Abstract
A low ratio of nonsynonymous and synonymous substitution rates (dN/dS) at a codon is an indicator of functional constraint caused by purifying selection. Intuitively, the functional constraint would also be expected to prevent such a codon from being deleted. However, to the best of our knowledge, the correlation between the rates of deletion and substitution has never actually been estimated. Here, we use 8595 protein-coding region sequences from nine mammalian species to examine the relationship between deletion rate and dN/dS. We find significant positive correlations at the levels of both sites and genes. We compared our data against controls consisting of simulated coding sequences evolving along identical phylogenetic trees, where deletions occur independently of substitutions. A much weaker correlation was found in the corresponding simulated sequences, probably caused by alignment errors. In the real data, the correlations cannot be explained by alignment errors. Separate investigations on nonsynonymous (dN) and synonymous (dS) substitution rates indicate that the correlation is most likely due to a similarity in patterns of selection rather than in mutation rates.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The functional constraint on a genomic region is defined by its sensitivity to mutations, that is, the proportion of mutations that negatively affect its function (Graur 2016, pp. 116–120). Genomic regions subject to strong functional constraints are expected to perform important functions and to evolve relatively slowly. Mutations can take many forms, including nucleotide substitutions, insertions, and deletions (indels); as a result, functional constraint can be defined separately with respect to each type of mutation. One might expect functional constraints with respect to nucleotide substitutions and indels to be correlated—if the function of a genomic region can be disrupted by a substitution it can probably also be disrupted by an indel. However, this is not necessarily the case. For example, nucleotide substitutions at a fourfold degenerate site in a protein-coding gene may be selectively neutral because the protein product is not affected. If that fourfold degenerate site is deleted, however, it will cause a frameshift that will likely disrupt the function of that protein. Sites that only experience selection when they are deleted were referred to as “indifferent DNA” (Graur et al. 2015).
Deletions have been less extensively studied than substitutions for two reasons. First, indels are more difficult to detect than substitutions (Landan and Graur 2009; Nagy et al. 2012). Second, indels have not been modeled mathematically as well as substitutions (but see Lunter et al. 2006). Nevertheless, a few studies have attempted to compare the patterns of functional constraint arising from both kinds of mutations. Taylor et al. (2004) identified 1743 indel events in 1282 genes (out of a dataset of 8148 genes) from human–mouse–rat triple alignments. They compared indel rates in genes of different functions using Gene Ontology (Ashburner et al. 2000), and found that intracellular proteins and enzymes are less likely to have indels. When they compared variation in indel and substitution rates, they discovered similar distributions among categories (Waterston et al. 2002). These results indicate that functional categories that are more “important” to an organism tend to have both reduced amino acid replacement and reduced amino acid loss. One limitation of the study by Waterston et al. is that it focused on groups of genes rather than on individual genes, because substantial variation in selection can occur within a category.
Another study used a 28-vertebrate alignment to study coding-sequence conservation (Miller et al. 2007). The authors tested the hypothesis that more conserved amino acids are more likely to cause diseases when deleted. They analyzed the gene encoding the enzyme phenylalanine hydroxylase, mutations in which can cause phenylketonuria. The conservation levels of codons involved in disease-causing deletions turned out to be the same as for the gene overall. Miller et al. (2007) concluded that long-term selection against nonsynonymous mutations is consistent with short-term selection (as implied by diseases) against amino acid deletions. One strength of this study was the ability to identify deleterious mutations directly from clinical data. It was, however, based on only a single gene.
Chen et al. (2009) studied the ratio of nucleotide substitution to indel rates across mammalian and bacterial genomes. They interpreted the ratio as an indicator of the relative strengths of selection on the two types of mutations. They found that, within coding regions, more conserved genes have higher substitution to indel ratios than less conserved genes. This result suggests that indels (even non-frameshifting ones) are subject to relatively stronger selection than substitutions in conserved genes. However, as the comparison is focused on which type of mutation is more common, it does not directly help to resolve the correlation between the two types of changes.
In a population-level comparison between 179 human genomes, Montgomery et al. (2013) found that indel-based variations were highly localized: half of them were identified in only ~ 4% of the genome, likely due to mutation rate effects. The mutation rate heterogeneity was different between indels and substitutions; for example, recombination hotspots accompanied an increase of indels but not single nucleotide polymorphisms. As expected, the authors found evidence that indels in protein-coding sequences are subject to strong purifying selection. Indeed, even non-frameshift indel variants were found to have lower allele frequencies (a hallmark of purifying selection) than non-coding indels.
In a survey of seven species of eubacteria, and eight species of eukaryotes, Sung et al. (2016) showed that indel rates were strongly positively correlated with substitution rates among species. However, this study considered only genomic rates, not the rates at specific sites.
While the studies mentioned above are broadly consistent with the existence of a correlation between the rates of deletion and substitution, they do not provide conclusive evidence for it. Furthermore, they do not address the potential causes of the correlation, if it exists. There are two, not mutually exclusive hypotheses. First, a correlation in the strength of purifying selection against both types of mutations. This would be expected to cause a positive correlation between the deletion rate and dN, but not dS because the latter is not expected to result from purifying selection (Nei and Gojobori 1986; Price and Graur 2016). Therefore, there would also be a positive correlation between the deletion rate and the dN/dS ratio. Second, a positive correlation in the rates of both types of mutations would be expected to yield positive correlations between the deletion rate and both dN and dS, but not the dN/dS ratio.
Here, we use mammalian protein-coding sequences and simulated sequences to study the correlation between the rates of deletion and substitution, and to disentangle the causes of the correlation. We find that there is indeed a positive correlation between the rates of deletion and substitution, and that it is likely to be caused primarily by selection, rather than mutation.
Results
Deletion and Nonsynonymous Substitution Rates Per Site are Positively Correlated
We collected sequences of protein-coding genes from nine mammalian genomes (Fig. 1, Lindblad-Toh et al. 2011), and aligned them with PROBCONS (Do et al. 2005). Sequence evolution was simulated along a phylogeny with the same topology as that obtained by Lindblad-Toh et al. with parameters derived from the real data (See Supplementary Text and Fig. S1). In-frame deletions of length 1–8 amino acids were identified (Fig. 2; see “Materials and Methods” for details); deletion rate, dN, dS, and dN/dS were measured for each codon. Hereafter, we refer to dN, dS, and dN/dS collectively as “substitution measures.”
The commonly accepted phylogenetic relationship among the nine species used in this study. This tree will be called the external reference tree throughout the paper. Seven different colors denote seven pairs of branches/lineages (a–g) on which deletions were estimated. The black-colored branches are the root of the tree. The branch lengths of the nine-species tree are derived from UCSC human/hg19/GRCh37 46-way multiple alignment (Kent et al. 2002). These branch lengths are used as guidance for simulation and estimation of deletion rates
Illustration of how we identify deletion events and non-used sites in protein sequences for each pair of lineages. a Identified short deletion at sites 3 and 4 in taxon 1. b Excluded sites 3 and 4 because of gaps in the outgroup. c Excluded sites 3 and 4 because both ingroup taxa contain gaps at those positions, thus it is impossible to know whether it is an insertion or a deletion. d Excluded sites 3–12 because of long (> 8aa) deletion. e Excluded sites 2–5 because of unknown amino acids. f Excluded sites 5 and 6 because they are included in a terminal gap
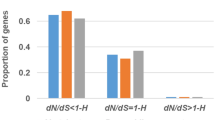
The correlations between deletion rate and the different substitution measures are summarized in Fig. 3. In the “All” dataset (in which all codons are included regardless of the presence or absence of gaps and substitutions), the deletion rate is positively correlated with both dN (Spearman correlation; ρ = 0.11) and the dN/dS ratio (ρ = 0.08) (Fig. 3a). The corresponding correlations in the simulated data are much lower (\(\bar {\rho }=0.01\) and 0.03 for dN/dS and dN, respectively, based on 1000 bootstrap replicates, see “Materials and Methods” for details; Z tests: Z = 57.87 for dN/dS and Z = 61.02 for dN, P < 0.0001 in both cases). The signal is even stronger when the true alignments from simulated data are used, indicating that alignment errors inflate dN/dS and dN estimates (Fig. S2). The deletion rate is also positively correlated with dS but the correlation is weaker than for dN (ρ = 0.04, Fig. 3a); however, this correlation is significantly stronger in the real data when compared to the simulated data (\(\bar {\rho }=0.01\); Z = 43.36, P < 0.0001).
Rates of deletion and substitution per site are positively correlated. Spearman correlations (ρ) between deletion rate and substitution measures (dN/dS, dN, and dS) in real and simulated data. a Based on the “All” dataset. b Based on the “NC-4+” dataset, where all sites without any substitutions or present in less than four species were removed. For the simulated data, the value shown is the mean of 1000 bootstrap replicates, and the error bars are 2.5–97.5% quantiles. Real data produce higher correlations than simulated data for all measures
To evaluate the robustness of the patterns summarized in Fig. 3a to uncertainty in the estimates of deletion and substitution rates, we repeated the analyses on the “NC-4+” dataset, containing only sites that have at least one nucleotide substitution and that are present (i.e., not gaps) in at least 4 species (see “Materials and Methods” for details). The patterns for dN and dN/dS are essentially unchanged (Fig. 3b). However, the correlation between deletion rate and dS disappears; indeed, the correlation is higher in the simulated data than in the real data (Z = − 10.69, P < 0.0001). Results for datasets with different thresholds of non-gap characters are similar to those for “NC-4+.” The exclusion of constant sites does not change the dN/dS-deletion correlation for simulated data (ρ = 0.01 for “NC”) but slightly increases it for real data (ρ = 0.09 for “NC”); higher thresholds for number of non-gap characters slightly reduces the correlation for both real and simulated data, often within the rounding of the numbers (real data, ρ = 0.08 for “4+,” 0.07 for “6+,” and 0.08 for “NC-6+”; simulated data, ρ = 0.01 for “4+”, “6+” and “NC-6+”).
Figure 4 shows the correlation with a density heatmap, where combinations of substitution measures and deletion rate are plotted. In the real data (Fig. 4a, b), counts of codons with non-zero deletion rate decline more slowly with increasing dN/dS (or dN) compared to simulated data (Fig. 4d, e) where the decline is parallel to that in codons with zero deletion rate. In contrast, dS (Fig. 4c, f) has a minimum of 0.1–0.2 due to a lower bound of local mutation rate, and the patterns are more similar between real and simulated data.
Considering that the interordinal relationship in Laurasiatheria is not entirely resolved, we re-calculated deletion rate using two alternative trees and calculated Spearman correlation coefficients with the same methods. The difference from calculations based on the main tree is negligible (Fig. S3).
We conclude that rates of deletion and nonsynonymous substitution per site are positively correlated. The correlation between deletion rate and dS is largely driven by sites with low substitution rate and/or uncertain estimates of rates of deletion and substitution. These results indicate that patterns of purifying selection against amino acid replacement and against amino acid loss are correlated to each other.
Deleted Sites Show Higher Rates of Nonsynonymous Substitution
If the rates of deletion and substitution are positively correlated then sites found to be deleted in at least one taxon would be expected to show a higher rate of substitution than sites that are present in all taxa. Figure 5 summarizes the results of an analysis testing this prediction. We used Cohen’s D, a measure of effect size (Cohen 1988). Cohen’s D is the ratio of the difference between two distributions’ means and their pooled standard deviation. D < 0.2 is considered a small effect size, while D > 0.5 is a medium or large effect size. Consistent with the results of the correlation analyses, both dN and dN/dS show medium to large effect sizes in both “All” and “NC-4+” datasets of the real data, whereas the simulated datasets show small effect sizes. The differences for dS are also statistically significant (Z = 50.93 for “All” and Z = 10.94 for “NC-4+,” P < 0.0001 in both cases), but much smaller in magnitude. Further investigation showed that removing alignment error by using the TRUE alignment data eliminates some but not all of the correlation (Fig. S4). While both dN and dS have a positive effect size in TRUE alignment, they are cancelled out when the ratio, dN/dS, is used; in that case, TRUE alignment shows non-significant effect size in both “All” and “NC-4+.”
Effect size (Cohen’s D) indicating the difference of substitution measures (dN/dS, dN, and dS) means between deleted and non-deleted sites. a Based on the “All” dataset. b Based on the “NC-4+” dataset, where all sites without any substitutions or present in less than four species were removed. For the simulated data, the shown value is the mean of 1000 bootstrap re-samplings, and the error bars are 2.5–97.5% quantiles
We compare the distributions of dN/dS in deleted and non-deleted sites in Fig. 6. The sharpest contrast is between the three leftmost columns: in the real data, 63.2% of deleted sites have dN/dS ≥ 0.2, while the number is 34.8% for non-deleted sites (Fig. 6a). The difference is negligible in the simulated data (Fig. 6b; 33.6 and 32.2% for deleted and non-deleted sites, respectively). A two-sample two-tailed Kolmogorov–Smirnov test (Kolmogorov 1933; Smirnov 1948) gives DKS = 0.2886, n = 7.2 × 104 and 3.6 × 106 for real data, and DKS = 0.0262, n = 4.7 × 105 and 2.2 × 107 for simulated data (both P < 0.0001). These results confirm that rates of deletion and non-synonymous substitution per site are positively correlated.
Histograms showing dN/dS distribution comparisons between sites with and without deletion, in both a real and b simulated data aligned with PROBCONS. Based on the “NC-4+” dataset. The axis marks the lower bound of each bin, i.e., the bin marked “0” indicates 0 ≤ dN/dS < 0.1. It can be observed that the distributions are much more different in real data than in simulated data: the non-deleted sites have a heavier left tail, while the deleted sites have a heavier right tail
Deletion and Nonsynonymous Substitution Rates Per Gene are Positively Correlated
To reduce the stochastic effects caused by limited number of mutations at each site, we decided to look at the same correlation at the gene level. We used the same statistical method with gene-averaged deletion rates and substitution measures.
As for the site data, the Spearman correlation coefficients between the deletion rate and substitution measures are significant and positive (Figs. 7, 8; all P < 0.0001). However, the strength of correlation depends on the substitution measure used. For both dN and dN/dS, the correlation is strong (ρ ≈ 0.5), but for dS it is weaker (ρ = 0.14). These correlations disappear completely in the simulated data (Fig. 8), and a negative but non-significant correlation is discovered with TRUE alignments of the simulated data (Fig. S5). Using a weighted deletion rate based on number of codons deleted and a rate based on number of deletion events does not seem to produce substantially different results (Fig. 8a, b), although the latter gives slightly higher correlation coefficients. We conclude that rates of deletion and nonsynonymous substitution per gene are positively and strongly correlated.
Gene-wise Spearman correlations between deletion rate and substitution (dN/dS, dN, and dS) in real and simulated data. In both dN/dS and dN, the correlation in real data is high (ρ ≈ 0.45) compared to simulated data (< 0.05); the difference is much less pronounced in dS. For the simulated data, the shown value is the mean of 1000 bootstrap re-samplings, and the error bars are 2.5–97.5% quantiles. The deletion rate was calculated based on a number of codons deleted, b number of deletion events
Deletion and Nonsynonymous Substitution Rates Within Genes are Positively Correlated
We also analyzed the correlation within genes, to see whether the site-wise correlation is entirely caused by the difference between genes. Figure 9 shows the distribution of within-gene correlation for both real and simulated data. In real data, we only used 463 genes in “All” and 454 in “NC-4+” (Fig. S6) that have an estimated ancestral length over 1500 aa and contains at least one deletion. In simulated data, 2062 genes in “All” and 2041 genes in “NC-4+” fit the same criteria and were used. In smaller genes, the sample size is too small to generate reliable correlation coefficients. In dN/dS, the real data give a slightly higher correlation compared to the simulated data (ρ ≈ 0.05 compared to ρ ≈ 0.02), although not to the level of genome-wide, site-wise correlation. dN produced a similar pattern. We used two-sample two-tailed Kolmogorov–Smirnov tests to examine the distributions’ differences. For “All” DKS = 0.3677 for dN/dS, 0.2658 for dN and 1 for dS, n = 463 (real data) & 2062 (simulated data); for “NC-4+” DKS = 0.3919 for dN/dS, 0.2343 for dN and 0.9995 for dS, n = 454 (real data) & 2041 (simulated data) (P < 0.0001 in all cases).
Histograms of distributions of within-gene Spearman correlation between substitution measures and deletion rate, using “All” dataset. Data are based on genes with an “ancestral” length of over 500 codons, and at least one deletion event. A total of 463 real genes and 2062 simulated genes were used. a Real data, dN/dS; b real data, dN; c real data, dS; d simulated data, dN/dS; e simulated data, dN; f simulated data, dS
Discussion
Implications on Protein Sequence Evolution
Our study shows that there is indeed a positive correlation between the probability of a codon being deleted and its dN/dS value (Figs. 3, 4, 5, 6, 7, 8). dN is also correlated with deletion rates, at a level similar to dN/dS. In contrast, the correlation is weak when dS is used. These results indicate that the correlation between rates of the two types of mutations (deletions and substitutions), if any, is negligible, because mutation rate heterogeneity is expected to affect dN and dS equally. Instead, a more plausible explanation for the results is that a common force, purifying selection, determines both replacement and deletion rates.
The simplest mechanism for this correlation is that both deletion and replacement disrupt the function of an amino acid residue in the protein, and reduce the fitness of individuals bearing the mutation. However, this mechanism alone cannot explain that the site-wise correlation is weak (ρ ≈ 0.1). One possible explanation for the weakness of the correlation is the existence of “indifferent DNA” (Graur et al. 2013, 2015). Indifferent DNA refers to sequences that are subject to strong purifying selection against deletions but not against substitutions. This is because its functionality relies more on its length rather than the exact sequences. For example, it is possible that certain amino acids are required to maintain the spatial relationships between other amino acids in the protein and, therefore, cannot be deleted, but can be replaced by multiple amino acids with similar biochemical properties. Consistent with this idea, the scatter plot in Fig. 7 shows many genes with low deletion rate and high dN/dS, but few genes with high deletion rate and low dN/dS.
The potential non-independence between selection against substitutions and deletions has been neglected in models of sequence evolution. For example, INDELible, one of the most comprehensive and frequently used programs for simulating the evolution of protein sequences, does not allow variation of indel rates along the sequence (Fletcher and Yang 2009). On the other hand, programs like SIMPROT (Pang et al. 2005) implement an algorithm that chooses indel positions relative to their substitution rates. ROSE (Stoye et al. 1998) and indel-Seq-Gen (Strope et al. 2009) limit indels to less conserved regions of sequences. A possible future approach to reflect the real-life correlation between deletion and amino acid replacements is to have variable deletion rates per site (or per gene) with the level of correlation with dN, dS, or dN/dS specified by the user.
Difference Between Site-Wise, Gene-Wise, and Within-Gene Analyses
We found that the Spearman correlations between deletion rate and dN, dS, and dN/dS are higher in gene-wise comparisons (Fig. 8) than in site-wise comparisons (Fig. 3). Site-wise and gene-wise analyses on evolutionary parameters often yield different results (e.g., Wang et al. 2013). dN/dS values vary in a much larger range in site-wise than gene-wise analyses (Lindblad-Toh et al. 2011). Elevated deletion rates and dN in our data are most likely due to relaxed purifying selection, although it is possible that a small minority of sites (individual amino acids) have undergone positive selection (dN/dS > 1). Thus, site-wise studies can in principle provide a higher resolution on the patterns of selection; however, they also suffer from a higher sampling error and, therefore, risk being over-parameterized (Rodrigue et al. 2010).
The difference in the magnitudes of the gene-wise and site-wise correlations indicates that the gene-wise correlation is not entirely explained by site-wise correlations within genes. A likely mechanism for this discrepancy is heterogeneity in levels of selective constraint between proteins. Such differences would be expected to cause a positive correlation among genes that would not be detectable within genes. An earlier study showed that most indels occur in intrinsically disordered regions of proteins, which are fast-evolving compared to structured regions (Light et al. 2013). Since proteins vary in the proportion of sequence occupied by disordered regions, this could explain our results (Fig. 9).
Artifactual Correlation Caused by Alignment Errors
Aside from the biological insights into protein sequence evolution, this study also provides information about consequences of alignment errors. There is no pre-determined correlation between indels and dN/dS in the simulated sequences, and thus all estimated correlation is due to artifacts. The correlation between dN/dS and deletion in true alignments of simulated sequences is indistinguishable from zero, which confirmed this point. The same correlations estimated from inferred alignment, on the other hand, are consistently and significantly higher than zero. The only difference between them is the presence of alignment error, and therefore, we can conclude that the small correlation observed in simulated reconstructed alignments is caused by alignment errors.
Multiple sequence alignment is a mathematically difficult (NP-complete) problem. The number of possible alignments from a set of sequences is exceedingly large; while one or more of them must be globally optimal, it is impossible to search through the solution space in reasonable time (Slowinski 1998). All current multiple sequence alignment algorithms use heuristic methods. These algorithms typically produce alignments that are shorter than the true alignment due to preferring mismatches over gaps, and gives mathematically optimal placements while the real process is sub- or co-optimal (Landan and Graur 2008, 2009). Regions that are rich in insertions and deletions are difficult to align due to co-optimal placement of gaps, thus putting gaps and mismatches together more often than it should be.
On the other hand, there is a correlation between dN and deletion as well as dS and deletion in simulated sequences that cannot be explained by alignment errors. This phenomenon appears in both true and inferred alignments, and in both site-wise and gene-wise analyses. There are a few possible hypotheses to explain this phenomenon: (1) Different rates of evolution (tree length) among different genes, because dN, dS, and deletion rate are all indicators of total evolutionary change along the entire tree. (2) An internal bias of the simulation algorithm which causes mutations more likely to occur in deleted sites than expected. (3) A bias of FUBAR’s estimates of dN and dS, where codons with less non-gap species are more likely to produce higher estimates.
Phase-1 and Phase-2 Deletions
A phase-1 or phase-2 codon deletion (deletions that start and end after the first or second codon position) can cause an amino acid mismatch without nucleotide substitutions. They are also called non-conservative deletions because they do not conserve the undeleted amino acids (de la Chaux et al. 2007). However, past studies demonstrated that such events are less common than expected by chance. In a study on pairwise indel event between mouse and rat, 12% of indels found are non-conservative, in contrast with a simulation expectation of 29% (Taylor et al. 2004); another study (de la Chaux et al. 2007) gave an even lower estimate that 4% of all deletions are non-conservative from three-primate alignments.
Unfortunately, with the simulation and alignment methods we used, we could not account for the effects for such deletions, nor could we mimic them by simulation. Nevertheless, the mismatch caused by non-conservative deletions usually does not happen in the same site as the gap. For example, if ACGCAT (Thr-His) became A—AT (Asn), the Asn residue will be aligned into one of the sites, while the gap occupies the other. The elevated dN/dS would thus only occur in the non-gap site. It is possible that the presence of such a mismatch complicates the alignment process and attracts other alignment errors, but we are not able to quantify this effect.
Long Deletions
Our study limited the length of deletion to eight amino acids (24 nucleotides) or fewer. There are several reasons for excluding longer deletions. First, long indels in protein sequences usually accompany large changes in the protein’s function or structure. Repeatable protein structures such as alpha helix (Scholtz and Baldwin 1992) and zinc finger (Klug and Rhodes 1987) are usually ten amino acids or longer. Such large-scale changes in protein structure usually result in strong fitness effects and must be studied with a case-by-case basis and integrated with biochemical experiments. While short indels can have consequences in protein structural domains, they are usually preserved only in regions with weak purifying selection and do not change the protein’s function drastically (Zhang et al. 2011). Second, long gaps that can be interpreted as long deletions can co-occur with alignment difficulties. This includes, again, two situations: (1) Real long deletions can cause alignment errors because of unrealistic values of gap-extending penalties. (2) When highly diverged or non-homologous regions are aligned with each other, long gaps can occur as algorithmic artifacts. Non-homologous sections can exist in corresponding regions of orthologous proteins if structural mutations such as translocation occurred.
Caveats
When we calculated site-wise and gene-wise deletion rates, we treated each deletion individually and did not consider how close together two deletions in a gene can be. In a neutral indel model by Lunter et al. (2006), the length of intergap segments (IGSs), gap-free regions of an alignment between two indel events, was identified as a good predictor for purifying-selected regions. If indels were randomly distributed, IGS lengths would have a geometric distribution; instead, from a human–mouse comparison, this is only true for segments shorter than 50 bp. However, long IGSs (100 bp or more) are highly overrepresented relative to the null expectation, indicating the existence of blocks that are resistant to indels, possibly due to purifying selection.
We assumed a particular evolutionary history for Mammalia (Lindblad-Toh et al. 2011). This phylogeny places the horse (Perissodactyla) and the dog (Carnivora) together as sister groups, while the cow (Cetartiodactyla) is a sister group for the horse + dog clade. This hypothesis of Laurasiatherian evolution, known as Pegasoferae, is supported by a phylogenetic study using molecular data (Nishihara et al. 2006). However, the evolutionary relationship among horse, dog, and cow is still under debate. A rival hypothesis groups the horse and the cow together (Perissodactyla + Cetartiodactyla = Euungulata), to the exclusion of the dog (Prasad et al. 2008). To test whether a different phylogenetic hypothesis changed our results, we re-calculated deletion rates using alternative trees. We found that the changes in the results are negligible (Fig. S3). Therefore, we have arbitrarily chosen the Pegasoferae hypothesis. Another related potential complication is that individual genes may not have the same evolutionary history as the species that carry them, a phenomenon known as incomplete lineage sorting (ILS) (Maddison 1997). ILS is more likely to occur when two or more speciation events occur in quick succession. In our nine-species tree, the group that is most likely to suffer from such effect is the Laurasiatheria (Hallström et al. 2011). However, the use of alternative trees within the Laurasiatheria did not change our results appreciably.
We used only protein-coding sequences in our study, because dN/dS, a reliable indicator of functional constraint, can only be calculated for protein-coding sequences. Selection against indels and substitutions in non-coding regions can be more efficiently studied in population-level analyses or between closely related species but this would be beyond the scope of this study. While it is possible to estimate selection in non-coding DNA from multiple sequence alignments by comparing them to purportedly neutral synonymous sites (Wong and Nielsen 2004), it requires reliable alignments, which is complicated by rearrangements and large amount of indels among mammalian orders. Extending our study to non-coding DNA sequences, such as RNA genes, remains a challenge for the future.
Materials and Methods
Data Collection and Analysis of dN, dS, and dN/dS
A list of aligned mammalian protein sequences was taken from Lindblad-Toh et al. (2011). To make sure that only good-quality genome sequences were used, we only included data from nine mammalian species (Fig. 1): human (Homo sapiens), chimpanzee (Pan troglodytes), macaque (Macaca mulatta), rat (Rattus norvegicus), mouse (Mus musculus), guinea pig (Cavia porcellus), dog (Canis lupus familiaris), cow (Bos taurus), and horse (Equus caballus). We retained 8605 alignments. Coding DNA sequences that correspond to these sequences were retrieved from ENSEMBL 2011 archive (Flicek et al. 2011).
All protein sequences were aligned with PROBCONS with default parameters (Do et al. 2005), and DNA sequences were aligned using the protein alignments as guides. Maximum likelihood trees were produced with RAxML (Stamatakis 2006) from the alignments, with a GTR + Gamma model and tree topology restricted to that of Fig. 1, and all other parameters set to default (standard hill-climbing algorithm). To reduce bias caused by unrealistic trees, ten genes that produced a total tree length above five were discarded. (In the 8605 genes, the mean tree length is 0.744 and standard deviation is 1.289. The shortest removed tree length is 6.897 and longest retained is 4.038.) Throughout the study, we used the remaining 8595 genes. This corresponds to ~ 42% of all human protein-coding genes. We decided on these nine species because they represent all main branches of Boreoeutheria, which contains the vast majority of mammalian species; these are also among the best annotated and highest quality genomes.
The DNA alignments were processed through the program HyPhy using the FUBAR script (Murrell et al. 2013), which estimated the dN and dS of each site using an approximate Bayesian algorithm, a Markov chain Monte Carlo process that compares a large number of site classes to identify and estimate selection.
Deletion Identification and Statistical Analysis
Deletions of one to eight amino acids were identified along seven pairs of branches (Fig. 1). These branch pairs are as follows: (A) human and chimpanzee lineages (red branches, macaque as outgroup); (B) ape and macaque lineages (green branches, cow as outgroup); (C) rat and mouse lineages (indigo branches, guinea pig as outgroup); (D) murid and guinea pig lineages (orange branches, human as outgroup); (E) primates and rodents lineages (purple branches, cow as outgroup); (F) dog and horse lineages (yellow branches, cow as outgroup); (G) (dog + horse) and cow lineages (cyan branches, human as outgroup). The outgroup was used to determine whether a gap in the alignment is caused by an insertion or a deletion (Fig. 2a). In branch pair (B), the closest outgroup is a rodent, but cow was chosen because rodents have long branch lengths. For a lineage containing multiple species (e.g., apes), only the branch before the divergence (e.g., divergence between human and chimpanzee) was analyzed. This was done by combining multiple sequences into an “ancestral” sequence: any site that is a gap in all combined species is a gap site in the “ancestor,” and if the site is not a gap in at least one of these sequences, it is considered non-gap in the “ancestor.” In this way, every branch in the nine-species tree, excluding the root branch, was searched for deletions without repetition. The root branch (the branch separating the primates-rodents group and other mammals) was not searched for deletions because the directions of its indels could not be determined.
A fraction of amino acid sites is excluded from analysis because of ambiguity and difficulties in detecting deletions or substitutions. These sites include gaps in the outgroup (Fig. 2b), gaps in both ingroup taxa (Fig. 2c), deletions over 8 amino acids long (Fig. 2d), ambiguous amino acids (Fig. 2e), and terminal gaps (Fig. 2f). In some cases, we excluded a site in the analysis of one lineage pair but not another.
The weighted deletion rate of an amino acid site, D, is calculated as \(D=\frac{{\mathop \sum \nolimits_{{i=1}}^{7} {D_i}}}{{\mathop \sum \nolimits_{{i=1}}^{7} {L_i}{V_i}}}\). We define Di = 1 if that site is part of a deletion in the ith lineage pair, and 0 otherwise; Vi = 1 if that site is not excluded in that lineage pair, and 0 otherwise; Li is the sum of branch lengths of the ith lineage pair, based on the placental tree (without chromosome X) from human/hg19/GRCh37 46 species multiple alignment (http://genomewiki.ucsc.edu/index.php/Human/hg19/GRCh37_46-way_multiple_alignment, Kent et al. 2002; Fig. 1).
Site-wise weighted deletion rates were re-calculated using two alternative trees that differ from the main tree in the relationship within Laurasiatheria; in one tree, the horse and the cow were considered sister groups (Euungulata) and in the other the dog and the cow were considered sister groups. Because the branch lengths were not available for alternative trees, we used an ad hoc approach that kept the length of terminal branches and used the length of the internal branch (the one separating the horse–dog ancestor from the Laurasiatheria ancestor) for the new internal branches. This has minimal effects on deletion rate estimation because this branch is short.
The weighted deletion rate of a gene, DG, is calculated as \({D_{\text{G}}}=\frac{{\mathop \sum \nolimits_{{j=1}}^{n} \mathop \sum \nolimits_{{i=1}}^{7} {D_{ij}}}}{{\mathop \sum \nolimits_{{j=1}}^{n} \mathop \sum \nolimits_{{i=1}}^{7} {L_{ij}}{V_{ij}}}}\), where n is the number of codons in the gene, Dij is Di in the jth codon in that gene, and Lij is Li in the jth codon in that gene.
An alternative gene-wise deletion rate is calculated as \({D_{{\text{GN}}}}=\frac{N}{{\mathop \sum \nolimits_{{j=1}}^{n} \mathop \sum \nolimits_{{i=1}}^{7} ({L_{ij}} \times {V_{ij}})}}\) where N is the number of deletion events identified in any lineage in that gene. DGN is called the event-number deletion rate of a gene.
For each amino acid site in each alignment, its deletion rate and three substitution measures (dN, dS, and dN/dS) were obtained. For each alignment method, Spearman correlation coefficients were calculated between the weighted deletion rate, D, and the three substitution measures. This dataset uses all sites and is thus named “All.” See Table 1 for summary statistics on this dataset.
To reduce the effects of spuriously high or low values of dN/dS due to “gappy” sites, the correlation coefficients were re-calculated for (1) sites that have not experienced a gap event in at least four sequences, and (2) sites that have not experienced a gap event in at least six sequences. These datasets are referred to as “4+” and “6+,” respectively. Many sites have not experienced any nucleotide substitution, and their dN/dS is indeterminate due to division by 0, only approximated using extrapolation from other sites. Therefore, we generated sub-datasets in which these constant sites were excluded. These datasets were named “NC-All,” “NC-4+,” and “NC-6+,” where “NC” stands for “no constant”.
Coding-Sequence Simulation and Analysis
We simulated coding DNA sequences using INDELible (Fletcher and Yang 2009). INDELible evolves nucleotide sequences along the input tree based on a nucleotide substitution model. These substitutions are subject to selection as determined by dN/dS, randomly drawn from an input distribution for each site. Insertions and deletions, always multiples of three nucleotides, are independently modeled and have a uniform rate among sites; however, the number of indels is proportional to the branch length.
We simulated a total of 8595 genes × 5 replicates. For each gene, the ancestral gene length and level of divergence were based on the values derived from the corresponding real gene (see Supplementary Text and Fig. S1 for details). The distribution of dN/dS was a gamma distribution with a shape parameter of α = 0.5 (approximated from real data) and a mean calculated from its real data counterpart. The distribution was discretized into 50 bins between 0 and 1 (0–0.02, 0.02–0.04 …), 20 bins between 1 and 2 (1–1.05, 1.05–1.1 …) and 1 bin above 2. In each bin, the dN/dS value used was the median. If a bin (usually the ones with highest dN/dS) has a probability below 10−6 in the gamma distribution, it was not used. The absolute deletion rate for each gene was drawn from a gamma distribution with a shape parameter of α = 0.6 (approximated from real data) and mean = 0.79 (the mean SAI from the real data), so that it is independent from substitution rate (see Supplementary Text); the relative indel rate was calculated based on absolute indel rate and branch lengths. Indel length was modeled with a power law distribution with the maximal length of 40 codons (Cartwright 2009).
The simulated protein sequences were aligned with PROBCONS (alternative alignment tools give identical results), and then nucleotide alignments were threaded through the protein alignments. We estimated deletion rates and substitution measures based on these alignments, as well as for the “true” alignment (as control for alignment error), as described above for real data. See Table 1 for summary statistics on the simulated data.
We used bootstrapping to generate plausible ranges of values of the sequence statistics to compare with the ones obtained from real data. We generated 1000 bootstrap subsets of the simulated data. In each subset, one random replicate was chosen from the five for each of the 8595 genes. Spearman correlation coefficients were calculated for each subset. Each subset was processed as described for real data to generate datasets of each type (“All,” “4+,” “6+,” “NC-All,” “NC-4+,” and “NC-6+”). For the Spearman correlation coefficients, the mean, standard deviation, and 2.5 and 97.5% quantiles were calculated. We used Z tests to compare the Spearman coefficients derived from real and simulated data.
Distribution of dN/dS in Deleted Sites
All real mammalian protein sites that have undergone at least one deletion in any lineage were extracted from the data set and their distributions of estimated dN/dS are computed. The distributions were compared with those from deletion-free sites with χ2 tests. Effect sizes (Cohen’s D, Cohen 1988) were calculated between dN/dS distributions in deletion and non-deletion codons. These analyses were only done on “All” and “NC-4+” datasets as representative of all six datasets. These procedures were repeated for the simulated data. Similar to the previous section, 1000 bootstrap subsets were used, and the mean, standard deviation, and 2.5 and 97.5% quantiles were calculated. Z tests were used to compare real to simulated data.
Analysis of Gene-Wise and Within-Gene Correlations
For both real and simulated data, we calculated gene-wise dN, dS, dN/dS, and deletion rate. Gene-wise dN and dS are the mean of corresponding values of “4+” sites over the whole gene. We did not use “NC-4+” because excluding substitution-free sites is likely to lead to overestimation of the substitution measures. Gene-wise dN/dS is gene-wise dN divided by gene-wise dS. The calculation of two alternative gene-wise deletion rates, DG and DGN, is described in a previous sub-section.
We calculated the Spearman correlation between gene-level deletion rate and substitution measures in both real and simulated data. Similar to previous sections, in the simulated data bootstrapping is used. Each subsample includes only one replicate for every simulated gene. The mean, standard deviation, and 2.5 and 97.5% quantiles were calculated. Z tests were used to compare real to simulated data.
We calculated within-gene Spearman correlation between deletion rates and substitution measures, using 466 real genes and 466 × 5 = 2330 simulated genes that have the derived “ancestral gene length” longer than 1500 amino acids. The correlation coefficients are calculated for both “All” and “NC-4+” datasets. For the real data, genes (three such genes in “All” and twelve in “NC-4+”) that do not have any deletions identified were removed from the data, while the rest (463 in “All” and 454 in “NC-4+”) were used to calculate the mean and standard deviation.
Data Availability
We uploaded our real and simulated alignments as well as Perl scripts of key steps on GitHub project “Mammal-Protein-Selection” (https://github.com/y-zheng/Mammal-Protein-Selection).
References
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA (2000) Gene ontology: tool for the unification of biology. Nat Genet 25(1):25
Cartwright R (2009) Problems and solutions for estimating indel rates and length distributions. Mol Biol Evol 26:473–480
Chen J-Q, Wu Y, Yang H, Bergelson J, Kreitman M, Tian D (2009) Variation in the ratio of nucleotide substitution and indel rates across genomes in mammals and bacteria. Mol Biol Evol 26:1523–1531
Cohen J (1988) Statistical power analysis for the behavioral sciences, 2nd edn. Lawrence Erlbaum Associates, Hillsdale, p 67
de la Chaux N, Messer PW, Arndt PF (2007) DNA indels in coding regions reveal selective constraints on protein evolution in the human lineage. BMC Evol Biol 7:19
Do CB, Mahabhashyam MSP, Brudno M, Batzoglou S (2005) ProbCons: probabilistic consistency-based multiple sequence alignment. Genome Res 15:330–340
Fletcher W, Yang Z (2009) INDELible: a flexible simulator of biological sequence evolution. Mol Biol Evol 26:1879–1888
Flicek P, Amode MR, Barrell D, Beal K, Brent S, Chen Y, Clapham P, Coates C, Fairley S, Fitzgerald S et al (2011) Ensembl 2011. Nucleic Acids Res 39:D800–D806
Graur D (2016) Molecular and genome evolution. Sinauer Associates, Sunderland
Graur D, Zheng Y, Price N, Azevedo RBR, Zufall RA, Elhaik E (2013) On the immortality of television sets: “function” in the human genome according to the evolution-free gospel of ENCODE. Genome Biol Evol 5:578–590
Graur D, Zheng Y, Azevedo RBR (2015) An evolutionary classification of genomic function. Genome Biol Evol 7:642–645
Hallström BM, Schneider A, Zoller S, Janke A (2011) A genomic approach to examine the complex evolution of laurasiatherian mammals. PLoS ONE 6(12):e28199
Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D (2002) The human genome browser at UCSC. Genome Res 12:996–1006
Klug A, Rhodes D (1987) Zinc fingers: a novel protein fold for nucleic acid recognition. Cold Spring Harb Symp Quant Biol 52:473–482
Kolmogorov A (1933) Sulla determinazione empirica di una legge di distribuzione. G Ist Ital Attuari 4:83–91
Landan G, Graur D (2008) Local reliability measures from sets of co-optimal multiple sequence alignments. Pac Symp Biocomput 13:15–24
Landan G, Graur D (2009) Characterization of pairwise and multiple sequence alignment errors. Gene 441:141–147
Light S, Sagit R, Ekman D, Elofsson A (2013) Long indels are disordered: a study of disorder and indels in homologous eukaryotic proteins. Biochim Biophys Acta 1834(5):890–897
Lindblad-Toh K, Garber M, Zuk O, Lin MF, Parker BJ, Washietl S, Kheradpour P, Ernst J, Jordan G, Mauceli E et al (2011) A high-resolution map of human evolutionary constraint using 29 mammals. Nature 478:476–482
Lunter G, Ponting CP, Hein J (2006) Genome-wide identification of human functional DNA using a neutral indel model. PLoS Comp Biol 2(1):e5
Maddison WP (1997) Gene trees in species trees. Syst Biol 46(3):523–536
Miller W, Rosenbloom K, Hardison RC, Hou M, Taylor J, Raney B, Burhans R, King DC, Baertsch R, Blankenberg D et al (2007) 28-way vertebrate alignment and conservation track in the UCSC Genome Browser. Genome Res 17:1797–1808
Montgomery SB, Goode DL, Kvikstad E, Albers CA, Zhang ZD, Mu XJ, Ananda G, Howie B, Karczewski KJ, Smith KS, Anaya V (2013) The origin, evolution, and functional impact of short insertion–deletion variants identified in 179 human genomes. Genome Res 23(5):749–761
Murrell B, Moola S, Mabona A, Weighill T, Sheward D, Pond SLK, Scheffler K (2013) FUBAR: a fast, unconstrained bayesian approximation for inferring selection. Mol Biol Evol 30:1196–1205
Nagy LG, Kocsubé S, Csanádi Z, Kovács GM, Petkovits T, Vágvölgyi C, Papp T (2012) Re-mind the gap! Insertion–deletion data reveal neglected phylogenetic potential of the nuclear ribosomal internal transcribed spacer (ITS) of fungi. PLoS ONE 7:e49794
Nei M, Gojobori T (1986) Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol 3:418–426
Nishihara H, Hasegawa M, Okada N (2006) Pegasoferae, an unexpected mammalian clade revealed by tracking ancient retroposon insertions. Proc Natl Acad Sci USA 103:9929–9934
Pang A, Smith AD, Nuin PAS, Tillier ERM (2005) SIMPROT: using an empirically determined indel distribution in simulations of protein evolution. BMC Bioinform 6:236
Prasad AB, Allard MW, NISC Comparative Sequencing Program, Green ED (2008) Confirming the phylogeny of mammals by use of large comparative sequence data sets. Mol Biol Evol 25:1795–1808
Price N, Graur D (2016) Are synonymous sites in primates and rodents functionally constrained? J Mol Evol 82:51–64
Rodrigue N, Philippe H, Lartillot N (2010) Mutation-selection models of coding sequence evolution with site-heterogeneous amino acid fitness profiles. Proc Natl Acad Sci USA 107(10):4629–4634
Scholtz JM, Baldwin RL (1992) The mechanism of alpha-helix formation by peptides. Annu Rev Biophys Biomol Struct 21(1):95–118
Slowinski JB (1998) The number of multiple alignments. Mol Phylogenet Evol 10(2):264–266
Smirnov N (1948) Table for estimating the goodness of fit of empirical distributions. Ann Math Stat 19:279–281
Stamatakis A (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690
Stoye J, Ever D, Meyer F (1998) Rose: generating sequence families. Bioinformatics 14:157–163
Strope CL, Abel K, Scott SD, Moriyama EN (2009) Biological sequence simulation for testing complex evolutionary hypotheses: indel-Seq-Gen version 2.0. Mol Biol Evol 26:2581–2593
Sung W, Ackerman MS, Dillon MM, Platt TG, Fuqua C, Cooper VS, Lynch M (2016) Evolution of the insertion-deletion mutation rate across the tree of life. G3: Genes Genomes Genetics 6(8):2583–2591
Taylor MS, Ponting CP, Copley RR (2004) Occurrence and consequences of coding sequence insertions and deletions in mammalian genomes. Genome Res 14:555–566
Wang H, Susko E, Roger AJ (2013) The site-wise log-likelihood score is a good predictor of genes under positive selection. J Mol Evol 76:280–294
Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P et al (2002) Initial sequencing and comparative analysis of the mouse genome. Nature 420:520–562
Wong WS, Nielsen R (2004) Detecting selection in noncoding regions of nucleotide sequences. Genetics 167(2):949–958
Zhang Z, Huang J, Wang Z, Wang L, Gao P (2011) Impact of indels on the flanking regions in structural domains. Mol Biol Evol 28(1):291–301
Acknowledgements
We used the Maxwell cluster from the Center of Advanced Computing and Data Systems (CACDS) at the University of Houston. CACDS staff provided technical support. We would like to thank Sarah Parks and her colleagues at EMBL-European Bioinformatics Institute for their help in running the SLR program on part of our data. R.B.R.A. was funded by NIH R01GM101352. We would also like to thank Jaanus Suurväli and Jan Gravemeyer at University of Cologne for their help in manuscript editing.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zheng, Y., Graur, D. & Azevedo, R.B.R. Correlated Selection on Amino Acid Deletion and Replacement in Mammalian Protein Sequences. J Mol Evol 86, 365–378 (2018). https://doi.org/10.1007/s00239-018-9853-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-018-9853-9