. penguin. We are grateful to Dr. James Weaver, Dr. Steve Weiner, and Dr. Lia Addadi for providing the shells of L. gigantea. This work was supported by Grants-in-Aid for Scientific Research (17GS0311, 22248037, and 22228006) from the Japan Society for the Promotion of Science (JSPS). M.S. was supported by a Research Fellowship of JSPS for young scientists.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. 1a
Sequence strategy for determination of the complete cDNA sequence for each Pif homologues. (a) pmPif. (b) pmxPif. (c) ppPif. The sequence of each primer number is shown in supplementary Table 1. (JPEG 39 kb)
Fig. 1b
(JPEG 36 kb)
Fig. 1c
(JPEG 35 kb)
Fig. 2a
The complete cDNA and deduced amino acid sequence of pmPif. (b) The complete cDNA and deduced amino acid sequence of pmxPif. (c) The complete cDNA and deduced amino acid sequence of ppPif. Orange box is a signal peptide. Blue box indicates the VWA domain. Yellow box is the Peritrophin A-type chitin-binding domain. Purple box is BXBB (B means a basic amino acid residue) that is a Kex2-like protease cleavage site. Green box is the sequence that has the similarity of aragonite-binding protein (Pif 80). The underlines indicate the polyadenylation signal sequences. (JPEG 241 kb)
Fig. 2b
(JPEG 228 kb)
Fig. 2c
(JPEG 476 kb)
Table 1
(DOCX 22 kb)
Rights and permissions
About this article
Cite this article
Suzuki, M., Iwashima, A., Kimura, M. et al. The Molecular Evolution of the Pif Family Proteins in Various Species of Mollusks. Mar Biotechnol 15, 145–158 (2013). https://doi.org/10.1007/s10126-012-9471-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10126-012-9471-2