Abstract
Land plants possess some of the most unusual mitochondrial genomes among eukaryotes. However, in early land plants these genomes resemble those of green and red algae or early eukaryotes. The question of when during land plant evolution the dramatic change in mtDNAs occurred remains unanswered. Here we report the first completely sequenced mitochondrial genome of the hornwort, Megaceros aenigmaticus, a member of the sister group of vascular plants. It is a circular molecule of 184,908 base pairs, with 32 protein genes, 3 rRNA genes, 17 tRNA genes, and 30 group II introns. The genome contains many genes arranged in the same order as in those of a liverwort, a moss, several green and red algae, and Reclinomonas americana, an early-branching eukaryote with the most ancestral form of mtDNA. In particular, the gene order between mtDNAs of the hornwort and Physcomitrella patens (moss) differs by only 8 inversions and translocations. However, the hornwort mtDNA possesses 4 derived features relative to green alga mtDNAs—increased genome size, RNA editing, intron gains, and gene losses—which were all likely acquired during the origin and early evolution of land plants. Overall, this genome and those of other 2 bryophytes show that mitochondrial genomes in early land plants, unlike their seed plant counterparts, exhibit a mixed mode of conservative yet dynamic evolution.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The mitochondrial genomes of land plants (embryophytes)—compared with those of animals and fungi, which generally show a stable and conservative mode of evolution despite having decreased gene content (Gray et al. 2004)—exhibit several features highlighting their dynamic evolution. These features include significant size expansion, frequent intra-genomic rearrangement, formation of subgenomic circles in addition to the master genome circle, frequent losses and transfers of genes to the nucleus, RNA editing, large-scale intron gains, trans-splicing of group II introns, and replacement of some tRNA genes by their chloroplast counterparts (Knoop 2004; Kubo and Newton 2008; Palmer et al. 2000; Schuster and Brennicke 1994). Because land plants represent a lineage of eukaryotes that spans an evolutionary time of approximately 500 million years (Wellman et al. 2003), and because most of the previously mentioned features were initially observed in angiosperms, it is not clear when these features began to appear during land plant evolution.
Since the early 1990s and especially during the last 10 years, 15 mitochondrial genomes from land plants have been sequenced. These include a liverwort (Marchantia polymorpha) (Oda et al. 1992a), a moss (Physcomitrella patens) (Terasawa et al. 2007), a gymnosperm (Cycas taitungensis) (Chaw et al. 2008), and 12 angiosperms (Arabidopsis thaliana, Beta vulgaris subsp. vulgaris, Brassica napus, Nicotiana tabacum, Oryza sativa, Sorghum bicolor, Tripsacum dactyloides, Triticum aesitvum, Zea luxurians, Z. mays subsp. mays, Z. mays subsp. parviglumis, and Z. perennis) (Allen et al. [unpublished]; Clifton et al. 2004; Handa 2003; Kubo et al. 2000; Ogihara et al. 2005; Sugiyama et al. 2005; Tian et al. 2006; Unseld et al. 1997). In addition, 6 green algal relatives of land plants have been sequenced for their mitochondrial DNAs (mtDNAs): four charophytes (Chaetosphaeridium globosum, Chara vulgaris, Chlorokybus atmophyticus, and Mesostigma viride) (Turmel et al. 2002a, 2002b, 2003, 2007) and two prasinophycean green algae (Nephroselmis olivacea and Ostreococcus tauri) (Robbens et al. 2007; Turmel et al. 1999). Comparative analyses of these genomes have shown that the mitochondrial genomes of Physcomitrella, Marchantia, Chara, Chaetosphaeridium, Chlorokybus, Nephroselmis, and Ostreococcus possess similar gene order and content to those of the red alga Cyanidioschyzon merolae and the early-branching eukaryote Reclinomonas americana (Gray et al. 2004; Ohta et al. 1998; Robbens et al. 2007; Terasawa et al. 2007; Turmel et al. 2007). The mtDNA of Reclinomonas represents the most ancestral form of mitochondrial genomes in eukaryotes (Gray et al. 2004; Lang et al. 1997). Furthermore, the above-mentioned species cover a well-sampled continuum of photosynthetic organisms on the eukaryote phylogeny from the origin of eukaryotes, to the emergence of algae possessing plastids of primary endosymbiosis, and to the evolution of land plants (Baldauf 2003; Rodriguez-Ezpeleta et al. 2007). Thus, it can be inferred that the basic genome structure, order, and content of the mitochondrial genomes have been maintained rather stably since their origin from bacterial endosymbionts until the early stage of land plant evolution. Moreover, several of the derived features of land plant mitochondrial genomes, e.g., frequent intra-genomic rearrangement, formation of subgenomic circles in addition to the master genome circle, trans-splicing of introns, and replacement of some tRNA genes by their chloroplast counterparts, are only restricted to seed plants. These features most likely evolved after liverworts and mosses had diverged from the common ancestor of all other land plants. To date, no mitochondrial genome has been sequenced in hornworts, lycophytes, or monilophytes, three groups that represent critical stages of early land plant evolution (Qiu 2008). Hence, there is a major gap in our current knowledge of mitochondrial genome evolution in land plants.
In this study, we sequenced the mitochondrial genome of the hornwort Megaceros aenigmaticus. Hornworts represent one of the three clades of bryophytes, which make up a paraphyletic group at the base of the land plant phylogeny. Several recent studies have suggested that they are sister to vascular plants (Groth-Malonek et al. 2005; Kelch et al. 2004; Qiu et al. 2006, 2007). Hornworts have approximately 300 living species (Duff et al. 2007) and are far less diverse than other two clades of bryophytes, liverworts and mosses. Thus, sequencing a hornwort mitochondrial genome should provide important insights into the transition from the somewhat ancestral type of mitochondrial genomes in liverworts and mosses to the derived type in seed plants.
Materials and Methods
Approximately 10 g fresh thallus material of M. aenigmaticus R. M. Schuster was collected in the field from Tennessee and brought to the laboratory for cleaning under a dissecting scope. A voucher specimen numbered Qiu 06044 was deposited at the University Herbarium in the University of Michigan, Ann Arbor, MI.
Total cellular DNA was extracted using the cetyltrimethyl–ammonium bromide (CTAB) method (Doyle and Doyle 1987) and purified with phenol extraction to remove proteins. A fosmid library was constructed using the CopyControl kit (EPICENTRE Biotechnologies, Madison, WI). Eight overlapping clones containing mitochondrial DNA fragments were identified through Southern hybridizations using mitochondrial gene probes with the HRP chemiluminescent blotting kit (KPL, Gaithersburg, MD). The inserts were sequenced for both forward and reverse strands through primer-walking on an ABI 3100 genetic analyzer (Applied Biosystems, Foster City, CA). Sequences were assembled using Sequencher (Gene Codes, Ann Arbor, MI).
The genome was annotated in four steps. First, genes for known mitochondrial proteins and rRNAs were identified by Basic Local Alignment Search Tool (BLAST) searches (Altschul et al. 1990) (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi) of the nonredundant database at the National Center for Biotechnology Information (NCBI). The exact gene and exon/intron boundaries as well as putative RNA editing sites were predicted by alignment of orthologous genes from annotated plant mitochondrial genomes available at the organelle genomic biology website at NCBI (http://www.ncbi.nlm.nih.gov/genomes/ORGANELLES/organelles.html). Second, genes for hypothetical proteins were identified using the web-based tool Open Reading Frames Finder (orf finder; http://www.ncbi.nlm.nih.gov/projectes/gorf/) with the standard genetic code. Third, genes for tRNAs were found using tRNAscan-SE (Lowe and Eddy 1997) (http://lowelab.ucsc.edu/tRNAscan-SE/). Finally, repeated sequences were searched using REPuter (Kurtz et al. 2001) (http://bibiserv.techfak.uni-bielefeld.de/reputer/).
To determine genomic origins of tRNA genes, sequences of the same tRNA gene from both mitochondrial and chloroplast (if available) genomes of 9 selected eukaryotes (see later text) were obtained from the same organelle genomic biology website of NCBI as previously indicated. Sequences were aligned using CLUSTAL_X (Thompson et al. 1997). The genomic origins of tRNA genes in Megaceros and other land plants were determined by visual inspection of aligned mitochondrial and/or chloroplast tRNA gene sequences from these 10 organisms.
For comparison of gene order and content as well as determination of genomic origins of tRNA genes, nine species that have the most ancestral form of mitochondrial genome of their respective clades and occupy key positions on the eukaryote phylogeny (Baldauf 2003; Qiu 2008; Rodriguez-Ezpeleta et al. 2007) were selected from the same organelle genomic biology website at NCBI as shown previously. These include the following: R. americana for jakobids (early-branching eukaryotes; NC_001823), C. merolae for red algae (NC_000887, NC_004799 [the latter accession number is for the chloroplast genome]), N. olivacea for prasinophyceaen algae (early-branching green algae; NC_008239, NC_000927), C. vulgaris for charophytes (NC_005255, NC_008097), M. polymorpha for liverworts (NC_001660, NC_001319), P. patens for mosses (NC_007945, NC_005087), C. taitungensis for gymnosperms (NC_010303, NC_009618), O. sativa for monocots (NC_007886, NC_001320), and B. napus (NC_008285) for eudicots. Other red and green algae and angiosperms at that Web site were also examined for their gene order and content, but were not included here due to either space limitation or their possession of many genomic features unique to themselves.
Results
General Features of the Hornwort Mitochondrial Genome
The mitochondrial genome of Megaceros has the size of 184,908 base pairs (bp) with 54.0% AT content. Exons, introns, and intergenic spacers account for 16%, 34%, and 50% of the genome, respectively. The entire genome is assembled as a single large circular molecule (Fig. 1). Three long repeats were detected, 1 direct (A) and 2 inverted (B and C). The repeats A and B both involve 2 copies of identical sequences, which are 506- and 536-bp long, respectively. The repeat C has 2 copies of sequences that are 93% identical, and they differ by a 3-bp deletion in a total length of 172 bp. Although 1 copy of all 3 repeats is located in intergenic regions, the other either spans a portion of trnMf and atp8 (repeats A and B, respectively) or is located in a nad9 group II intron (repeat C). In addition, 2 families of tetra-nucleotide microsatellite sequences were detected. The TATG sequence iterates 31 times in an intergenic region upstream of trnPugg, whereas the AAGG sequence iterates 29 times and is located in a group II intron in the pseudogene sdh3. The largest intergenic spacer, between rps4 and 26S rRNA gene, is 11,486 bp long, and there are three orfs >100 codons in this spacer. The largest non–orf-containing intergenic spacer is 5,578 bp long and is between 26S rRNA gene and atp9. There are 3 pairs of overlapping genes: rps14-rpl5 by 1 bp, cox1-atp4 by 4 bp, and rps12-rps7 by 4 bp.
Gene map of M. aenigmaticus mitochondrial genome. Genes (exons are shown as closed boxes) shown on the outside of the circle are transcribed clockwise, whereas those on the inside are transcribed counter-clockwise. Genes with group II introns (open boxes) are labeled with asterisks. Pseudogenes are indicated with the prefix “ψ”. The map was drawn using OGDRAW (Lohse et al. 2007)
Use of the standard genetic code and consideration of a small number of RNA-editing events allowed annotation of all protein-coding genes. Hence, the hornwort mitochondrial genome almost certainly uses the standard genetic code and experiences RNA editing when its genes are expressed. Because evolutionary divergence may affect the prediction of editing sites based on alignment or use of computer software, and because hornworts are notorious for possessing a high and idiosyncratic level of divergence at the molecular and morphologic levels, no attempt was made to predict RNA-editing sites explicitly without carrying out cDNA analysis. Putative RNA-editing sites were only suggested for the sake of removing internal stop codons and creating appropriate start and stop codons. Genes requiring a minimal number of editing events to be translated are listed in Table 1.
Gene Content and Order
The Megaceros mitochondrial genome contains 32 protein genes, 3 rRNA genes, and 18 tRNA genes (Table 2). The 32 protein genes include 8 genes for NADH:ubiquinone oxidoreductase (complex I of the respiratory chain as designated in Gray et al. 1999; nad1-6, 4L, 9), 2 genes for succinate:ubiquinone oxidoreductase (complex II; sdh3, 4), 1 gene for ubiquinol:cytochrome c oxidoreductase (complex III; cob), 3 genes for cytochrome c oxidase (complex IV; cox1-3), 5 genes for adenosine triphosphate synthase (complex V; atp1, 4, 6, 8, 9), 1 gene for cytochrome c biogenesis (ccmFC), 10 genes for ribosomal proteins, and 2 genes for other functions (tatC, orf-bryo1). Of these protein genes, 12 are pseudogenes, 9 of them being ribosomal protein genes. Alignments of these pseudogenes with their orthologs from Marchantia and Physcomitrella, documenting multiple indels that disrupt reading frames, are provided in Supplementary Fig. 1. Twenty-eight orfs >100 codons are also present in the genome. All except 2 of them are between 100 and 134 codons long and may not represent real genes. One orf, designated as orf_bryo1 (159 codons), likely represents a gene with undetermined function because it is present in all 3 bryophyte mitochondrial genomes sequenced so far (Oda et al. 1992a; Terasawa et al. 2007). The other orf is 196 codons long and may be a candidate for another uncharacterized gene.
The 17 tRNA genes in the Megaceros mitochondrial genome (Table 2) represent a significantly smaller set than those in the mtDNAs of Marchantia and Physcomitrella (27 and 24, respectively). The tRNAs for 4 amino acids—N, R, S, and V—are not among the ones encoded by these genes, and they are thus likely nucleus-encoded and imported into the mitochondrion. As in other land plants and charophytes, e.g., Chara (Turmel et al. 2003), Cycas (Chaw et al. 2008), and Z. mays ssp. mays (Clifton et al. 2004), the trnIcau encodes a tRNA whose anticodon UAU is likely obtained through post-transcriptional modification. In terms of genomic origin, our comparative analyses of tRNA genes from both mitochondrial and chloroplast (when available) genomes of the 10 species included in this study show that all 17 tRNA genes in Megaceros mtDNA are of mitochondrial origin (Table 2).
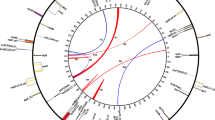
The gene order in Megaceros mtDNA is similar to those in mitochondrial genomes of Physcomitrella, Marchantia, Chara, Nephroselmis, Cyanidioschyzon, and even Reclinomonas but quite different from those of Cycas, Oryza and Brassica mtDNAs (Fig. 2). In particular, the gene orders between Megaceros and Physcomitrella mtDNAs are strikingly similar (Fig. 3). Because moss represents the closest ancestral outgroup of the hornwort (Qiu et al. 2006, 2007), a detailed comparison was performed to identify hypothetical genomic changes that could bring the 2 genomes to complete collinearity. This exercise may underestimate the number of changes because the evolutionary distance between the 2 taxa is quite large, and the 2 genomes by no means represent the ancestral genomes of mosses and hornworts. Still, it was done to obtain an idea of how similar the 2 genomes are in terms of gene order. Overall, 4 syntenic blocks can be recognized. Only 2 major inversions, 1 each for a block, need to be proposed to bring the 2 genomes largely collinear. In addition, 6 local inversions and translocations involving individual genes within the 4 blocks can account for all remaining differences in the gene order. Finally, 15 deletions and 1 insertion are required to explain the gene content difference between the 2 genomes (Fig. 3).
Evolution of gene order in the mitochondrial genomes of selected protists and land plants. The genes are presented in the order as they appear in the genome, with 26S rRNA gene chosen as the standard beginning of the genome. They are shown in 4 different colors, which indicate 4 blocks of the mitochondrial genome of the reference species P. patens. The coloring system is designed to facilitate tracking of gene order changes across the 10 genomes. Physcomitrella mtDNA is chosen as the reference genome because on the phylogeny it is immediately below the genome under study, that of M. aenigmaticus. The genes unique to only one genome or of chloroplast origin are not colored. If a gene is not present in Physcomitrella mtDNA, its color is determined based on its general presence in the Physcomitrella block in other genomes. The genes on the forward and reverse strands are shown above and below the line, respectively. The full names of abbreviated gene names used here are listed in Table 2. The taxa are arranged according to a putative phylogeny of protistan eukaryotes and land plants (Baldauf 2003; Qiu 2008; Rodriguez-Ezpeleta et al. 2007)
Gene order comparison between M. aenigmaticus and P. patens mitochondrial genomes. The genomes are shown in the same colored blocks as in Fig. 2. For Physcomitrella mtDNA, the blue block has been rearranged to facilitate comparison. Two arrowed brackets indicate two major inversions involving the blocks va-cb and ti-t10, respectively. Solid lines mark local inversions involving n3 and tc, whereas dashed lines indicate local translocations involving t9, r26, t5, and n1. Small vertical arrows denote gene deletions/insertions t8, t15, t2, t13, tr, tv, n7, l2, s19, s3, l16, s2, mb, mc, mz, and s8* (*s8 actually represents a deletion in Physcomitrella mtDNA because it is present at that location in the mtDNAs of Marchantia, Nephroselmis, Cyanidioschyzon, and Reclinomonas [see Fig. 2])
Intron Content
The Megaceros mitochondrial genome contains 30 group II introns and no group I intron, according to the definitions of these mobile genetic elements (Michel and Dujon 1983). They are located in these 15 genes: atp1, atp6, atp9, ccmFC, cob, cox1, cox2, nad1, nad2, nad3, nad4, nad5, nad6, nad9, and sdh3. Twelve of these introns are newly discovered in this study. They and nad1i348 and nad5i881, which were discovered in 2 earlier studies (Beckert et al. 1999; Dombrovska and Qiu 2004), make up the 14 group II introns that are unique to hornworts (Supplementary Table 1). The gene ccmFC in Megaceros is a pseudogene; however, its intron ccmFCi829, which was likely gained in the common ancestor of mosses, hornworts, and vascular plants, is still present. Likewise, sdh3i100 is still in the pseudogenized sdh3. In both cases, the 5’ exon-intron boundaries have been eroded because of length mutations; however, the alignments show that the introns in Megaceros and Physcomitrella are most likely orthologous (Supplementary Fig. 2). All 30 group II introns in the hornwort mitochondrial genome are cis-spliced. Only four of them—atp6i80, nad1i287, nad1i348, and NAD9I246—are <800 bp long. Group II introns longer than this length usually contain orfs with various domains involved in intron splicing and mobility (Mohr et al. 1993). Hence, most introns in Megaceros mtDNA likely contain protein-coding orfs.
Discussion
Genome Size
The mitochondrial genome of Megaceros, at 184,908-bp long, is almost of the same size as that of Marchantia (186,609 bp) (Oda et al. 1992a) but significantly larger than that of Physcomitrella (105,340 bp) (Terasawa et al. 2007). It is also smaller than the smallest seed plant mitochondrial genome sequenced so far, that of Brassica (221,853 bp) (Handa 2003). The percentages of introns and intergenic spacers in this genome are generally similar to those of two other bryophyte mtDNAs, whereas that of exons in Megaceros mtDNA is lower (Table 3).
With the exception of Chlorokybus mtDNA, which likely represents an isolated case of genome size increase in that species (Turmel et al. 2007), it appears that the mitochondrial genomes of Reclinomonas (Lang et al. 1997), red algae (Burger et al. 1999; Ohta et al. 1998), prasinophycean green algae (Robbens et al. 2007; Turmel et al. 1999), and charophytes (Turmel et al. 2002a, 2002b, 2003) are generally small (<70 kb). A significant size increase occurred during the origin of land plants, mostly caused by expansion of intergenic spacers according to our analysis of the percentages of exons, introns, and intergenic spacers in mitochondrial genomes of Chara and three bryophytes (Table 3). Since then, the mitochondrial genomes have remained in the size range of 100 to 200 kb in bryophytes (Oda et al. 1992a; Terasawa et al. 2007) and have increased to >200 kb in seed plants (Allen et al. [unpublished]; Chaw et al. 2008; Clifton et al. 2004; Handa 2003; Kubo et al. 2000; Ogihara et al. 2005; Sugiyama et al. 2005; Tian et al. 2006; Unseld et al. 1997). Among all major lineages of eukaryotes, land plants are the only ones that possess such large mitochondrial genomes (Gray et al. 1999, 2004). Two questions are certainly worth exploring in future studies as more land plant mtDNAs are sequenced: Is there any reason for this genome size increase? If so, what caused it?
Gene Content
This is the most gene-poor genome among all land plant mitochondrial genomes sequenced to date. Compared with those of Physcomitrella, Marchantia, and Chara, the hornwort mtDNA has lost or is in the process of losing most genes encoding enzymes for cytochrome c biogenesis (ccm) and ribosomal proteins (rpl and rps). The genes for succinate:ubiquinone oxidoreductase (sdh) and tRNAs are also on the way out (Table 2). The only nad gene that has been reported to be pseudogenized in the land plant mitochondrial genomes, nad7 (Groth-Malonek et al. 2007), is also absent in this genome. The rest of the gene repertoire of Megaceros mtDNA is highly similar to that of Physcomitrella, Marchantia, and Chara, and even Nephroselmis and Cyanidioschyzon (Table 2). Hence, it seems that with the exception of those four categories of genes that were or are being lost in the hornwort, the rest of the mitochondrial genes have been stably inherited since the shared common ancestor of rhodophytes (red algae) and viridiplants (green algae and land plants). Because the seed plant mitochondrial genomes do contain ccm, rpl, rps, and sdh genes, some tRNA genes, and other “standard” mitochondrial genes found in green and red algae (Table 2), gene content reduction in the hornwort mitochondrial genome represents an isolated case on its own rather than the loss of these genes in the common ancestor of hornworts and vascular plants. The loss of so many genes in Megaceros mtDNA actually represents a derived feature of this otherwise relatively ancestral type of mitochondrial genome as indicated by the conserved gene order.
The loss of these four categories of genes from Megaceros mtDNA also fits the pattern that has been previously observed. In a large survey of angiosperm mitochondrial genes, rpl, rps, and sdh genes were shown to be most prone to be lost from the mitochondrial genomes of some angiosperm lineages (Adams et al. 2001, 2002). The dramatically reduced mitochondrial genomes of some green and red algae (e.g., Chlamydomonas eugametos and Porphyra purpurea) have also lost some or most genes in these four categories (Burger et al. 1999; Denovan-Wright et al. 1998). Likewise, highly reduced mitochondrial genomes of animals and most fungi have lost almost all sdh, ccm, rpl, and rps genes (Gray et al. 1999). In land plants, the loss of ccm and tRNA genes has been less well known until now. In particular, the loss of tRNA genes may be related to the phenomenon of some mitochondrial tRNA genes being replaced by their chloroplast counterparts at some stage of vascular plant evolution.
Chloroplast-originated tRNA genes that replace their mitochondrial counterparts were first reported to be present in the mitochondrial genome of potato (Marechal-Drouard et al. 1990), and then found in all sequenced mtDNAs of angiosperms (Clifton et al. 2004; Handa 2003; Kubo et al. 2000; Ogihara et al. 2005; Sugiyama et al. 2005; Tian et al. 2006; Unseld et al. 1997). Most recently, the sequenced gymnosperm Cycas mtDNA has also been shown to contain several tRNA genes of chloroplast origin (Chaw et al. 2008). In contrast, the liverwort Marchantia mtDNA contains no such alien tRNA genes (Oda et al. 1992b). The analyses in this study further confirm that replacement of tRNA genes has not occurred in mitochondrial genomes of Reclinomonas, Cyanidioschyzon, Nephroselmis, Chara, and Physcomitrella (Table 2). The lack of chloroplast tRNA genes now shown in the mtDNA of the hornwort suggests that this intriguing molecular evolutionary phenomenon most likely occurred during or after the origin of vascular plants. One interesting observation made in this study is that for different tRNA genes, replacement seems to have happened at different evolutionary time points. For example, trnHgug replacement likely took place in the common ancestor of seed plants or earlier because all three seed plants examined in this study have only the chloroplast copy in their mitochondrial genomes. In contrast, trnWcca might have had its replacement in the common ancestor of all angiosperms because only the two angiosperms among all examined land plants have the chloroplast copy, whereas all other land plants have their native mitochondrial copy (Table 2). To determine whether replacement can happen for all tRNA genes in the land plant mitochondrial genome as well as when it happens for each tRNA gene, a large study may be required to sample plants that represent major clades of angiosperms, gymnosperms, monilophytes, and lycophytes.
Gene Order
The fact that 8 inversions and translocations can make the mitochondrial genomes of Megaceros and Physcomitrella completely collinear (Fig. 3) shows a surprising extent of gene order conservation. This level of conservation between mtDNAs of a hornwort and a moss, which diverged >400 million years ago (Kenrick and Crane 1997), can be better appreciated when placed in the broad perspective of mitochondrial genome evolution in land plants. In seed plants, intragenomic rearrangement frequency is much higher. For example, the mtDNAs of Cycas, Oryza, and Brassica show virtually no collinearity (Fig. 2). In Z. mays, mtDNAs of 2 fertile cytotypes within a single species have experienced 16 rearrangements (Allen et al. 2007)! On the other hand, the slow pace of change in the hornwort mitochondrial gene order is not unexpected because previous studies have reported that the mitochondrial genomes from Chara to Marchantia to Physcomitrella show largely conserved gene order (Terasawa et al. 2007; Turmel et al. 2003). The result from this study extends the phylogenetic breadth of conservative mitochondrial gene order evolution to a more derived lineage of early land plants, i.e., hornworts, right before evolution of vascular plants (Fig. 2). A broader comparison of these early land plant and Chara mtDNAs with those of phylogenetically more ancestral lineages—such as the prasinophycean green alga Nephroselmis, the red alga Cyanidioschyzon, and the early branching eukaryote Reclinomonas—shows that the mitochondrial gene order has not changed dramatically from the origin of mitochondria to the emergence of hornworts (Fig. 2). These data also suggest that the rapid intragenomic rearrangement seen in seed plant mitochondrial genomes began after hornworts had diverged from the common ancestor of vascular plants. It will now be interesting to see whether mitochondrial genomes of early vascular plants, such as lycophytes and ferns, have conserved or reshuffled gene order.
The conservation of gene order in Megaceros and two other bryophyte mtDNAs is also reflected by the lack of any trans-spliced group II intron in these genomes (Oda et al. 1992a; Terasawa et al. 2007). In contrast, the highly rearranged mitochondrial genomes of seed plants have several trans-spliced group II introns (Bonen 2008; Chaw et al. 2008; Dombrovska and Qiu 2004; Malek et al. 1997; Malek and Knoop 1998), which often render exons of a single gene scattered over the entire genome on both strands. Although most trans-spliced introns are bipartite (i.e., the intron being broken into two pieces) and evolved from a single ancient event during seed plant evolution (Dombrovska and Qiu 2004; Malek et al. 1997; Malek and Knoop 1998), two extreme cases of trans-splicing have been reported. One is tripartite trans-splicing of nad5i1477 in Oenothera berteriana (Knoop et al. 1997). The other involves nad1i728, which has evolved from cis- to trans-splicing 15 times independently among >400 angiosperms investigated. Further, these trans-splicing events occurred at different evolutionary time points (some quite ancient and others more recent) and involve 2 loci within the intron (Qiu and Palmer 2004). These 2 extreme cases highlight the dynamic nature of seed plant mtDNAs in terms of intragenomic rearrangement.
Whatever the causative agents may be, their invasion of the mitochondrial genome during post–bryophyte evolution of land plants are mostly likely responsible for the gene order disruption and breakup of cis-spliced introns seen in seed plant mtDNAs. Sequencing of mitochondrial genomes from lycophytes and monilophytes will be of urgent need for identifying such genome rearrangement agents through comparison with the available bryophyte and seed plant mtDNAs.
This study was designed as part of a larger project to determine the starting point when mitochondrial genomes in land plants embarked on the journey of volatile evolution, in particular showing reshuffled gene order. The significance of gene order change and, more precisely, intragenomic rearrangement seen so far in mitochondrial genomes of seed plants will be better understood when one considers the number of species involved and the profound nature of change on the genome. Seed plants represent a clade of multicellular eukaryotes with approximately 250,000 species that are primary producers of the terrestrial ecosystems (Mabberley 1987). All indications suggest that the type of large, rearranged mitochondrial genomes seen in a dozen or so sequenced seed plants are likely to exist in all seed plants and possibly all vascular plants (Dombrovska and Qiu 2004; Knoop 2004; Malek et al. 1997; Palmer et al. 1992). Furthermore, the extent of change in seed plant mtDNAs is more profound than simple gene-order reshuffling. It may also involve significant genome size increase, alien DNA invasion, formation of subgenomic circles in addition to the master genome circle, intron trans-splicing, tRNA gene replacement, and lineage-specific dramatic rate acceleration of sequence evolution (Gray et al. 2004; Knoop 2004; Kubo and Newton 2008; Palmer et al. 2000; Schuster and Brennicke 1994). Although dramatic reshuffling of gene order and sometimes gene losses have been found in mtDNAs of some red and green algae, e.g., P. purpurea (Burger et al. 1999), Chondrus crispus (Richard et al. 1998), C. eugametos (Denovan-Wright et al. 1998), and M. viride (Turmel et al. 2002b), the number of species involved and the extent of genome changes impacted are probably not anywhere near what happened in seed plant mtDNAs. Red and green algae have only approximately 5000 and 8000 species, respectively (van den Hoek et al. 1995), and some of them still have the ancestral type of mitochondrial genomes, e.g., C. merolae (Ohta et al. 1998), Prototheca wickerhamii (Wolff et al. 1994), and Pseudendoclonium akinetum (Pombert et al. 2004). A logic and intriguing question to ask is this: Why do seed plants or probably vascular plants have such unique mitochondrial genomes?
Introns
Fourteen hornwort-specific group II introns, 12 of them newly discovered here, of 32 present in hornwort mitochondrial genomes, as found in this and earlier studies (Beckert et al. 1999; Qiu et al. 1998) (Supplementary Table 1), represent a significant wave of intron gain. This is another derived aspect of an otherwise ancestral type of mitochondrial genome in the hornwort (the other being gene loss; see previous text). Physcomitrella (7 of 23), Marchantia (16 of 25), and even Chara (8 of 13) all have experienced major waves of intron invasion (Supplementary Table 1). Most of the introns found in Cycas, Oryza, and Brassica mtDNAs (Supplementary Table 1) actually extend down to lycophytes (Dombrovska and Qiu 2004; Malek and Knoop 1998); thus the common ancestor of vascular plants might have also gained many introns. Before evolution of Chara, group II introns were rare in mtDNAs of Mesostigma (Turmel et al. 2002b) and Chaetosphaeridium (Turmel et al. 2002a). However, Chlorokybus is an exception; its unusually large mitochondrial genome has 14 group II introns, and 10 of them are present in trn genes (Turmel et al. 2007). In all other mitochondrial genomes mentioned previously, introns tend to occur more frequently in genes encoding proteins involved in respiration than in those encoding ribosomal proteins, tRNAs, and rRNAs. Given that the sufficient diversity of charophytes, bryophytes, and seed plants has been sampled in mitochondrial genome sequencing, this intron distribution pattern is unlikely to change significantly. It is then natural to ask whether this pattern has any evolutionary functional significance.
Because group II intron distribution in organellar genomes has been exploited for resolving difficult phylogenetic problems (Manhart and Palmer 1990; Qiu et al. 1998, 2006), the intron distribution pattern in the Megaceros mtDNA can also be compared with those in charophytes, Marchantia, Physcomitrella, and seed plants to infer relations among three bryophyte lineages and vascular plants. With a small amount of homoplasy (intron losses or independent gains), shared presence of six introns (ccmFCi829, cox2i373, cox2i691, nad1i728, nad4i461, and nad5i230) in mosses, hornworts, and vascular plants supports liverworts as the earliest diverging lineage of land plants. If no homoplasy is allowed, there are three introns each supporting either hornworts (nad2i1282, nad5i1455, and nad5i1477) or mosses (nad2i156, nad7i140, and nad7i209) as the sister to vascular plants. If some homoplasy is permitted, two more introns, nad2i709 and nad4i976, can join the three to support the hypothesis of hornworts being sister to vascular plants (Supplementary Table 1). Hence, there seems to be slightly more evidence for hornworts than mosses to be sister to vascular plants. The intron distribution data overall also support the paraphyly of bryophytes. These results are consistent with those obtained from analyses of multigene supermatrices (Qiu et al. 2006, 2007) and chloroplast genomic structural characters (Kelch et al. 2004). Because there is no sequenced lycophyte or monilophyte mitochondrial genome, we prefer to wait until those genomes are available to carry out explicit phylogenetic analyses of intron distribution, gene order, and whole-genome sequence data as have been done in previous studies (Kelch et al. 2004; Qiu et al. 2006).
In conclusion, the mitochondrial genome of the hornwort M. aenigmaticus represents a largely ancestral type, in having rather conserved gene order and content that can be traced back to the beginning of eukaryotes and the common ancestor of rhodophytes and viridiplants, respectively. On the other hand, it also has two derived features, i.e., loss of some genes and gain of many introns. Its expanded size compared with most green and red algal mtDNAs and possession of RNA editing also represent derived features that were likely acquired during the origin of land plants. This genome does not resemble the mitochondrial genomes of seed plants, which have highly rearranged gene order, trans-spliced introns, and chloroplast-originated tRNA genes. The hornwort mtDNA represents a transitional state of mitochondrial genome evolution in charophytes and land plants, but it does so with some of its own unique characteristics. Together with those of Marchantia and Physcomitrella, Megaceros mtDNA shows that mitochondrial genome evolution in early land plants is highly conservative yet dynamic in certain aspects.
References
Adams KL, Qiu YL, Stoutemyer M, Palmer JD (2002) Punctuated evolution of mitochondrial gene content: high and variable rates of mitochondrial gene loss and transfer to the nucleus during angiosperm evolution. Proc Natl Acad Sci USA 99:9905–9912
Adams KL, Rosenblueth M, Qiu Y-L, Palmer JD (2001) Multiple losses and transfers to the nucleus of two mitochondrial respiratory genes during angiosperm evolution. Genetics 158:1289–1300
Allen JO, Fauron CM, Minx P, Roark L, Oddiraju S, Lin GN, Meyer L, Sun H, Kim K, Wang CY et al (2007) Comparisons among two fertile and three male-sterile mitochondrial genomes of maize. Genetics 177:1173–1192
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Baldauf SL (2003) The deep roots of eukaryotes. Science 300:1703–1706
Beckert S, Steinhauser S, Muhle H, Knoop V (1999) A molecular phylogeny of bryophytes based on nucleotide sequences of the mitochondrial nad5 gene. Plant Syst Evol 218:179–192
Bonen L (2008) Cis- and trans-splicing of group II introns in plant mitochondria. Mitochondrion 8:26–34
Burger G, Saint-Louis D, Gray MW, Lang BF (1999) Complete sequence of the mitochondrial DNA of the red alga Porphyra purpurea: cyanobacterial introns and shared ancestry of red and green algae. Plant Cell 11:1675–1694
Chaw SM, Shih ACC, Wang D, Wu YW, Liu SM, Chou TY (2008) The mitochondrial genome of the gymnosperm Cycas taitungensis contains a novel family of short interspersed elements, Bpu sequences, and abundant RNA editing sites. Mol Biol Evol 25:603–615
Clifton SW, Minx P, Fauron CMR, Gibson M, Allen JO, Sun H, Thompson M, Barbazuk WB, Kanuganti S, Tayloe C et al (2004) Sequence and comparative analysis of the maize NB mitochondrial genome. Plant Physiol 136:3486–3503
Denovan-Wright EM, Nedelcu AM, Lee RW (1998) Complete sequence of the mitochondrial DNA of Chlamydomonas eugametos. Plant Mol Biol 36:285–295
Dombrovska O, Qiu Y-L (2004) Distribution of introns in the mitochondrial gene nad1 in land plants: phylogenetic and molecular evolutionary implications. Mol Phylogenet Evol 32:246–263
Doyle JJ, Doyle JS (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Duff RJ, Villarreal JC, Cargill DC, Renzaglia KS (2007) Progress and challenges toward developing a phylogeny and classification of the hornworts. Bryologist 110:214–243
Gray MW, Burger G, Lang BF (1999) Mitochondrial evolution. Science 283:1476–1481
Gray MW, Lang BF, Burger G (2004) Mitochondria of protists. Annu Rev Genet 38:477–524
Groth-Malonek M, Pruchner D, Grewe F, Knoop V (2005) Ancestors of trans-splicing mitochondrial introns support serial sister group relationships of hornworts and mosses with vascular plants. Mol Biol Evol 22:117–125
Groth-Malonek M, Wahrmund U, Polsakiewicz M, Knoop V (2007) Evolution of a pseudogene: exclusive survival of a functional mitochondrial nad7 gene supports Haplomitrium as the earliest liverwort lineage and proposes a secondary loss of RNA editing in Marchantiidae. Mol Biol Evol 24:1068–1074
Handa H (2003) The complete nucleotide sequence and RNA editing content of the mitochondrial genome of rapeseed (Brassica napus L.): comparative analysis of the mitochondrial genomes of rapeseed and Arabidopsis thaliana. Nucleic Acids Res 31:5907–5916
Hiesel R, Combettes B, Brennicke A (1994) Evidence for RNA editing in mitochondria of all major groups of land plants except the Bryophyta. Proc Natl Acad Sci USA 91:629–633
Kelch DG, Driskell A, Mishler BD (2004) Inferring phylogeny using genomic characters: a case study using land plant plastomes. In: Goffinet B, Hollowell V, Magill R (eds) Molecular systematics of bryophytes. Missouri Botanical Garden Press, St. Louis, MO, pp 3–11
Kenrick P, Crane PR (1997) The origin and early diversification of land plants: a cladistic study. Smithsonian Institution Press, Washington, DC
Knoop V (2004) The mitochondrial DNA of land plants: peculiarities in phylogenetic perspective. Curr Genet 46:123–139
Knoop V, Altwasser M, Brennicke A (1997) A tripartite group II intron in mitochondria of an angiosperm plant. Mol Gen Genet 255:269–276
Kubo T, Newton KJ (2008) Angiosperm mitochondrial genomes and mutations. Mitochondrion 8:5–14
Kubo T, Nishizawa S, Sugawara A, Itchoda N, Estiati A, Mikami T (2000) The complete nucleotide sequence of the mitochondrial genome of sugar beet (Beta vulgaris L.) reveals a novel gene for tRNA(Cys)(GCA). Nucleic Acids Res 28:2571–2576
Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R (2001) REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res 29:4633–4642
Lang BF, Burger G, Okelly CJ, Cedergren R, Golding GB, Lemieux C, Sankoff D, Turmel M, Gray MW (1997) An ancestral mitochondrial DNA resembling a eubacterial genome in miniature. Nature 387:493–497
Lohse M, Drechsel O, Bock R (2007) OrganellarGenomeDRAW (OGDRAW)—a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet 52:267–274
Lowe TM, Eddy SR (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25:955–964
Mabberley DJ (1987) The plant book. Cambridge University Press, Cambridge, MA
Malek O, Brennicke A, Knoop V (1997) Evolution of trans-splicing plant mitochondrial introns in pre-Permian times. Proc Natl Acad Sci USA 94:553–558
Malek O, Knoop V (1998) Trans-splicing group II introns in plant mitochondria: the complete set of cis-arranged homologs in ferns, fern allies, and a hornwort. RNA 4:1599–1609
Manhart JR, Palmer JD (1990) The gain of two chloroplast transfer-RNA introns marks the green algal ancestors of land plants. Nature 345:268–270
Marechal-Drouard L, Guillemaut P, Cosset A, Arbogast M, Weber F, Weil J-H, Dietrich A (1990) Transfer RNAs of potato (Solanum tuberosum) mitochondria have different genetic origins. Nucleic Acids Res 18:3689–3696
Michel F, Dujon B (1983) Conservation of RNA secondary structures in two intron families including mitochondrial-, chloroplast- and nuclear-encoded members. EMBO J 2:33–38
Mohr G, Perlman PS, Lambowitz AM (1993) Evolutionary relationships among group II intron-encoded proteins and identification of a conserved domain that may be related to maturase function. Nucleic Acids Res 21:4991–4997
Oda K, Yamato K, Ohta E, Nakamura Y, Takemura M, Nozato N, Akashi K, Kanegae T, Ogura Y, Kohchi T et al (1992a) Gene organization deduced from the complete sequence of liverwort Marchantia polymorpha mitochondrial DNA—a primitive form of plant mitochondrial genome. J Mol Biol 223:1–7
Oda K, Yamato K, Ohta E, Nakamura Y, Takemura M, Nozato N, Akashi K, Ohyama K (1992b) Transfer RNA genes in the mitochondrial genome from a liverwort, Marchantia polymorpha—the absence of chloroplast-like transfer-RNAs. Nucleic Acids Res 20:3773–3777
Ogihara Y, Yamazaki Y, Murai K, Kanno A, Terachi T, Shiina T, Miyashita N, Nasuda S, Nakamura C, Mori N et al (2005) Structural dynamics of cereal mitochondrial genomes as revealed by complete nucleotide sequencing of the wheat mitochondrial genome. Nucleic Acids Res 33:6235–6250
Ohta N, Sato N, Kuroiwa T (1998) Structure and organization of the mitochondrial genome of the unicellular red alga Cyanidioschyzon merolae deduced from the complete nucleotide sequence. Nucleic Acids Res 26:5190–5198
Palmer JD, Adams KL, Cho YR, Parkinson CL, Qiu YL, Song KM (2000) Dynamic evolution of plant mitochondrial genomes: mobile genes and introns and highly variable mutation rates. Proc Natl Acad Sci USA 97:6960–6966
Palmer JD, Soltis D, Soltis P (1992) Large size and complex structure of mitochondrial DNA in two nonflowering land plants. Curr Genet 21:125–129
Pombert JF, Otis C, Lemieux C, Turmel M (2004) The complete mitochondrial DNA sequence of the green alga Pseudendoclonium akinetum (Ulvophyceae) highlights distinctive evolutionary trends in the Chlorophyta and suggests a sister-group relationship between the Ulvophyceae and Chlorophyceae. Mol Biol Evol 21:922–935
Pruchner D, Beckert S, Muhle H, Knoop V (2002) Divergent intron conservation in the mitochondrial nad2 gene: signatures for the three bryophyte classes (Mosses, liverworts, and hornworts) and the lycophytes. J Mol Evol 55:265–271
Qiu Y-L (2008) Phylogeny and evolution of charophytic algae and land plants. J Syst Evol 46:287–306
Qiu Y-L, Cho YR, Cox JC, Palmer JD (1998) The gain of three mitochondrial introns identifies liverworts as the earliest land plants. Nature 394:671–674
Qiu Y-L, Li LB, Wang B, Chen ZD, Dombrovska O, Lee J, Kent L, Li RQ, Jobson RW, Hendry TA et al (2007) A nonflowering land plant phylogeny inferred from nucleotide sequences of seven chloroplast, mitochondrial, and nuclear genes. Int J Plant Sci 168:691–708
Qiu Y-L, Li LB, Wang B, Chen ZD, Knoop V, Groth-Malonek M, Dombrovska O, Lee J, Kent L, Rest J et al (2006) The deepest divergences in land plants inferred from phylogenomic evidence. Proc Natl Acad Sci USA 103:15511–15516
Qiu Y-L, Palmer JD (2004) Many independent origins of trans splicing of a plant mitochondrial group II intron. J Mol Evol 59:80–89
Richard O, Bonnard G, Grienenberger JM, Kloareg B, Boyen C (1998) Transcription initiation and RNA processing in the mitochondria of the red alga Chondrus crispus: convergence in the evolution of transcription mechanisms in mitochondria. J Mol Biol 283:549–557
Robbens S, Derelle E, Ferraz C, Wuyts J, Moreau H, Van de Peer Y (2007) The complete chloroplast and mitochondrial DNA sequence of Ostreococcus tauri: organelle genomes of the smallest eukaryote are examples of compaction. Mol Biol Evol 24:956–968
Rodriguez-Ezpeleta N, Brinkmann H, Burger G, Roger AJ, Gray MW, Philippe H, Lang BF (2007) Toward resolving the eukaryotic tree: the phylogenetic positions of jakobids and cercozoans. Curr Biol 17:1420–1425
Schuster W, Brennicke A (1994) The Plant mitochondrial genome—physical structure, information content, RNA editing, and gene migration to the nucleus. Annu Rev Plant Physiol Plant Mol Biol 45:61–78
Sugiyama Y, Watase Y, Nagase M, Makita N, Yagura S, Hirai A, Sugiura M (2005) The complete nucleotide sequence and multipartite organization of the tobacco mitochondrial genome: comparative analysis of mitochondrial genomes in higher plants. Mol Genet Genomics 272:603–615
Terasawa K, Odahara M, Kabeya Y, Kikugawa T, Sekine Y, Fujiwara M, Sato N (2007) The mitochondrial genome of the moss Physcomitrella patens sheds new light on mitochondrial evolution in land plants. Mol Biol Evol 24:699–709
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Tian XJ, Zheng J, Hu SN, Yu J (2006) The rice mitochondrial genomes and their variations. Plant Physiol 140:401–410
Turmel M, Lemieux C, Burger G, Lang BF, Otis C, Plante I, Gray MW (1999) The complete mitochondrial DNA sequences of Nephroselmis olivacea and Pedinomonas minor: two radically different evolutionary patterns within green algae. Plant Cell 11:1717–1729
Turmel M, Otis C, Lemieux C (2002a) The chloroplast and mitochondrial genome sequences of the charophyte Chaetosphaeridium globosum: insights into the timing of the events that restructured organelle DNAs within the green algal lineage that led to land plants. Proc Natl Acad Sci USA 99:11275–11280
Turmel M, Otis C, Lemieux C (2002b) The complete mitochondrial DNA sequence of Mesostigma viride identifies this green alga as the earliest green plant divergence and predicts a highly compact mitochondrial genome in the ancestor of all green plants. Mol Biol Evol 19:24–38
Turmel M, Otis C, Lemieux C (2003) The mitochondrial genome of Chara vulgaris: insights into the mitochondrial DNA architecture of the last common ancestor of green algae and land plants. Plant Cell 15:1888–1903
Turmel M, Otis C, Lemieux C (2007) An unexpectedly large and loosely packed mitochondrial genome in the charophycean green alga Chlorokybus atmophyticus. BMC Genomics 8:137
Unseld M, Marienfeld JR, Brandt P, Brennicke A (1997) The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366, 924 nucleotides. Nat Genet 15:57–61
van den Hoek C, Mann DG, Jahns HM (1995) Algae: an introduction to phycology. Cambridge University Press, Cambridge, MA
Vangerow S, Teerkorn T, Knoop V (1999) Phylogenetic information in the mitochondrial nad5 gene of pteridophytes: RNA editing and intron sequences. Plant Biol 1:235–243
Wellman CH, Osterloff PL, Mohiuddin U (2003) Fragments of the earliest land plants. Nature 425:282–285
Wolff G, Plante I, Lang BF, Kuck U, Burger G (1994) Complete sequence of the mitochondrial DNA of the chlorophyte alga Prototheca wickerhamii: gene content and genome organization. J Mol Biol 237:75–86
Acknowledgments
We thank David K. Smith and Ken D. McFarland for helping obtaining the plant material; Jeffrey D. Palmer for making useful suggestions; and Jiayu Xue for assisting with data analyses. This work was supported by a National Science Foundation Early Career Award DEB 0332298 to Y.-L.Q. The M. aenigmaticus mitochondrial genome sequence reported in this study has been deposited in GenBank under accession number EU660574.
Author information
Authors and Affiliations
Corresponding author
Additional information
Libo Li and Bin Wang contributed equally to this work.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Li, L., Wang, B., Liu, Y. et al. The Complete Mitochondrial Genome Sequence of the Hornwort Megaceros aenigmaticus Shows a Mixed Mode of Conservative Yet Dynamic Evolution in Early Land Plant Mitochondrial Genomes. J Mol Evol 68, 665–678 (2009). https://doi.org/10.1007/s00239-009-9240-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-009-9240-7