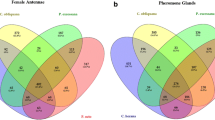
Abstract
How new mate recognition systems evolve when changes are required in both the male and female components remains a conundrum. Here, we investigated the molecular basis of pheromone reception in two species of tortricid (leafroller) moth, Ctenopseustis obliquana and C. herana. Male C. obliquana are attracted to a 90:10 blend of (Z)-8-tetradecenyl acetate (Z8-14:OAc) and (Z)-5-tetradecenyl acetate (Z5-14:OAc), whereas C. herana males are attracted to Z5-14:OAc alone. We used a transcriptome sequencing approach from adult male and female antennae to identify 47 olfactory receptors (ORs) from each species and assessed their expression levels in male and female antennae using RNA-Seq counting and quantitative RT-PCR. Three male-biased and one female-biased OR were identified in C. obliquana by quantitative RT-PCR, and four male-biased and one female-biased receptor in C. herana. The male-biased receptors, CoblOR7, CoblOR30, CherOR7, CherOR30, CherOR1a and CherOR1b were tested for their ability to respond to sex pheromone components in a HEK293 cell calcium assay. CoblOR7 and CherOR7 responded to Z8-14:OAc, however, no receptor for Z5-14:OAc was identified. In addition to Z8-14:OAc, CherOR7 also responded to Z7-14:OAc, indicating that this receptor may be under relaxed constraint. Of the 29 amino acid differences between CoblOR7 and CherOR7, significantly more are located in the third and the sixth transmembrane domain regions. Overall, these findings are consistent with studies revealing the presence of neurons tuned to both Z8-14:OAc and Z5-14:OAc in both species, but that for C. herana males, the ability to detect Z8-14:OAc is currently not required.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Changes in both the female and the male components are required for new mating systems to evolve. Pheromone-based mating systems offer a discrete communication system to understand how such systems evolve and are becoming increasingly studied for this purpose (Niehuis et al. 2013; Smadja and Butlin 2009; Shirangi et al. 2009; Symonds and Elgar 2008; Albre et al. 2013; Lassance and Lofstedt 2009). In moths, males locate females through upwind flight along a concentration gradient of the sex pheromone (Bradbury and Vehrencamp 1998). Compounds that make up the sex pheromone are mainly fatty-acid derivatives such as acetates, alcohols, and aldehydes that are typically 10, 12, 14, 16 or 18 carbons in length with one or two unsaturated positions along the chain (Linn and Roelofs 1995). Typically, pheromone blends are composed of one major component and one or more minor components. The composition of these sex pheromone blends is often highly specific, with little variation within populations and species. Species specificity in pheromone production and reception forms a robust mate recognition system that limits incompatible mating events (Linn and Roelofs 1995).
How new sex pheromones and pheromone blends are biosynthesised has been investigated in several species of moths, revealing changes in both desaturases or fatty acid reductases (Shirangi et al. 2009; Greenberg et al. 2003; Sakai et al. 2009; Albre et al. 2012; Xue et al. 2007). Sex pheromones are produced from simple fatty acids in specialised glands in the abdomen of females where the components are synthesised. Double bonds are introduced into fatty acids through specialised fatty-acyl desaturases (Knipple et al. 2002), often with several desaturases present possessing distinct affinities for different fatty acids substrates (Lienard et al. 2008). Differential expression of the genes encoding these desaturase enzymes can be crucial for producing new pheromone components. In the Ostrinia species, O. nubilalis and O. furnacalis, for example, the activation of ancestral desaturase genes, together with gene duplication and retroposon fusion has produced novel desaturase activities (Roelofs et al. 2002; Roelofs and Rooney 2003; Xue et al. 2007; Fujii et al. 2011). In O. scapulalis and O. furnacalis, the differential expression of desaturase genes is responsible for the production of species-specific pheromone components (Sakai et al. 2009). In addition to desaturases, altered fatty-acyl reductases (FARs) can produce distinct pheromones through differences in affinities for desaturated precusors in their conversion from fatty acids to alcohols. For example, the pheromones of the E and Z strains of O. nubilalis are produced by distinct alleles of the pheromone FAR. The alleles differ in the substrate specificity which leads to differentially reduced ratios in the final pheromone blend (Lassance et al. 2010).
How new pheromones are derived has been studied extensively but to understand the evolution of mating systems as a whole it is necessary to understand the role of the receiver and how the evolution of sex pheromone perception proceeds especially at the molecular level (Symonds and Elgar 2008). It seems difficult for mating systems that rely on chemical cues to evolve rapidly because purifying selection should prevent changes in either pheromone production or perception. A hypothesis that attempts to solve this dilemma is the “asymmetric tracking hypothesis”, which suggests that in a species where females are the limiting sex, greater variation may be tolerated in the male’s pheromone perception system to provide a scenario where rare males may exist that are able to sense a novel pheromone blend (Phelan 1992; Domingue et al. 2007). In males, changes in the preference for certain pheromones may well depend on alterations in a multigene family of receptors responsible for detecting the sex pheromone components (Gould et al. 2010; Wanner et al. 2010; Miura et al. 2010; Leary et al. 2012). It has been shown that even single mutations in the sequence of olfactory receptor (OR) genes can change the specificity of the receptor protein (Leary et al. 2012).
Moths perceive odorants through receptors located within sensilla on their antennae. These ORs contain seven transmembrane regions with a cytoplasmic N terminus and an external C terminus (Smart et al. 2008; Benton et al. 2006; Lundin et al. 2007). Together with the olfactory receptor co-receptor (Orco), they form a ligand-gated cation channel. While Orco is highly conserved (Guo and Kim 2007), ligand-binding ORs are more variable, with the C terminus being more highly conserved than the N terminus (Tunstall et al. 2007; Carraher et al. 2012). ORs involved in sex pheromone perception in moths are located in specialised long hair-like sensilla called sensilla trichodea (Rumbo 1981, 1983). These very long sensilla are typically more abundant on male antennae compared with females (Heinbockel and Kaissling 1996; Jordan et al. 2008; Mitsuno et al. 2008; Sakurai et al. 2004; Krieger et al. 2004). Receptors associated with sex pheromone perception in moths to date all fall into a separate phylogenetic clade and typically show higher levels of gene expression in the antennae of males compared with females (Bengtsson et al. 2012, Grosse-Wilde et al. 2011, Mitsuno et al. 2008).
Many olfactory receptors and pheromone receptors have been identified from a handful of moth species using a range of techniques including whole genome sequencing and RNA seq. From Heliothis virescens, 21 candidate OR genes have been identified in whole genome assemblies by BLAST searches using Drosophila ORs (Krieger et al. 2002). Sixty-six ORs were identified from the genome of the silkworm, Bombyx mori (Tanaka et al. 2009), 43 were identified from the antennal transcriptome of the codling moth, Cydia pomonella (Bengtsson et al. 2012), 47 from the antennal transcriptome of the tobacco hornworm, Manduca sexta (Grosse-Wilde et al. 2011) and 70 in the light-brown apple moth, Epiphyas postvittana, also identified from the antennal transcriptome (Corcoran 2014). From these and a range of other moths, sex pheromone receptors (PRs) have been identified, initially through evidence of male-biased expression or phylogenetic position and then through functional studies in a variety of assay systems, including HEK293 cells (Grosse-Wilde et al. 2007; Forstner et al. 2009), Xenopus oocytes (Sakurai et al. 2004; Nakagawa et al. 2005; Mitsuno et al. 2008; Miura et al. 2010; Wang et al. 2011; Wanner et al. 2010) and also in Drosophila olfactory receptor neurons (Syed et al. 2006; Kurtovic et al. 2007; Montagne et al. 2012).
The endemic New Zealand genera, Ctenopseustis (brown-headed leafroller, five species) and Planotortrix (green headed leafroller, seven species) (Tortricidae: Lepidoptera) include widespread and highly localised species (Dugdale 1990; Newcomb et al. 2014 in press). Adult wing patterning is highly variable within and between species, making many of the species within these genera morphologically cryptic (Dugdale 1990; Wearing et al. 1991). Many of the speciation events are thought to be very recent within the last million years, with some species not being able to be resolved using variation in neutral molecular markers due to incomplete lineage sorting (Langhoff et al. 2009). Within the two genera there are examples of highly polyphagus and some monophagus species on plants such as mangrove and fern. The polyphagus species C. obliquana, C. herana, P. octo and P. excessana together with the light-brown apple moth, Epiphyas postvittana, form a complex of horticultural pests in New Zealand (Wearing et al. 1991) and therefore have received more attention than the other species in Ctenopseustis and Planotortrix in terms of research.
Females of the two genera produce sex pheromone blends consisting of tetradecenyl acetates unsaturated at either of the 5, 7, 8 or 9 positions in the cis (Z) conformation (Roelofs and Brown 1982; Newcomb and Gleeson 1998; Foster et al. 1986). These double bonds are introduced through specialised fatty-acyl desaturases found in female pheromone glands (Foster and Roelofs 1988, 1996). Recently studies have been undertaken to investigate the regulation of desaturase genes in the pest species (Albre et al. 2012, 2013). Expression levels a Δ10-desaturase of C. obliquana and C. herana as well as P. octo and P. excessana are concordant with the presence (C. obliquana and P. octo) or absence (C. herana and P. excessana) of the pheromone component (Z)-8-tetradecenyl acetate (Z8-14:OAc) in the blend of respective species (Albre et al. 2012). Further crossing experiments conducted in the laboratory between P. octo and P. excessana revealed that the difference in expression of desat5, which encodes the Δ10-desaturase, is controlled by a trans-acting repressor and requires a cis-regulatory mutation in the desat5 promoter (Albre et al. 2013).
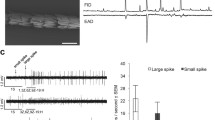
As well as sex pheromone production, the male’s ability to perceive pheromone compounds has also been investigated in the closely related species, C. obliquana and C. herana. These species have overlapping distributions, with C. obliquana found throughout New Zealand and C. herana restricted to the South Island. While C. obliquana males are attracted to a 90:10 blend of Z8-14:OAc and (Z)-5-tetradecenyl acetate (Z5-14:OAc) (Foster et al. 1986; Young et al. 1985), C. herana males are attracted to Z5-14:OAc alone (Foster and Roelofs 1987). Electrophysiological studies showed that trichoid sensilla of C. obliquana contain a large spike amplitude cell that responds strongly to the main component, Z8-14:OAc, and a small spike amplitude cell responding to the minor component, Z5-14:OAc. In C. herana the opposite is the case, with the large spike amplitude cell responding to Z5-14:OAc and a small spike amplitude cell responding to Z8-14:OAc (Hansson et al. 1989). In wind tunnel experiments, males of each species responded very selectively to pheromone blends produced by con-specific females and only a few males were attracted by blends deviating from those produced by pure-bred females (Foster et al. 1997). Field cage experiments showed that males of both species are only attracted to con-specific females (Clearwater et al. 1991) and genetic analysis provide evidence for no interbreeding in the wild between the two species (Langhoff et al. 2009; Newcomb et al. 2014 and references therein).
Here, we produce transcriptome databases constructed from the antennae of male and female C. obliquana and C. herana and mine them for olfactory receptors. In the New Zealand, native leafroller moths antennae show a strong sexual dimorphism with male antennae possessing an abundance of sensilla trichoidea type I, whereas females possess none (unpublished datain prep.). We identify candidate pheromone receptors through phylogenetic analysis and through the determination of differences in the levels of gene expression between male and female antennae. From a short list of candidate receptors, functional studies in HEK293 cells identify receptors that respond to pheromone.
Methods
Insects
Insects were reared in the Plant & Food Research insect rearing facility at the Mt Albert Research Centre, Auckland, New Zealand. The history of the laboratory strains of C. obliquana and C. herana has been reported previously (Gleeson et al. 2000). Eggs were collected and kept in a humid environment until larvae hatched. Larvae were reared separately in small glass tubes containing a general purpose diet (Clare and Singh 1988) at 18 °C. Pupae and adults were kept at 20 °C on a 16:8 light cycle. Adult moths were provided with cotton cloth soaked in water.
Nucleic Acid Isolation
RNA for transcriptome sequencing was isolated from 100 male and female antennae each dissected from 2 to 3 days old adults. RNA was extracted and purified using 800 µl Trizol (Invitrogen, Carlsbad, CA, USA) following the TRIzol Plus RNA Purification Kit protocol. DNase treatment was conducted on 10 µg of total RNA using the TURBO DNA-free Kit (Life technologies) following the manufacturer’s instructions on antennal RNA of C. herana.
RNA for quantitative RT-PCR (qPCR) experiments was isolated from male and female antennae dissected from 2 to 3 days old individuals, as well as whole bodies, using 800 µl Trizol (Invitrogen) following the manufacturer’s instructions. The initial screening for male-biased expression of odorant receptors was conducted with pools of ten antennae pairs; however, for subsequent qPCR experiments RNA extracted from single antennae pairs was used. One microgram of extracted RNA was treated with DNase (DNaseI amplification grade, Invitrogen) following the manufacturer’s instructions. cDNA was synthesised using iScript cDNA Synthesis Kit (Bio-Rad, Herts, United Kingdom) from 1 µg of total RNA, incubated at 25 °C for 5 min, 42 °C for 30 min and 85 °C for 5 min.
Next-Generation Sequencing and Bioinformatics
RNAseq libraries were constructed from both male and female adult antennae of C. obliquana and C. herana using Illumina’s standard protocols and sequenced at Macrogen (Seoul, South Korea). Quality score analysis on the read pairs for each library was undertaken using FastQC (FastQC 2008). In-house Perl scripts were used to trim all reads by 13 bases at their 5′ ends and remove any read pairs containing Ns and mononucleotides. Mitochondrial contamination was removed by mapping RNA-Seq read pairs to a reference mitochondrial genome of C. obliquana assembled from a draft genome. Mapping was performed using bowtie (version 1.0.0) (Langmead et al. 2009) with the reads mapping to the mitochondrial genome being removed. Thereafter, read pairs were trimmed to a minimum quality threshold of 20 using fastq-mcf from the ea-utils package (Aronesty 2011). Duplicates within the read files for C. obliquana were removed using in-house Perl scripts prior to assembly, while the redundancy in the C. herana sequences was removed after the assembly using cd-hit (Li and Godzik 2006). De novo assembly of the processed reads was performed for each of the individual libraries (Online Resource 1) with trans-ABySS (version 1.3.2) (Robertson et al. 2010), where a k-mer series of 31 to 75 with an increment of two bases was used for the libraries of C. obliquana, whereas k-mer values from 31 to 71 were used for the C. herana libraries.
Candidate ORs were identified using tblastn (Altschul et al. 1990) with the amino acid sequences of 70 ORs from the leafroller moth Epiphyas postvittana (Corcoran 2014) used as queries. Full length open reading frames of ORs were acquired using tblastn against the assembled contigs of male and female antennae from C. obliquana and C. herana. Where necessary, a draft genome assembly of C. obliquana was used to extend sequences (unpublished data). The post-processed RNA-Seq reads were mapped onto a constructed set of OR sequences using bowtie (version 2.1.0) (Langmead and Salzberg 2012). The resulting alignment was then used to obtain expected read counts using multiBamCov from the bedtools package (v2.16.2) (Quinlan and Hall 2010) and cufflinks (v2.1.1) (Trapnell et al. 2010) for Fragments Per Kilobase of transcript per Million reads (FPKM).
Sequence data were edited and aligned in Geneious (Kearse et al. 2012) using ClustalW with E. postvittana OR sequences. Maximum-likelihood trees were generated in Mega (Hall 2013) with a model chosen by ModelTest (Posada and Crandall 1998). The dN and dS rates were estimated using codon-based substitution models in PAML version 4.7 (Yang 2007) with the M3 model (Yang and Nielsen 2000), which has three categories of site with a free ω ratio for each site class (ω = dN/dS, the ratio of non-synonymous/synonymous substitution rates). Transmembrane domains were predicted using SPLIT 4.0 (Juretic et al. 2002) at the transmembrane prediction server (http://split4.pmfst.hr/split/4/). The topology diagram was constructed using TOPO2 Transmembrane Protein Display (Johns 2005) by the server at (http://www.sacs.ucsf.edu/TOPO2).
Quantitative Real-Time PCR
The expression levels of ORs in male and female antennae, as well as bodies, were determined by quantitative real-time PCR relative to α-tubulin, β-actin and elongation factor 1α (Turner et al. 2006). Primers were designed to the C. obliquana receptors and tested on the orthologous receptors from C. herana (Online Resource 2) and genomic DNA in both species. Reactions (10 µL) included 20 ng of cDNA, 5 µL 2 × SYBR Green Mix (Bio-Rad) and 200 nM of each forward and reverse primer. Quantitative real-time PCR cycling conditions were set up as following, 2 min at 95 °C followed by 45 cycles of 15 s at 95 °C, 30 s at 60 °C and 30 s at 72 °C. A final dissociation curve analysis was added with 15 s at 95 °C, 15 s at 60 °C and a gradual heating to 95 °C at 0.01 °C/s. Experiments were carried out with three biological replicates and three technical replicates per biological replicate, with negative controls for each replicate. Thermocycling was conducted on a LightCycler480 Real-Time instrument (Roche Diagnostics, Basel, Switzerland). To determine the amplification efficiency and the cycle threshold values for each reaction the software LinRegPCRv11 was used (Ramakers et al. 2003). The relative expression levels were calculated following a modified version of the ∆Cp method (Pfaffl 2001; Livak and Schmittgen 2001; Albre et al. 2013) as described in Corcoran et al. (2014).
Cell-Based Assays
Primers were designed to the 5′ and 3′ ends of the predicted open reading frames of ORs that showed male-biased expression in antennae or were members of the so-called pheromone receptor clade (Online Resource 3 for primer sequences). Standard PCR amplifications were carried out in 50 µL reaction volumes containing 0.5 U Platinum Taq polymerase (Invitrogen), 1 x reaction buffer, 1.25 mM magnesium chloride, 0.2 mM dNTP mix and 0.2 µM of each primer, with 2 µL of cDNA as template. For the PCR amplifications, a GeneAmp 9700 (Applied Biosystems, Carlsbad, CA, USA) PCR machine was used with an initial denaturation step of 5 min at 94 °C, followed by 35 cycles (94 °C for 10 s, 56 °C for 30 s, 72 °C for 45 s – 1.5 min) and a final elongation step at 72 °C for 7 min. PCR products were resolved on a 0.7 % TAE gel at 80 V for 60 min. Gel pieces were extracted using the QIAquick Gel Extraction Kit (QIAGEN, Venlo, Netherlands). Extracted PCR products were cloned into pCR8/GW/TOPO vectors (Invitrogen) and sequenced at Macrogen, Seoul, South Korea, using M13 forward and reverse primers. Once at least two clones of identical sequence were identifed; those plasmids were used for cloning into the expression vector. Full length clones were used as template to amplify full length sequences containing restriction sites for transformation into the pcDNA 5/TO expression vector (Invitrogen) (see Online Resource 4 for primer sequences). Again several clones were sequenced to check that acquired genes contained the correct sequence. Candidate genes were transfected into isogenic TREx HEK293 cell lines containing the E. postvittana OR co-receptor, EposOrco, which has 99.6 % amino acid identity to the Orco orthologue in both Ctenopseustis species, following the protocol described in Corcoran et al. (2014), except that no single-cell sorting was conducted.
Functional assays were carried out as described in (Corcoran et al. 2014). Briefly, 25,000 cells were plated into each well of a poly-d-lysine-coated, black-walled 96-well cell culture plate (Becton–Dickinson, Franklin Lakes, NJ, USA) and grown overnight (37 °C, 5 % CO2). The next day, the cell culture medium was replaced with fresh medium with 1 µg/ml doxycycline for induction and without doxycycline as control. Cells were grown again under the above conditions for 16–24 h before functional testing. Before the assay, the cell culture medium was removed and rinsed with assay buffer (DPBS containing 1 mM probenicid, pH 7.1). Fifty microliter of loading buffer (assay buffer containing 1 µM Fluor4-AM (Life Technologies) and 0.2 % pluronic acid) was added to each well and incubated for 30 min at room temperature in the dark. Then wells were rinsed twice with assay buffer, 100 µl of assay buffer was added followed by incubation for another 30 min at room temperature in the dark. Cell assays were conducted on an Omega FluoStar plate reader system. Baseline fluorescence was determined by exciting wells at 485 nM and reading emitted fluorescence at 535 nM. Receptors were screened with 30 µM mono-unsaturated tetradecenylacetates (in assay buffer with 0.5 % DMSO) that are used most commonly as sex pheromone components in species within the New Zealand endemic leafroller moth genera Ctenopseustis and Planotortrix (Newcomb and Gleeson 1998), including saturated tetradecenyl acetate (14:OAc), (Z)-5-tetradecenyl acetate (Z5-14:OAc), (Z)-7-tetradecenyl acetate (Z7-14:OAc) and (Z)-8-tetradecenyl acetate (Z8-14:OAc) (Pherobank, Netherlands; 95–99 % purity). As a vehicle control 0.5 % DMSO in assay buffer and as a control for Orco expression 50 µM of the insect Orco agonist VUAA1 (Jones et al. 2011) were used. After baseline determination, compounds were added to three non-induced and three induced wells. Change in fluorescence was measured for a period of 60 s after addition of the compound, vehicle control or VUAA1.
Experiments for concentration–response curves for compound-receptor combinations where positive responses were observed followed the same protocol for preparing plates and cells as for the screening experiments. In concentration–response experiments increasing concentrations from 0.014 µM up to 300 µM of compound were used for the compounds Z7-14:OAc and Z8-14:OAc in CoblOR7 and CherOR7. The mean response (±SEM) from each of the three induced and non-induced wells were used to construct concentration–response curves using the non-linear regression function of GraphPad data analysis software (GraphPad Software Inc, La Jolla, CA, USA).
Results
Four RNA-seq libraries were generated from the antennae of adult male and female C. obliquana and C. herana, generating from 39,823,878 to 71,083,823 raw sequence reads (Online Resource 5). RNA-seq data have been deposited into a Sequence Read Archive (SRA) database online (http://www.ncbi.nlm.nih.gov/) under the Accession Numbers 236626 and 236627. Resulting assemblies ranged from 102,817 to 226,501 contigs (Online Resource 1). Differences between species in the numbers of contigs may be associated with differences in DNAse treatment of RNA and/or removal of duplicates at different stages of the filtering process. Blast sets were established for the four transcriptomes and queried using the 70 ORs available from E. postvittana (Corcoran 2014). A draft genome of C. obliquana (unpublished data) was also employed to find additional sequences within ORs identified with the transcriptomes if missing from the antennal transcriptome assembles; we did not use the draft genome to explicitly mine OR genes.
Transcripts of 47 OR genes from C. obliquana and 47 from C. herana were identified in antennae and predicted protein sequences derived and the numbering of identified receptors follows largely the numbering of the orthologous E. postvittana ORs. Except for one OR from outside the sex pheromone receptor clade of each species (CoblOR66 and CherOR37), all receptors were represented by orthologous pairs (Fig. 1a). The orthologous ORs share from 89 to 99 % amino acid identity (Online Resource 6). The clade predicted to contain the sex pheromone receptors of many moth species is well supported by bootstrap analysis (93.3 % from 1000 bootstrap replicates; for detailed bootstrap values see Online Resource 7). However, orthology among the ORs of the two Ctenopseustis species and E. postvittana within this pheromone receptor clade is less clear (Fig. 1b), unlike other receptors that show male-biased expression from outside this clade in E. postvittana and the Ctenopseustis species (see later Fig. 1c).
Maximum-likelihood tree of odorant receptors from Ctenopseustis obliquana and C. herana a Circle tree of odorant receptors from C. obliquana and C. herana, together with those from Epiphyas postvittana. The tree is rooted with the odorant receptor co-receptor, Orco. The positions of OR01, OR07 and OR30 are indicated with stars. The sex pheromone receptor clade is highlighted in grey b Phylogeny of the sex pheromone receptor clade containing odorant receptor genes of E. postvittana, C. obliquana and C. herana. Odorant receptors displaying higher levels of expression in adult male compared with female antennae are highlighted. These include OR07 and OR01, showing the two versions of OR01 in C. herana. c Phylogeny of the second OR clade also containing receptors that are more highly expressed in male than female antennae from adult E. postvittana, C. obliquana and C. herana
Gene Expression of ORs
Because in moths, sex pheromone receptors are often more highly expressed in male than in female antennae (Krieger et al. 2004; Grosse-Wilde et al. 2010), we identified receptors that showed this pattern of sexual dimorphic expression between the sexes in antennae. Using RNA-seq counting with a two-fold cut-off criteria, six OR genes were identified as having male-biased expression in both species. Two ORs were identified as male biased in both species, OR7 and OR30, whereas in C. herana two similar genes (CherOR1a and CherOR1b; 86.9 % identical at the amino acid level), also showed male-biased expression. CherOR1b is homologous to CoblOR1 in C. obliquana, however, no related gene was found for OR01a in C. obliquana. Of the ORs that show male-biased expression, OR01 and OR07 are located in the sex pheromone receptor clade, whereas OR30 is located outside this clade in the phylogeny (Fig. 1a). RNA-seq count also revealed ORs that are more highly expressed in female than in male antennae (Online Resource 8 and 9). A two-fold cut-off on RNA-seq count revealed six female-biased ORs in C. obliquana (CoblOR14, CoblOR22, CoblOR25, CoblOR49, CoblOR58 and CoblOR63) and nine in C. herana (CherOR12, CherOR22, CherOR25, CherOR45, CherOR49, CherOR52, CherOR58, CherOR63 and CherOR64).
Thirty two genes in C. obliquana and 21 in C. herana were assessed for their levels of gene expression using qPCR. These included the three male-biased receptors by RNA-seq count and another four members from the pheromone receptor clade of C. obliquana and four male-biased receptors and the remaining four in the pheromone receptor clade of C. herana. Of those tested, three OR genes were significantly male biased in their expression in the antennae of both species (Online Resource 10 and 11). As found with RNA-seq count, OR7 and OR30 were significantly male biased (CoblOR7 P = 0.044; CherOR7 P = 0.014; CoblOR30 P = 0.002; CherOR30 P = 0.005), as was OR1 (CoblOR1 P = 0.004; CherOR1 P = 0.005) (Fig. 2). However, it should be noted that the primers designed for the qPCR screening for the two versions of OR1 in C. herana were located in regions were the paralogous genes are very similar and therefore, the results for the two OR1 genes from C. herana may be confounded. Attempts to design primers in regions that are distinct in the two genes failed to result in primers useful for qPCR. In C. obliquana where there is only one version of OR1, the transcript could be detected as male biased by qPCR. Quantitative PCR failed to confirm female-biased expression of tested receptors from C. obliquana (CoblOR14 P = 0.119, CoblOR22 P = 0.098, CoblOR25 P = 0.263, CoblOR49 P = 0.140) and C. herana (CherOR12 P = 0.315, CherOR22 P = 0.110, CherOR25 P = 0.565). Overall, of the 13 receptors that were identified by RNA-seq as potentially male or female biased and successfully tested in qPCR experiments, six were confirmed to show sex-biased gene expression. All of the male-biased ORs were expressed at significantly higher levels when tested in qPCR and potentially female-biased ORs showed a trend in this direction, although no receptor was statistically significant. This suggests that use of RNA-seq is a reasonable method to identify candidate OR genes for further testing.
Relative expression of the odorant receptors OR01, OR07 and OR30 in the male (blue) and female (red) antennae of adult a Ctenopseustis obliquana and b C. herana. CT values are the mean ± SE normalised to the housekeeping genes α-tubulin, β-actin and elongation factor 1α (BDL = below limits of detection)
Cell Assays
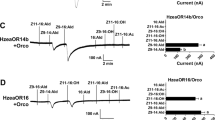
The full length cDNA sequences of three receptors from C. obliquana (OR1, OR7 and OR30) and their orthologues from C. herana, plus one additional OR (CherOR1a) were recovered by PCR from antennal cDNA. These seven OR genes were cloned into expression vectors, confirmed by sequencing and tested in cell assays against four mono-unsaturated tetradecenyl acetates. Of the receptors tested only OR7 from C. obliquana and C. herana responded to any of the compounds tested. While CoblOR7 responded to Z8-14:OAc (EC50 = 651 ± 33 nM) alone (Fig. 3a, b), CherOR7 responded to Z7-14:OAc (EC50 = 2012 ± 32 nM) and Z8-14:OAc (EC50 = 4022 ± 30 nM) (Fig. 3c, d). The remaining receptors showed no response to any of the tested components, with no receptor found that responded to Z5-14:OAc. It cannot be ruled out that these receptors (CoblOR1, CoblOR30, CherOR1a, CherOR1b, CherOR30) are capable of responding to pheromone components but are just not properly expressed or trafficked to the membrane in HEK293 cells.
Concentration-response curves of OR07 from Ctenopseustis obliquana and C. herana induced (red) and un-induced (blue) 10 s after injection of the test compound a CoblOR07 response to (Z)-7-tetradecenyl acetate b CoblOR07 response to (Z)-8-tetradecenyl acetate c CherOR07 response to (Z)-7-tetradecenyl acetate d. CherOR07 response to (Z)-8-tetradecenyl acetate
Sequence Comparison of CoblOR7 and CherOR7
Sequence comparisons among orthologues of OR7 from C. obliquana and C. herana, together with two species from the sister genus Planotortrix (P. octo and P. excessana; data not shown) revealed a dN/dS ratio of ω = 0.963 for C. obliquana (29.1 non-synonymous and 10.9 synonymous changes) and ω = 0.1311 for C. herana (1.8 non-synonymous and 5.1 synonymous changes) (Fig. 4a). A likelihood ratio test (LRT) of M0 (with one fixed ω ratio) versus M3 (with three categories of site with a free ω ratio for each site) was significant (Table 1). The M3 model is a better fit to the data than M0, indicating variability of the ω ratio at sites across the coding sequence of OR7. Potentially selected sites identified by M3 included amino acid positions 85, 276 and 300 (Table 2). The more stringent comparisons of M7 (“beta” neutral model) with M8 (“beta plus ω), and M8 with M8a (M8 with a fixed ω at 1), which are typically used as indicators of positive selection, were not significant.
a Maximum-likelihood tree of OR7 orthologues from Ctenopseustis obliquana (CoblOR7), C. herana (CherOR7), Planotortrix octo (PoctOR7) and P. excessana (PexcOR7). dN, dS and dN/dS values were generated with the M3 model b Predicted transmembrane topology of OR7 with variable sites highlighted. Amino acid substitutions in C. obliquana compared to a predicted common ancestor are in black, whereas amino acid substitutions in C. herana are in grey. Sites predicted to be under selection by M3 are indicated with an arrow. The double line indicates the membrane region, with extracellular and cytoplasmic sides labelled
To identify potential substitutions encoding the selectivity differences between CoblOR7 and CherOR7, as well as to investigate evidence for relaxed constraint, we compared the sequence of the two receptors. CoblOR7 and CherOR7 differ by 45 nucleotide substitutions, comprising 13 synonymous and 29 non-synonymous substitutions. The 29 non-synonymous substitutions are distributed across the predicted regions of the OR (Fig. 4b). A test for equal distribution of identified non-synonymous substitutions in the 15 regions (N terminal region, the seven transmembrane regions, the three intracellular loops, the three extracellular loops, and the C terminal region) was rejected (X 2 = 24.34, P = 0.042). There was a higher than expected proportion of amino acid substitutions in the third and sixth transmembrane regions (TM3 P = 0.02, TM6 P = 0.005). However, the contingency table included a number of empty cells violating an assumption of the X 2 test. Therefore, the non-parametric Fisher’s exact test was also conducted using 1,000,000 simulations to estimate a P value (P = 0.033), which was also significant. None of the selected sites identified in the M3 model fall in either TM3 or TM6.
Discussion
Here, we identify 47 odorant receptors expressed in adult antennae of each of the two closely related New Zealand endemic leaftroller moths, Ctenopseustis obliquana and C. herana using a transcriptome approach. Considering the number of OR genes found in the transcriptome of other species, including 43 in C. pomonella, 47 in M. sexta and 70 in E. postvittana (Bengtsson et al. 2012; Grosse-Wilde et al. 2011; Corcoran 2014), the number of ORs identified in the two Ctenopseustis species could be regarded as representative of the majority of the receptors from each of these species. Additionally to the 47 ORs identified and expressed in the antennae seven orthologous, ORs of E. postvittana were found in the genome but could not be detected in the transcripts of C. obliquana and C. herana (EposOR8, EposOR13, EposOR17, EposOR21, EposOR23, EposOR40 and EposOR55). Unfortunately, no information regarding the number of glomerili in these species is available. In addition to the mining OR genes, the transcriptomes described here will provide a useful resource for isolating and comparing other genes involved in chemosensory perception in these species, including odorant-binding proteins and odour-degrading enzymes.
Outside the pheromone receptor clade all OR genes described in Ctenopseustis are orthologous to each other and an E. postvittana OR. These include orthologues of Orco and the citral receptor EposOR3 that have been described previously (Jordan et al. 2009; Carraher et al. 2012). However, within the pheromone receptor clade this level of orthology breaks down with orthologues identified between C. obliquana and C. herana, but typically no counterpart in E. postvittana. The male-biased ORs, CoblOR1 and CoblOR7 in C. obliquana, as well as CherOR1a, CherOR1b and CherOR7 in C. herana, are found within this clade where pheromone receptors from several lepidopteran species also reside (Sakurai et al. 2004; Nakagawa et al. 2005; Grosse-Wilde et al. 2007; Wang et al. 2011). CoblOR7 and CherOR7 were both found to bind the pheromone component Z8-14:OAc, whereas neither of the other male-biased receptors in the pheromone receptor clade responded to any of the pheromone components presented in the HEK293 cell system. It would be interesting to test the receptors in other systems like Xenopus oocytes (Mitsuno et al. 2008; Nakagawa et al. 2005; Sakurai et al. 2004; Miura et al. 2010; Wang et al. 2010; Wanner et al. 2010) to verify our results. Furthermore, the presence of pheromone-binding proteins could have an effect on the ligand specificity of ORs (Grosse-Wilde et al. 2006). Although not strictly orthologous, EposOR7 was not found to bind any of E. postvittana’s pheromone components or any other compounds tested (Corcoran 2014). Similar to EposOR7, no direct orthologous OR could be found for EposOR1 in either of the two Ctenopseustis species. This OR from E. postvittana has been shown to bind a range of terpenoids and benzoates responding best to methyl salicylate as a ligand (Jordan et al. 2009) and shows a higher rate of molecular evolution than ORs outside the sex pheromone receptor clade (Carraher et al. 2012). More recently, EposOR1 has been shown to be capable of responding to the minor pheromone component for E. postvittana, (E,E)-9,11-tetradecenyl acetate (Corcoran 2014). The non sex-biased ORs, CoblOR6 and CherOR6, together with CherOR45 and CoblOR45a and CoblOR45b are the most closely related ORs to EposOR1. It would be interesting to understand whether these receptors have a similar function to EposOR1, but unfortunately to date efforts in amplify full length versions of the genes encoding CoblOR6, CherOR6, CherOR45 and both versions of CoblOR45 have been unsuccessful. The receptors binding Z5-14:OAc in C. obliquana or C. herana have not yet been identified. The remaining receptors that fall into the sex pheromone receptor clade are possible candidates for being able to respond to this compound, even if these receptors could not be detected as male biased in RNA-seq count or qPCR. Similarly, receptors from E. postvittana that reside in this clade, but do not show male-biased expression, have also been shown to respond to pheromone components (Corcoran 2014). OR30 shows higher levels of expression in male compared with female antennae, but falls outside the pheromone receptor clade. In the related species, E. postvittana EposOR30 and EposOR34, like CoblOR30 and CherOR30, also display male-biased expression and do not seem to respond to pheromone components (Corcoran 2014). Further efforts are required to identify ligands for these male-biased receptors that reside outside the pheromone receptor clade, and their orthologues. Apart from OR30, no orthologues of other male-biased receptors in E. postvittana (EposOR6, EposOR07, EposOR34) could be found in either of the two Ctenopseustis species.
Of the OR genes that tend towards being female biased in their expression, OR25, OR49, OR58 and OR63 show similar expression differences in both Ctenopseustis species, suggestive of a similar role in both species. Notwithstanding this, we should mention that none of these genes remained female biased in our qPCR analysis. While the female-biased ORs in E. postvittana are largely located in a distinct clade separate from the pheromone receptor clade (EposOR31, EposOR36 and EposOR40), in C. obliquana and C. herana, they are dispersed throughout the phylogeny. No ligands have been identified for any of the female-biased ORs in E. postvittana and none are closely related to other female-biased receptors from B. mori, which respond to linalool and benzoic acid (Anderson et al. 2009). Further research is required to identify the ligands for these female-biased receptors in Ctenopseustis and Epiphyas species, which may be tuned to volatile compounds produced by host plants or male-produced pheromones. If these receptors are involved in host finding then with the presence of both specialists and generalists in these genera, this system may be useful in understanding the evolution of host specificity (Dugdale 1990). Furthermore, it has been shown in other species that males can also produce pheromones that are used in proximity to the female and are an important factor during courtship for mate acceptance (Davie et al. 2010; Hillier and Vickers 2004, 2011). Males in the sister genus, Planotortrix, have been observed to present hair-pencil-like structures at the tip of the abdomen to the female during courtship (unpublished data). Such structures typically produce male pheromones in moths.
Electrophysiological investigations of male antennae of C. obliquana and C. herana have revealed that both species can perceive the sex pheromone components, Z5-14:OAc and Z8-14:OAc, even though Z8-14:OAc is not required to attract C. herana males behaviourally (Hansson et al. 1989). However, the neurophysiological response is different in each species, with C. obliquana possessing a neuron with a large spike amplitude that responds strongly to the major component Z8-14:OAc and another neuron with a small spike amplitude responding weakly to the minor component Z5-14:OAc. Neurons in C. herana show an opposite response to these compounds (Hansson et al. 1989). As such identifying a receptor in C. herana, as well as C. obliquana, that responds to Z8-14:OAc (namely OR7) is consistent with these electrophysiological results. Presumably, once the receptor responding to Z5-14:OAc is identified it too will be found to be present and expressed in the antennae of males of both species. Therefore, the genetic differences explaining the changes in either neuron size or the expression of the receptor in either the large or small neuron are likely to be encoded at a locus/loci distinct to the pheromone receptors.
Interestingly, the pharmacology of the two OR7 orthologues is different. CoblOR7 is an order of magnitude more sensitive to the C. obliquana sex pheromone component Z8-14:OAc compared with CherOR7. Furthermore, of the compounds tested, CoblOR7 responds only to Z8-14:OAc, while in addition to this compound, CherOR7 also responds to Z7-14:OAc. This compound is not a sex pheromone component used by either of these two species, but is used as a pheromone component in the ancestral species with the genus, C. servana (Foster and Dugdale 1988) and species within the sister genus Planotortrix (Foster et al. 1986; Foster and Dugdale 1988). Considering that C. herana uses Z5-14:OAc solely as it’s sex pheromone, it would suggest that CherOR7 is under relaxed constraint compared with its orthologue CoblOR7. That is, mutations in CherOR7 that alter sensitivity and selectivity in this receptor are less likely to have any impact on the ability of males to locate female C. herana. In the closely related species of corn borer, the European and the Asian corn borer, a single mutation in the sequence of an OR gene changes the specificity of that receptor to the compound it is binding. This mutation is found in the third transmembrane region (TM3) in position 148 where a threonine to alanine substitution is responsible for an alteration of sensitivity to the pheromone component E11-14:OAc (Leary et al. 2012). Interestingly, in CoblOR7 there are also amino acid differences to CherOR7 in TM3 at positions 147 (alanine to glycine), 148 (serine to leucine) and 149 (phenylalanine to cysteine). Another highly variable region is located in TM6 where five amino acid differences in positions 322, 323, 324, 327 and 328 are located. Although comparison of M7/M8 and M8/M8a did not provide evidence of positive selection in OR7, the M0/M3 comparison revealed substantial variability among amino acid sites especially at positions 85 (TM2), 276 (L4) and 300 (TM5) (Table 2). However, we note that these likelihood-based methods can produce false positives (Suzuki and Nei 2001, 2002) and that the posterior probabilities at these predicted sites under selection, while over 0.75 did not reach 0.95. Together the amino acid substitutes from these two analyses become candidates responsible for the changes in specificity and selectivity of CherOR7, except for position 276 where CherOR7 and CoblOR7 do not differ.
The pheromone component Z8-14:OAc is produced in the pheromone gland by a Δ10-desaturase (desat5) from palmitic acid and is followed by a round of β-oxidation before reduction and acetylation. The basal species within the genus Ctenopseustis, C. servana, does not use pheromone components that contain double bonds in an even position. This would mean that in the evolution of the genus, presumably there must have been a gain of expression of desat5 in their pheromone glands after the split from C. servana (Newcomb and Gleeson 1998; Albre et al. 2012). C. servana uses a blend containing Z5-14:OAc and Z7-14:OAc and it has been discussed that in the evolution of pheromone production, C. obliquana gained the use of Z8-14:OAc as a pheromone component (Albre et al. 2012). It has been hypothesised that South Island populations of a C. obliquana ancestor lost the expression of desat5 in the pheromone gland that gave rise to C. herana (Albre et al. 2012). Our results suggest a different model, the high substitution rate in the lineage leading to the Z8-14:OAc receptor, CoblOR7, and low substitution rate in the unspecific Z7/Z8-14:OAc orthologue, CherOR7, could suggest that C. obliquana and C. herana ancestors could have split from C. servana creating a scenario where positive selection maintained mutations that increased the specificity and sensitivity towards Z8-14:OAc in C. obliquana ancestors and a lack of selection in C. herana slowly decreased the specificity of OR7 to Z7-14:OAc. Positive selection has previously been found to act on odorant receptor orthologues in Drosophila (Tunstall et al. 2007). It has been suggested that positive selection may act on some Drosophila ORs, especially on amino acid sites that could be responsible for altering the sensitivity of receptors towards odorants (Tunstall et al. 2007).
Changes in the sensitivity and selectivity of pheromone receptors during periods of relaxed constraint might allow a currently ‘unused’ receptor to evolve to be able to perceive a future novel component produced by a variant female. Such changes may contribute to the formation of a new species, perhaps by producing ‘rare males’ that perceive and therefore are able to ‘track’ divergent female pheromones as suggested in the “asymmetric tracking hypothesis”(Phelan 1992). In Ostrinia nubilalis, it has been shown that a few ‘rare males’ are capable of responding to the pheromone blend of O. furnacalis (Domingue et al. 2007). Differences in the neurophysiological response were responsible for these rare males flying upwind to the pheromone blend of the closely related species, revealing that males can perceive a larger range of compounds, but only a few of these trigger a behavioural response depending on how the male’s olfactory receptor neurons are tuned (Domingue et al. 2007). In C. obliquana and C. herana, it also has been shown that males in both species can perceive the compound Z8-14:OAc, even though it is only used in C. obliquana as a pheromone component, but the neurophysiological response is different (Hansson et al. 1989). Ostrinia scapulalis males possess pheromone receptors that are very specific and some that are broadly tuned also to pheromones of congeners (Miura et al. 2010). A more broadly tuned receptor as found in C. herana could provide the ability to produce males with a preference for more pheromone components when such broadly tuned receptor genes are expressed in large spike amplitude neurons or perhaps to be able to identify females moths of closely related species and respond negatively towards them. In the wasp Nasonia, it has been shown that pheromone components can exist in a population without being initially perceived by the responder and a preference for this component can evolve to an additional pheromone component leading to the evolution of a new species (Niehuis et al. 2013). Finally, a further possibility is that the receptor remains unused and becomes a pseudogene, losing its ability to produce a functional receptor as has been found in receptor genes in Drosophila (Guo and Kim 2007).
References
Albre J, Lienard MA, Sirey TM, Schmidt S, Tooman LK, Carraher C, Greenwood DR, Lofstedt C, Newcomb RD (2012) Sex pheromone evolution is associated with differential regulation of the same desaturase gene in two genera of leafroller moths. PLoS Genet 8(1):e1002489
Albre J, Steinwender B, Newcomb RD (2013) The evolution of desaturase gene regulation involved in sex pheromone production in leafroller moths of the genus Planotortrix. J Hered 104(5):627–638
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215(3):403–410
Anderson AR, Wanner KW, Trowell SC, Warr CG, Jaquin-Joly E, Zagatti P, Robertson H, Newcomb RD (2009) Molecular basis of female-specific odorant responses in Bombyx mori. Insect Biochem Mol Biol 39(3):189–197
Aronesty E (2011) ea-utils: Command-line tools for processing biological sequencing data; Expression Analysis. http://code.google.com/p/ea-utils/wiki/FastqMcf. Accessed August 2013
Bengtsson JM, Trona F, Montagne N, Anfora G, Ignell R, Witzgall P, Jacquin-Joly E (2012) Putative chemosensory receptors of the codling moth, Cydia pomonella, identified by antennal transcriptome analysis. PLoS ONE 7(2):e31620
Benton R, Sachse S, Michnick SW, Vosshall LB (2006) Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol 4(2):240–257
Bradbury JW, Vehrencamp SL (1998) Principles of animal communication. Sinauer Associates Inc, Sunderland
Carraher C, Authier A, Steinwender B, Newcomb RD (2012) Sequence comparisons of odorant receptors among tortricid moths reveal different rates of molecular evolution among family members. PLoS ONE 7(6):e38391
Clare GK, Singh P (1988) Laboratory rearing of Ctenopseustis obliquana (Walker) (Lepidoptera: Tortricidae) on an artificial diet. N Z J Zool 15:435–438
Clearwater JR, Foster SP, Muggleston SJ, Dugdale JS, Priesner E (1991) Intraspecific variation and interspecific differences in sex-pheromones of sibling species in Ctenopseustis obliquana complex. J Chem Ecol 17(2):413–429
Corcoran JA (2014) The identification of pheromone receptors from the lightbrown apple moth, Epiphyas postvittana. Unpublished PhD thesis, The University of Auckland, New Zealand
Corcoran JA, Jordan MD, Carraher C, Newcomb RD (2014) A novel method to study insect olfactory receptor function using HEK293 cells. Insect Biochem Mol Biol (in press)
Davie LC, Jones TM, Elgar MA (2010) The role of chemical communication in sexual selection: hair-pencil displays in the diamondback moth Plutella xylostella. Anim Behav 79(2):391–399
Domingue MJ, Musto CJ, Linn CE, Roelofs WL, Baker TC (2007) Altered olfactory receptor neuron responsiveness in rare Ostrinia nubilalis males attracted to the O. furnacalis pheromone blend. J Insect Physiol 53(10):1063–1071
Dugdale JS (1990) Reassessment of Ctenopseustis Meyrick and Planotortrix Dugdale with descriptions of two new genera (Lepidoptera: Tortricidae). N Z J Zool 17(3):437–465
FastQC (2008). http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc/. Accessed August 2013
Forstner M, Breer H, Krieger J (2009) A receptor and binding protein interplay in the detection of a distinct pheromone component in the silkmoth Antheraea polyphemus. Int J Biol Sci 5(7):745–757
Foster SP, Dugdale JS (1988) A comparison of morphological and sex-pheromone differences in some New Zealand Tortricinae moths. Biochem Syst Ecol 16(2):227–232
Foster SP, Roelofs WL (1987) Sex-pheromone differences in populations of the brown-headed leafroller, Ctenopseustis obliquana (Lepidoptera, Tortricidae, Tortricinae). J Chem Ecol 13(3):623–629
Foster SP, Roelofs WL (1988) Sex-pheromone biosynthesis in the leafroller moth Planotortrix excessana by delta 10 desaturation. Arch Insect Biochem Physiol 8(1):1–9
Foster SP, Roelofs WL (1996) Sex pheromone biosynthesis in the tortricid moth, Ctenopseustis herana (Felder & Rogenhofer). Arch Insect Biochem Physiol 33(2):135–147
Foster SP, Clearwater JR, Muggleston SJ, Dugdale JS, Roelofs WL (1986) Probable sibling species complexes within two described NZ leafroller moths. Naturwissenschaften 73(3):156–158
Foster SP, Muggleston SJ, Lofstedt C, Hansson B (1997) A genetic study on pheromonal communication in two Ctenopseustis moths. In: Carde RT, Minks AK (eds) Insect pheromone research: new directions. Chapman & Hall, New York, pp 514–524
Fujii T, Ito K, Tatematsu M, Shimada T, Katsuma S, Ishikawa Y (2011) Sex pheromone desaturase functioning in a primitive Ostrinia moth is cryptically conserved in congeners’ genomes. Proc Natl Acad Sci U S A 108(17):7102–7106
Gleeson D, Holder P, Newcomb R, Howitt R, Dugdale J (2000) Molecular phylogenetics of leafrollers: application to DNA diagnostics. N Z Plant Prot 53:157–162
Gould F, Estock M, Hillier NK, Powell B, Groot AT, Ward CM, Emerson JL, Schal C, Vickers NJ (2010) Sexual isolation of male moths explained by a single pheromone response QTL containing four receptor genes. Proc Natl Acad Sci U S A 107(19):8660–8665
Greenberg AJ, Moran JR, Coyne JA, Wu CI (2003) Ecological adaptation during incipient speciation revealed by precise gene replacement. Science 302(5651):1754–1757
Grosse-Wilde E, Svatos A, Krieger J (2006) A pheromone-binding protein mediates the bombykol-induced activation of a pheromone receptor in vitro. Chem Senses 31(6):547–555
Grosse-Wilde E, Gohl T, Bouche E, Breer H, Krieger J (2007) Candidate pheromone receptors provide the basis for the response of distinct antennal neurons to pheromonal compounds. Eur J Neurosci 25(8):2364–2373
Grosse-Wilde E, Kuebler LS, Bucks S, Vogel H, Wicher D, Hansson BS (2011) Antennal transcriptome of Manduca sexta. Proc Natl Acad Sci U S A 108(18):7449–7454
Grosse-Wilde E, Stieber R, Forstner M, Krieger J, Wicher D, Hansson BS (2010) Sex-specific odorant receptors of the tobacco hornworm Manduca sexta. Front Cell Neurosci 4:22
Guo S, Kim J (2007) Molecular evolution of Drosophila odorant receptor genes. Mol Biol Evol 24(5):1198–1207
Hall BG (2013) Building phylogenetic trees from molecular data with MEGA. Mol Biol Evol 30(5):1229–1235
Hansson BS, Lofstedt C, Foster SP (1989) Z-linked inheritance of male olfactory response to sex-pheromone components in 2 species of tortricid moths, Ctenopseustis obliquana and Ctenopseustis sp. Entomol Exp Appl 53(2):137–145
Heinbockel T, Kaissling K-E (1996) Variability of olfactory receptor neuron responses of female silkmoths (Bombyx mori L.) to benzoic acid and (±)-linalool. J Insect Physiol 42(6):565–578
Hillier NK, Vickers NJ (2004) The role of heliothine hairpencil compounds in female Heliothis virescens (Lepidoptera: Noctuidae) behavior and mate acceptance. Chem Senses 29(6):499–511
Hillier NK, Vickers NJ (2011) Hairpencil volatiles influence interspecific courtship and mating between two related moth species. J Chem Ecol 37(10):1127–1136
Johns SJ (2005) Transmembrane protein display software. http://www.sacs.ucsf.edu/TOPO2. Accessed January 6 2014
Jones PL, Pask GM, Rinker DC, Zwiebel LJ (2011) Functional agonism of insect odorant receptor ion channels. Proc Natl Acad Sci U S A 108(21):8821–8825
Jordan MD, Stanley D, Marshall SDG, de Silva D, Crowhurst RN, Gleave AP, Greenwood DR, Newcomb RD (2008) Expressed sequence tags and proteomics of antennae from the tortricid moth Epiphyas postvittana. Insect Mol Biol 17(4):361–373
Jordan MD, Anderson A, Begum D, Carraher C, Authier A, Marshall SDG, Kiely A, Gatehouse LN, Greenwood DR, Christie DL, Kralicek AV, Trowell SC, Newcomb RD (2009) Odorant Receptors from the light brown apple moth (Epiphyas postvittana) recognize important volatile compounds produced by plants. Chem Senses 34(5):383–394
Juretic D, Zoranic L, Zucic D (2002) Basic charge clusters and predictions of membrane protein topology. J Chem Inf Comput Sci 42(3):620–632
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A (2012) Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28(12):1647–1649
Knipple DC, Rosenfield CL, Nielsen R, You KM, Jeong SE (2002) Evolution of the integral membrane desaturase gene family in moths and flies. Genetics 162(4):1737–1752
Krieger J, Raming K, Dewer YM, Bette S, Conzelmann S, Breer H (2002) A divergent gene family encoding candidate olfactory receptors of the moth Heliothis virescens. Eur J Neurosci 16(4):619–628
Krieger J, Grosse-Wilde E, Gohl T, Dewer YME, Raming K, Breer H (2004) Genes encoding candidate pheromone receptors in a moth (Heliothis virescens). Proc Natl Acad Sci U S A 101(32):11845–11850
Kurtovic A, Widmer A, Dickson BJ (2007) A single class of olfactory neurons mediates behavioural responses to a Drosophila sex pheromone. Nature 446(7135):542–546
Langhoff P, Authier A, Buckley TR, Dugdale JS, Rodrigo A, Newcomb RD (2009) DNA barcoding of the endemic New Zealand leafroller moth genera Ctenopseustis and Planotortrix. Mol Ecol Res 9(3):691–698
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9(4):357–359
Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10(3):R25
Lassance JM, Lofstedt C (2009) Concerted evolution of male and female display traits in the European corn borer Ostrinia nubilalis. BMC Biol 7:10
Lassance JM, Groot AT, Lienard MA, Antony B, Borgwardt C, Andersson F, Hedenstrom E, Heckel DG, Lofstedt C (2010) Allelic variation in a fatty-acyl reductase gene causes divergence in moth sex pheromones. Nature 466(7305):486–487
Leary GP, Allen JE, Bunger PL, Luginbill JB, Linn CE Jr, Macallister IE, Kavanaugh MP, Wanner KW (2012) Single mutation to a sex pheromone receptor provides adaptive specificity between closely related moth species. Proc Natl Acad Sci U S A 109(35):14081–14086
Li W, Godzik A (2006) Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22(13):1658–1659
Lienard MA, Strandh M, Hedenstrom E, Johansson T, Lofstedt C (2008) Key biosynthetic gene subfamily recruited for pheromone production prior to the extensive radiation of Lepidoptera. BMC Evol Biol 8:270
Linn CE, Jr., Roelofs WL (1995) Pheromone communication in moths and its role in the speciation process. In: Lambert DM, Spencer HG (eds) Speciation and the recognition concept: theory and application. John Hopkins University Press, Baltimore and London, pp 263–300
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25(4):402–408
Lundin C, Kall L, Kreher SA, Kapp K, Sonnhammer EL, Carlson JR, Heijne G, Nilsson I (2007) Membrane topology of the Drosophila OR83b odorant receptor. FEBS Lett 581(29):5601–5604
Mitsuno H, Sakurai T, Murai M, Yasuda T, Kugimiya S, Ozawa R, Toyohara H, Takabayashi J, Miyoshi H, Nishioka T (2008) Identification of receptors of main sex-pheromone components of three Lepidopteran species. Eur J Neurosci 28(5):893–902
Miura N, Nakagawa T, Touhara K, Ishikawa Y (2010) Broadly and narrowly tuned odorant receptors are involved in female sex pheromone reception in Ostrinia moths. Insect Biochem Mol Biol 40(1):64–73
Montagne N, Chertemps T, Brigaud I, Francois A, Francois MC, de Fouchier A, Lucas P, Larsson MC, Jacquin-Joly E (2012) Functional characterization of a sex pheromone receptor in the pest moth Spodoptera littoralis by heterologous expression in Drosophila. Eur J Neurosci 36(5):2588–2596
Nakagawa T, Sakurai T, Nishioka T, Touhara K (2005) Insect sex-pheromone signals mediated by specific combinations of olfactory receptors. Science 307(5715):1638–1642
Newcomb RD, Gleeson DM (1998) Pheromone evolution within the genera Ctenopseustis and Planotortrix (Lepidoptera : Tortricidae) inferred from a phylogeny based on cytochrome oxidase I gene variation. Biochem Syst Ecol 26(5):473–484
Newcomb RD, Steinwender B, Albre J, Foster SP (2014) The endemic New Zealand genera Ctenopseustis and Planotortrix: A down-under story of leafroller moth sex pheromone evolution and speciation. In: Allison JD, Cardé RT (eds) Pheromone Communication in Moths: Evolution, Behavior and Application. University of California Press, Los Angeles (in press)
Niehuis O, Buellesbach J, Gibson JD, Pothmann D, Hanner C, Mutti NS, Judson AK, Gadau J, Ruther J, Schmitt T (2013) Behavioural and genetic analyses of Nasonia shed light on the evolution of sex pheromones. Nature 494(7437):345–348
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29(9):e45
Phelan PL (1992) Evolution of sex pheromones and the role of asymmetric tracking. In: Roitberg BD, Isman MB (eds) Insect chemical ecology: an evolutionary approach. Chapman & Hall, New York, pp 265–314
Posada D, Crandall KA (1998) Modeltest: testing the model of DNA substitution. Bioinformatics 14(9):817–818
Quinlan AR, Hall IM (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26(6):841–842
Ramakers C, Ruijter JM, Deprez RH, Moorman AF (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 339(1):62–66
Robertson G, Schein J, Chiu R, Corbett R, Field M, Jackman SD, Mungall K, Lee S, Okada HM, Qian JQ, Griffith M, Raymond A, Thiessen N, Cezard T, Butterfield YS, Newsome R, Chan SK, She R, Varhol R, Kamoh B, Prabhu AL, Tam A, Zhao Y, Moore RA, Hirst M, Marra MA, Jones SJ, Hoodless PA, Birol I (2010) De novo assembly and analysis of RNA-seq data. Nat Methods 7(11):909–912
Roelofs WL, Brown RL (1982) Pheromones and evolutionary relationships of Tortricidae. Ann Rev Ecol Evol Syst 13:395–422
Roelofs WL, Rooney AP (2003) Molecular genetics and evolution of pheromone biosynthesis in Lepidoptera. Proc Natl Acad Sci U S A 100(Suppl 2):14599
Roelofs WL, Liu WT, Hao GX, Jiao HM, Rooney AP, Linn CE (2002) Evolution of moth sex pheromones via ancestral genes. Proc Natl Acad Sci U S A 99(21):13621–13626
Rumbo ER (1981) Study of single sensillum responses to pheromone in the light brown apple moth, Epiphyas postvittana, using an averaging technique. Phys Entomol 6(1):87–98
Rumbo ER (1983) Differences between single cell responses to different components of the sex-pheromone in males of the light brown apple moth (Epiphyas postvittana). Phys Entomol 8(2):195–201
Sakai R, Fukuzawa M, Nakano R, Tatsuki S, Ishikawa Y (2009) Alternative suppression of transcription from two desaturase genes is the key for species specific sex pheromone biosynthesis in two Ostrinia moths. Insect Biochem Mol Biol 39(1):62–67
Sakurai T, Nakagawa T, Mitsuno H, Mori H, Endo Y, Tanoue S, Yasukochi Y, Touhara K, Nishioka T (2004) Identification and functional characterization of a sex pheromone receptor in the silkmoth Bombyx mori. Proc Natl Acad Sci U S A 101(47):16653–16658
Shirangi TR, Dufour HD, Williams TM, Carroll SB (2009) Rapid evolution of sex pheromone producing enzyme expression in Drosophila. PLoS Biol 7(8):e1000168
Smadja C, Butlin RK (2009) On the scent of speciation: the chemosensory system and its role in premating isolation. Heredity 102(1):77–97
Smart R, Kiely A, Beale M, Vargas E, Carraher C, Kralicek AV, Christie DL, Chen C, Newcomb RD, Warr CG (2008) Drosophila odorant receptors are novel seven transmembrane domain proteins that can signal independently of heterotrimeric G proteins. Insect Biochem Mol Biol 38(8):770–780
Suzuki Y, Nei M (2001) Reliabilities of parsimony-based and likelihood-based methods for detecting positive selection at single amino acid sites. Mol Biol Evol 18:2179–2185
Suzuki Y, Nei M (2002) Simulation study of the reliability and robustness of the statistical methods for detecting positive selection at single amino acid sites. Mol Biol Evol 19:1865–1869
Syed Z, Ishida Y, Taylor K, Kimbrell DA, Leal WS (2006) Pheromone reception in fruit flies expressing a moth’s odorant receptor. Proc Natl Acad Sci U S A 103(44):16538–16543
Symonds MRE, Elgar MA (2008) The evolution of pheromone diversity. Trends Ecol Evol 23(4):220–228
Tanaka K, Uda Y, Ono Y, Nakagawa T, Suwa M, Yamaoka R, Touhara K (2009) Highly selective tuning of a silkworm olfactory receptor to a key mulberry leaf volatile. Curr Biol 19:881–890
Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28(5):511–515
Tunstall NE, Sirey T, Newcomb RD, Warr CG (2007) Selective pressures on Drosophila chemosensory receptor genes. J Mol Evol 64(6):628–636
Turner CT, Davy MW, MacDiarmid RM, Plummer KM, Birch NP, Newcomb RD (2006) RNA interference in the light brown apple moth, Epiphyas postvittana (Walker) induced by double-stranded RNA feeding. Insect Mol Biol 15(3):383–391
Wang G, Carey AF, Carlson JR, Zwiebel LJ (2010) Molecular basis of odor coding in the malaria vector mosquito Anopheles gambiae. Proc Natl Acad Sci U S A 107(9):4418–4423
Wang G, Vasquez GM, Schal C, Zwiebel LJ, Gould F (2011) Functional characterization of pheromone receptors in the tobacco budworm Heliothis virescens. Insect Mol Biol 20(1):125–133
Wanner KW, Nichols AS, Allen JE, Bunger PL, Garczynski SF, Linn CE, Robertson HM, Luetje CW (2010) Sex pheromone receptor specificity in the European corn borer moth Ostrinia nubilalis. PLoS One 5(1):e8685
Wearing CH, Thomas WP, Dugdale JS (1991) Australian and New Zealand species. In: van der Geest LPS, Evenhuis HH (eds) Tortricoid pests, their biology, natural enemies and control. Elsevier, Amsterdam, pp 453–471
Xue B, Rooney AP, Kajikawa M, Okada N, Roelofs WL (2007) Novel sex pheromone desaturases in the genomes of corn borers generated through gene duplication and retroposon fusion. Proc Natl Acad Sci U S A 104(11):4467–4472
Yang Z (2007) PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24(8):1586–1591
Yang Z, Nielsen R (2000) Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol Biol Evol 17(1):32–43
Young H, Galbreath RA, Benn MH, Holt VA, Struble DL (1985) Sex-pheromone components in the New Zealand brown headed leafroller Ctenopseustis obliquana (Lepidoptera, Tortricidae). Zeitschrift Fur Naturforschung C-a Journal of Biosciences 40(3–4):262–265
Acknowledgments
We would like to thank Anne Barrington for supplying moths. Also, we would like to thank Melissa Jordan and Jacob Corcoran for early access to Epiphyas postvittana OR data and for establishing the HEK293 cell-assay. Funding was provided by the Allan Wilson Centre.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Steinwender, B., Thrimawithana, A.H., Crowhurst, R.N. et al. Pheromone Receptor Evolution in the Cryptic Leafroller Species, Ctenopseustis obliquana and C. herana . J Mol Evol 80, 42–56 (2015). https://doi.org/10.1007/s00239-014-9650-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-014-9650-z