Abstract
Key insight into the complexities of apoptosis may be gained from the study of its evolution in lower metazoans. In this study we describe two genes from a cnidarian, Aiptasia pallida, that are homologous to key genes in the apoptotic pathway from vertebrates. The first is a novel ancient caspase, acasp, that displays attributes of both initiator and executioner caspases and includes a caspase recruitment domain (CARD). The second, a Bcl-2 family member, abhp, contains a BH1 and BH2 domain and shares structural characteristics and phylogenetic affinity with a group of antiapoptotic Bcl-2s including A1 and Bcl-2L10. The breadth of occurrence of other invertebrate homologues across the phylogenetic trees of both genes suggests that the complexity of apoptotic pathways is an ancient trait that predates the evolution of vertebrates and higher invertebrates such as nematodes and flies. This paves the way for establishing new lower metazoan model systems for the study of apoptosis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Programmed cell death (PCD) is a highly conserved mechanism of cell deletion that destroys redundant, dysfunctional, damaged, and diseased cells. This intrinsic process is of fundamental importance in the development, growth, health, and tissue homeostasis of multicellular organisms. The term apoptosis was initially used to describe morphological characteristics of an intrinsic mammalian cell death pathway (Wyllie et al. 1980) in response to cytotoxic agents and is different from developmental PCD (Hengartner and Bryant 2000). Although apoptosis was initially attributed to mammals only, it was the discovery of the key PCD genes governing cell death in the nematode Caenorhabditis elegans, that launched the modern field of apoptosis and PCD research underscoring the ancestral nature of these pathways (Ellis et al. 1991; Yuan et al. 1993). Recently, identification of homologues to apoptotic genes and signaling pathways across a broad range of animals, plants, and fungi suggests that apoptosis predates the evolution of animal multicellularity (Lamkanfi et al. 2002; Uren et al. 2000).
Apoptosis occurs in three interconnected stages: initiation, execution, and cell deletion. Central to the entire complex process are the cysteine-dependent aspartate specific proteases or caspases, which are activated in an amplifying proteolytic cascade by cleaving and activating one another (Fuentes-Prior and Salvesen 2004). Once activated, they proceed to cleave other proteins in the cell. Caspases are synthesized as single inactive zymogens comprised of an N-terminal prodomain of varying length, followed by large and small subunit domains approximately 20 and 10 kDa in size, respectively (Donepudi and Grütter 2002; Nicholson 1999). To date, mammalian caspases have been identified in three distinct subfamilies: the initiator, executioner, and cytokine activator caspases. The initiator caspases, 2, 8, 9, and 10, have a large, 130–219-amino acid (aa) prodomain that contains either a caspase recruitment domain (CARD; in caspases 2 and 9) or death effector domains (DEDs; in caspases 8 and 10) that engage in protein-protein interactions important to the activation process. In contrast, executioner caspases, 3, 6 and 7, have a small prodomain, consisting of only 23–28 aa, and lack a CARD or DEDs (Srinivasula et al. 2001). The cytokine activator caspases, 1, 4, 5, 11, and 12, have an 80 to 121-aa prodomain that contains a CARD but these proteins do not function directly in apoptosis.
In mammals, the cell death cascade begins with initiator caspase activation in response to either extrinsic or intrinsic signals, via two distinct but interrelated pathways. The extrinsic pathway can begin with binding of an external messenger, such as tumor necrosis factor or CD95/Fas ligand, to a membrane-bound receptor, which in turn activates an initiator caspase through protein-protein interactions of CARDs or DEDs (Ashkenazi and Dixit 1998; Green and Ferguson 2001; Suryaprasad and Prindiville 2003). Alternatively, membrane-bound initiation can be bypassed through granzyme B perforin activation of executioner caspases (MacDonald et al. 1999). In vertebrates, intrinsically mediated initiation begins with mitochondrial membrane disruption resulting in cytochrome c (cyt c) release. Cyt c binds Apaf-1, which in turn binds initiator procaspase 9 in a CARD-CARD interaction causing activation (Droin and Green 2004). Once activated, the initiator caspases activate executioner caspases. Finally, the executioners cleave a variety of cytoskeletal elements and cell adhesion molecules throughout the cell and within the nucleus, including proteins such as the DNA repair enzyme poly(ADP)-ribose polymerase (Li and Darzynkiewicz 2000) and inhibitory domains of endonucleases such as ICAD/DFF45, which initiates cleavage of nuclear material (Woo et al. 2004).
Regulation of cell death initiation and activation is governed by the protein-protein interactions of anti- and pro-apoptotic members of the B-cell lymphoma 2 (Bcl-2) protein family (Adams and Cory 1998; Reed 1998). To date, 12 homologues have been described in mammals. All Bcl-2s contain various combinations of four highly conserved BH death domains depending on function (Droin and Green 2004). Some members are extrinsic membrane proteins and contain a transmembrane domain near the C-terminus. Pro- and anti-apoptotic Bcl-2 heterodimerization can be concentration-dependent and a controlling influence on the decision by a cell to survive or die (Itoh et al. 2003; Oltvai et al. 1993).
The phylogeny and classification of caspases and Bcl-2s, based on gene sequence, protein structure, and function, have focused almost entirely on genes and proteins from vertebrates, the arthropod Drosophila melanogaster and the nematode C. elegans. Recent genomic studies of cnidarians indicate that cnidarian genomes contain many genes previously considered to be vertebrate innovations because of their absence from the Drosophila or Caenorhabditis genomes (Kortschak et al. 2003; Kuo et al. 2004). Further, gene diversity in the genomes of lower metazoans is much higher than previously predicted and some derived lineages such as flies and nematodes have a lower gene family diversity than lower metazoans (Kusserow et al. 2005). Both of these new insights underscore the need for characterization of apoptotic genes and pathways in these more ancestral groups. Such studies would provide insight into the evolution of these complex pathways.
The description of apoptosis and its encoding genes in lower metazoans is in an early phase of discovery. Apoptotic cell death has been observed in two classes of cnidarians: the hydrozoans H. vulgaris (Cikala et al. 1999) and H. oligactis (Kuznetsov et al. 2002) and the anthozoans Haliplanella lineata (Mire and Venable 1999) and Aiptasia pallida (Dunn et al. 2002, 2004). Caspases have been described in the sponge Geodia cydonium (Wiens et al. 2003) and in the cnidarian H. vulgaris (Cikala et al. 1999). Bcl-2-like cDNAs and expressed sequences tags (ESTs) have also been identified in sponges (Wiens et al. 2000) and hydroid cnidarians, respectively (GenBank accession nos. gi31054993 and gi37511437). In this study we build on previous studies of apoptosis in A. pallida by characterizing a caspase and a Bcl-2 family member, and place them in an evolutionary context by performing phylogenetic analyses of both sequences.
Materials and Methods
Cloning of Aiptasia pallida Caspase acasp and Bcl-2 Family Member abhp Sequences from cDNA Using RT-PCR
To isolate acasp and abhp sequences from Aiptasia pallida cDNA, a series of degenerate primers was designed from known sequences from hydrozoans. For acasp, an initial RT-PCR with primers designed from the Hydra vulgaris caspase 3B sequence (GenBank accession no. AAF98012), 5′-ATTCAAGCATGTCGNGG-3′ and 5′-CATTAATAAACATGANCC-3′, produced a 269-bp product (nucleotides 925–1194) which was then ligated into a pGEMT vector (Promega). Following amplification in DH5α Escherichia coli cells (Invitrogen), the plasmid DNA was sequenced with a Terminator v3.1 cycle sequencing kit (BigDye) using a 3100 Genetic Analyzer (ABI Prism), 3100 data collection software v.1.1, and DNA sequencing analysis software v.3.7 (ABI Prism). Specific primers 5′-GACATCTTCGTCTCTT-3′ and 5′-ACAACTGCGTT GACTACGTG-3′ were then designed from the sequenced product and used in a nested RT-PCR in combination with a 5′RLM-RACE kit (Ambion) to generate a 1026 bp product (nucleotides 1–1026). The 3′ end (nucleotides 408–1482) was then amplified using a RT-PCR and primers 5′-GTCCATCCAAGGTCCGTTGA-3′ and 5′-GCCGAATTCT15-3′. Both the 5′ and the 3′ PCR products were cloned and sequenced as described above. A minimum of three clones was sequenced for each cloned fragment, and a single contiguous sequence of 1482 nucleotides was generated using MacDNASIS Pro V3.6 (Hitachi Software). Primers specifically designed from the contiguous sequence 5′-ATGGAACAAAT ACACAGAGACGC-3′ and 5′-TTAATTGTTGCCGTGGT GATGTT-3′ were used to check the generated sequence.
For abhp, an initial RT-PCR with degenerate primers 5′-AAYT GGGGNMGNGTNGTT-3′ and 5′-AAYTCGTCCCANCCNCC-3′ designed from the BH1 and BH2 domains of the Hydra magnipapillata putative Bax sequence (accession no. CD267166) produced a 159-bp product (nucleotides 293–452) which was amplified and sequenced using the methods as above. The 3′ end (nucleotides 379–609) was amplified using a nested RT-PCR with specifically designed forward primers 5′-CACACTTTCGGAG CAAAGGT-3′ and 5′-GCGAAAAAGTCCGACTGA-3′ and reverse primer 5′-GCCGAATTCT15-3′. Specific primers 5′-ATCC AGTCGGACTTTTTCG-3′ and 5′-ACCTTTGCTCCGAAAG TGTG-3′ designed from the sequenced product were used in a nested RT-PCR with a 5′RLM-RACE kit (Ambion) to generate a 335-bp product (nucleotides 1–335). The PCR products were cloned and sequenced as described above. A minimum of three clones was sequenced for each cloned fragment, and a single contiguous sequence was generated. Primers designed from the contiguous sequence 5′-ATGCCACCGATAATACCTGA-3′ and 5′-AGTGTTCATTCAAACTTACCA-3′ were used to check the generated sequence of the full open reading frame.
Phylogenetic and Structural Analyses of Sequences
The consensus sequences of the acasp caspase (large and small subunit combined), the acasp prodomain and CARD, and abhp were compared with known protein sequences using Blast and PSIBlast network databases at the National Center for Biotechnology Information (NCBI). The caspase, the caspase prodomain, and abhp sequences were submitted as separate queries to the NCBI database. Returned sequences against queries were accepted for alignment analysis on the basis of >25% of 80+ residue identity (Sander and Schneider 1991), in combination with an expect value (E) threshold lower than 10−4 (NCBI). Invertebrate caspases that were in the database but that did not meet these criteria were not included in the alignments or analyses. As part of the abhp analysis, the EST putative sequences for the Bcl-2 protein family members Bax and Bak from Hydra magnipapillata were added to give a cnidarian representation. Multiple alignments of predicted amino acid sequences were performed with ClustalX v. 1.81(Thompson et al. 1997), edited manually, and back translated with BioEdit v.5.06 (Hall 1999). Phylogenetic comparisons were performed using weighted parsimony (WP) and Bayesian analyses. For WP analyses, step matrices for the six bidirectional nucleotide transformations were constructed based on the −ln of pairwise base frequencies using the program STMatrix (Francois Lutzoni and Stefan Zoller, Department of Biology, Duke University). Analyses were performed using PAUP v.4.0b10 (Swofford 2004) on parsimony informative characters with 100 heuristic replicates of random sequence addition with tree bisection-reconnection branch swapping, MULTREES ON, and gaps treated as missing. Nodal support was estimated with 10,000 replicates of nonparametric bootstrapping with the same search options described above. Bayesian Metropolis coupled Markov chain Monte Carlo (B-MCMCMC) analyses were conducted using Mr. Bayes 3.0 (Huelsenbeck 2001). All searches were conducted using four chains for a total of 10,000,000 generations, with phylogenetic trees sampled every 100 generations. A total of three independent 10,000,000 generation analyses were conducted to verify likelihood convergence and burn-in parameter. In estimating the likelihood of each tree, we used the general time-reversible model with invariant sites and gamma distribution (GTR+I+Γ) and employed this model separately for each codon position resulting in three data partitions per analysis. Nodal support in B-MCMCMC analyses was estimated as posterior probabilities calculated from the posterior distribution of trees excluding burn-in trees (Hulsenbeck and Rannala 2004). To predict the secondary structure of the caspase, caspase CARD domain, and acasp, the amino acid sequences were analyzed by PSIPRED (Jones 1999; McGuffin et al. 2000) and GenTHREADER servers (McGuffin and Jones 2003). The predicted structures were then compared and aligned with known structures using the NCBI molecular modeling database (MMDB) in conjunction with Cn3D v4.1 (NCBI) software. Peptide mass calculations were performed with the Expert Protein Analysis System (ExPASY) (Wilkins et al. 1997).
Results
Caspase and CARD Domain Structure and Phylogeny
The Aiptasia pallida putative caspase, acasp, is comprised of 1482 nucleotides encoding 415 aa, with an approximate molecular weight of 46.3 kDa (Fig. 1; GenBank accession no. DQ218058). Sequence analysis of acasp reveals a prodomain and a caspase large and small subunit. The large prodomain includes a characteristic CARD domain sequence between aa 1 and aa 88, with an approximate molecular weight of 9.7 kDa (see ahead for detailed analysis of the CARD). The boundary between the prodomain and the large subunit is unknown. There is a possible prodomain linker between the CARD and the large subunit that would be cleaved during activation, but the cleavage site is not clear. The large subunit could therefore range in size from 15.4 to 23.3 kDa and ends at aa 300. There are similarities in the primary structure of acasp to executioner caspases including amino acids in acasp in similar positions to R179, H237, and C285 in humans, which are key to executioner activation (Fig. 1). C295 in acasp, corresponding to C285 in humans, occurs within the highly conserved QAC*G pentapeptide motif, critical to substrate binding and catalysis at the adjacent P1 Asp-X binding site (Donepudi and Grütter 2002; Fuentes-Prior and Salvesen 2004; Nicholson 1999; Srinivasula et al. 2001). A 14-aa linker region follows the large subunit and contains two potential Asp (P1) tetrapeptide maturation cleavage sites, DGVD or DATD. The small subunit, spanning aa 315–aa 415, with an approximate molecular weight of 23.4 kDa, contains the highly conserved SWR and GSWFI motifs important in substrate binding and typical of executioner caspase 3s (Nicholson and Thornberry 1997; Piana et al. 2003).
The nucleotide, predicted amino acid sequence, and secondary structure of Aiptasia pallida caspase, acasp (GenBank accession no. DQ218058). aa 1–89•, prodomain with CARD; aa 89•–300•, potential linker and large subunit region; aa 315•–415, small subunit;  , highly conserved QACxG (QACQR) caspase motif;
, highly conserved QACxG (QACQR) caspase motif;  , proposed maturation substrate binding site region;
, proposed maturation substrate binding site region;  , α-helix;
, α-helix;  , β strand as determined by PSIPRED. Key β-strands and α-helix to structure function are labeled a–e and 1–5, respectively.
, β strand as determined by PSIPRED. Key β-strands and α-helix to structure function are labeled a–e and 1–5, respectively.
PSIPRED and GenTHREADER analysis of acasp returned a predicted structure match to caspase 3/CPP32 (E = 3e-05 [Okamoto et al. 1999; Rotonda et al. 1996]), interleukin-1β (IL1-β/caspase 1; (E = 3e-05 [Okamoto et al. 1999]), and caspase 2 (E=0.0005 [Schweizer et al. 2003]). The acasp predicted secondary structure displayed five key parallel and one antiparallel β-strand and five helices in the same positions as mammalian caspase structures (Fig. 1), which are essential to the twisted β-sheet/helix tertiary structure and to the function of active caspases (Donepudi and Grütter 2002). Key residues of known caspase active binding sites, R179, H237, and C285 (Fuentes-Prior and Salvesen 2004), were located in similar positions within the acasp structure. R199 is in a reciprocated loop position between βa and α1, H 256 is between βc and α3, and C295 is part of the highly conserved catalytic peptide in the linker between βd and βe of the large and small subunit (Fuentes-Prior and Salvesen 2004, Fig. 1).
Phylogenetic analyses of acasp and caspases from other organisms, including relevant caspase sequences from invertebrates, to represent the known higher-level diversity of animal caspases, are shown in Fig 2. Trees from the two different analyses were highly congruent except for placement of some of the invertebrate homologues (see below). Overall, orthologues from the major groups of animals sampled (e.g., invertebrates, mammals) grouped together within either a large caspase 3, 6, and 7 executioner clade, a cytokine clade of caspases 1, 4, and 11, or three initiator clades, one of caspase 9s, one of 8s and 10s, and one of 2s (Fig. 2). Both WP and B-MCMCMC analyses grouped acasp with caspase 3B from another cnidarian, Hydra vulgaris, on a basal lineage of the executioner clade.
Phylogenetic tree of metazoan caspases including A. pallida acasp (large and small subunit only without the CARD domain) showing B-MCMCMC topology and posterior probabilities. Weighted parsimony (WP) bootstrap values are shown in parentheses. Nodes receiving bootstrap values of <50% are indicated by (#). Sequences from invertebrates are shown in boldface. The thin arrow indicates alternative placement of Geodia cydonium in the WP analysis. The block arrow points to node in the executioner clade at which the proposed loss of the CARD domain occurred. GenBank accession numbers for the sequences used in the multiple alignments and phylogenetic analyses are as follows. Caspase 1s: Mus musculus 266322, Rattus norvegicus 31542341, Xenopus laevis 2493523. Caspase 2s: Gallus gallus 1490878, Homo sapiens 37889083, Mus musculus 6680848. Caspase 3s: Canis familiaris 20975412, Danio rerio 1888385, Gallus gallus 45382613, Geodia cydonium 27526602, Homo sapiens 1651687, 3a Hydra vulgaris 9754624, 3b Hydra vulgaris 9754622, Mus musculus 6753284, 3a Oryzias latipes 21623673, 3b Oryzias latipes 2162371, Rattus norvegicus 6978605. Caspase 3/7: Branchiostoma floridae 24078495. Caspase 4s: Homo sapiens 4502577, Mus musculus 38512079. Caspase 5: Homo sapiens 4757914. Caspase 6s: Branchiostoma floridae 24078497, Homo sapiens 14916483, Gallus gallus 45382021, Mus musculus 31981865, Oncorhynchus mykiss 8163768, Rattus norvegicus 13929092. Caspase 7s: Homo sapiens 1730093, Mus musculus 6680850, Xenopus laevis 7619904. Caspase 8s: Danio rerio 188583387, Homo sapiens 15718708, Mus musculus 4138211, Rattus norvegicus 11560103, Xenopus laevis 7619906. Caspase 9s: Gallus gallus 16555409, Homo sapiens 30584067, Rattus norvegicus 13928812, Xenopus laevis 7619908. Caspase 10s: Homo sapiens 47419841, Xenopus laevis 7619910. Caspase 11s: Mus musculus 2094804, Rattus norvegicus 16758556. Others: caspase Suberites domuncula 27528704, Ced 3 Caenorhabditis elegans 1753940, Ced 3 Pristionchus pacificus 5305337, DECAY Drosophila melanogaster 17137722, DREDD Drosophila melanogaster 24638925, DRICE Drosophila melanogaster 1929040, STRICA Drosophila melanogaster 19921676.
Other invertebrate homologues were distributed throughout the trees and in some cases were resolved differently by WP and B-MCMCMC analyses, highlighting the complexity and diversity of the system. Another homologue from H. vulgaris, caspase 3A, resolved as an isolated lineage that did not display any direct or close phylogenetic affinity to any clades of other caspases. In the B-MCMCMC analyses, caspase 3 from the sponge Geodia cydonium also resolved as an isolated lineage (Fig. 2) but grouped with the initiator caspase 9s in the WP analysis (data not shown). A caspase from another sponge, Suberites domuncula, resolved in the initiator caspase 8 and 10 clade in the B-MCMCMC analyses but resolved with two D. melanogaster homologues by WP with very weak support (data not shown). From the higher invertebrates, two sequences from nematodes grouped outside of the executioner clade. Ced3 from C. elegans grouped with caspase 2 orthologues, and ced3 from Pristionchus pacificus branched early off of the cytokine activator/caspase 2 initiator cluster. In contrast, all four caspase homologues from D. melanogaster grouped in a single cluster within the executioner clade and diverged from vertebrates after the cnidarian sequences according to the Bayesian analysis. WP provided poor resolution for the four fly homologues, with two occurring in a grouping with S. domuncula and the other two forming single branches (data not shown).
In stark contrast to results from the large and small subunit of acasp grouping with executioner caspases, all of the 30 significant PSIBlast hits returned against the acasp CARD sequence were from vertebrate initiator complex proteins and initiator caspases, the highest 12 of which are reported in Table 1. There were no significant returned hits associated with cytokine or executioner caspase prodomains, nor were there any from invertebrate sequences. Phylogenetic analysis of the acasp CARD domain with CARDs from other caspases was not performed due to the short length of the domain.
As with the results from the blast searches, the predicted structure of the acasp CARD domain suggests that its structure and function are similar to those of CARD domains in apoptosis initiator protein complexes and procaspases. PSIPRED and GenTHREADER analyses indicated a predicted acasp CARD structure of six helices closely resembling the seven-helix human Apaf-1 CARD (E = 0.0002 [Vaughn et al. 1999]), human Apaf-1 CARD in complex with caspase 9 (E = 0.0003 [Qin et al. 1999]), human IL1-β (caspase 1) CARD (E= 0.0003 [Humke et al. 2000]), and the six-helix human RAIDD CARD adaptor complex (E = 0.014 [Chou et al. 1998]). The structure of the Apaf-1 CARD in complex with procaspase 9 is different from that of the RAIDD CARD in that the Apaf-1 CARD helix 1 (H1) is bent to such a degree that the disruption of the bonds divides it into two small helices (H1a and b [Qin et al. 1999]). In the acasp CARD structure, this split in H1 is not evident, therefore H1 appears to be a single helix like the RAIDD CARD. The hydrophobic amino acids responsible for the globular central core of the structure and critical for CARD/CARD interactions were well conserved between the acasp CARD and the Apaf-1 CARD in complex with procaspase 9 and RAIDD CARDs (Chou et al. 1998; Qin et al. 1999) (supplementary Fig.1a). Specifically, identical amino acids important to structure and interactions, R13 L56, and L60 in the Apaf-1 CARD, are present in the acasp CARD (supplementary 1a), and likewise R10, R13, L20, L35, T48, R52, and G64 in the RAIDD CARD are found in the acasp CARD (supplementary 1b). The amino acids R10, R13, and R52 found in the acasp CARD are also known to be important to the CARD of procaspase 9 and its interaction with Apaf-1 (Qin et al. 1999).
Bcl-2 Family Member Structure and Phylogeny
The Aiptasia pallida putative Bcl-2 family member, abhp, is comprised of 609 nucleotides encoding 155 aa, with an approximate molecular weight of 17.6 kDa (Fig. 3; GenBank accession no. DQ211980). The most significant of the 118 returned PSIBlast hits against abhp were from vertebrate anti-apoptotic Bcl-2 family members—Mcl-1, A1, R1, Bcl212, Bcl-W, and Bcl-x —and the pro-apoptotic Bak (data not shown). Anti-apoptotic Bcl-2-like 10 (Bcl-2L10) returned the highest identity, 42%, and a similarity of 58%. Sequence analysis of abhp indicated the presence of two BH death domains which correspond to the BH1 and BH2 death domains of the Bcl-2 protein family. There was no evidence of a BH3, BH4, or transmembrane domain. Based on the presence or absence of the four BH domains and the transmembrane domain, abhp most closely resembled anti-apoptotic A1, which, unlike all other Bcl-2 family members, also contains only BH1 and BH2 domains (Fig. 4).
The homologous BH1 domain, aa 91–113, contains a highly conserved NWGRIV motif and EL and FG components which are present in the BH1 domain of many orthologues (Fig. 5; Prosite PS1080 [Cepero et al. 2001]). The BH2 domain, aa 136–153, contains the highly conserved WI***GGW motif but shares little identity and similarity with other regions of orthologous genes (Fig. 5; Prosite PS01256 [Cepero et al. 2001]). In structural analyses, the predicted abhp secondary structure of six helices (pointed out in Fig. 3) was closely aligned with the known structures of BclxL (E = 3e-05 [Muchmore et al. 1996]), Bax (E = 3e-05 [Suzuki et al. 2000]), ced9 (E = 1e-04 [Yan et al. 2004]), and a Bcl-2 homologue from the Kaposi sarcoma virus (E = 0.0004 [Huang et al. 2002]).
Phylogenetic analyses of abhp and Bcl-2s from other organisms are shown in Fig. 6. Overall the Bayesian analyses provided strong support for more terminal clades but weak support for deep branching nodes. The WP analysis replicated the topography of the Bayesian tree but with stronger support of the internal nodes. There were three well-supported clades, comprised of (1) pro-apoptotic Bax orthologues, (2) anti-apoptotic Bcl-2 and Bclx orthologues, and (3) pro-apoptotic Bak orthologues. The A. pallida abhp fell within a fourth, poorly supported group that included anti-apoptotic members. In the Bayesian analysis, abhp grouped with anti-apoptotic Bcl-2L10 from R. norvegicus but with weak support. However, in the WP analysis, abhp resolved as a single lineage within the same divergent anti-apoptotic cluster. Bcl-2 homologues from C. elegans and D. melanogaster were not included in the analysis because their alignment with abhp fell below the threshold for inclusion in the analysis (see Materials and Methods).
Phylogenetic tree of metazoan Bcl-2 family members, including A. pallida abhp, showing B-MCMCMC topology and posterior probabilities. WP bootstrap values are shown in parentheses. Nodes receiving bootstrap values of <50% are indicated by (#). Sequences from invertebrates are shown in boldface. GenBank accession numbers for the sequences used in the multiple alignments and phylogenetic analyses are as follows: A1 Gallus gallus 45382371, Bak1 Hydra magnipapillata 37511437, Bak2 Hydra magnipapillata 52859456, Bak Homo sapiens 4502363, Bak Ovis aries 9621788, Bak Mus musculus 6671612, Bak Rattus norvegicus 8050832, Bax1 Hydra magnipapillata 31054870, Bax 2 Hydra magnipapillata 31054993, Bax Canis familiaris 2732190, Bax Felis catus 19168496, Bax Homo sapiens 20631958, Bax Sus scrofa 38304060, Bcl2 Canis familiaris 40846417, Bcl2 Cricetulus longicaudatus 15213854, Bcl-2 Felis catus 25166611, Bcl2 Gallus gallus 231635, Bcl2 ‘like’ Danio rerio 18858349, Bclx Gallus gallus 539495, Bclx Oryctolagus cuniculus 9652565, Bclx Rattus norvegicus 1083601, Bclx Mus musculus 2493277, Bclxβ Mus musculus 2636676, Bclxγ Mus musculus 2636672, Bclx Ovis aries 9621786, Bclxl Felis catus 19570354, Bclx Homo sapiens 2118487, Bcl2L1 Homo sapiens 20336335, Bclx iso Homo sapiens 7438463, Bclxl Sus scrofa 69597767, BclW Homo sapiens 4757842, Bcl WEL Rattus norvegicus 32185285, Bcl2l2 Mus musculus 25955645, Bcl-2L10 Rattus norvegicus 16758550, BHP Suberites domuncula 9188120, GRS Homo sapiens 1694789, Mcl 1 Canis familiaris 23978940, Mcl 1 Danio rerio 35902998, Mcl1 Felis catus 32127949, Mcl1 Gallus gallus 4883769, Mcl1 Homo sapiens 476995, and Mcl1 Rattus norvegicus 453823.
Other invertebrate Bcl-2 sequences included in the analyses ranged widely across the tree. BHP from the sponge Suberites domuncula resolved as a single basal lineage in the Bayesian analyses but grouped in the poorly resolved anti-apoptotic clade containing abhp by WP analysis. Two Bak ESTs from the cnidarian Hydra magnipapillata grouped with other pro-apoptotic orthologues, however, two H. magnipapillata Bax ESTs fell into the weakly supported clade grouping closest to abhp and Bcl-2L10 from R. norvegicus but with very weak support.
Discussion
Acasp Has Characteristics of Both Executioner and Initiator Caspases
Acasp is an ancient caspase that has changed relatively little over time compared to homologues from other organisms (Fig. 2) and may more closely resemble a caspase precursor than caspases from other lower and higher metazoans. Acasp encodes a zymogen that contains a large and small subunit closely resembling an executioner caspase but a prodomain containing a CARD that resembles an initiator caspase. Phylogenetic analyses of the large and small subunit placed acasp within the executioner clade with strong support (Fig. 2).
In contrast, the acasp prodomain containing a CARD had the highest homology with initiator caspases 2 and 9 and not executioner caspases, which have short prodomains with no CARD (Table 1) (Nicholson and Thornberry 1997; Srinivasula et al. 2001). Further, the identity of key amino acids external to and within the multihelix structure between the acasp CARD and CARDs of Apaf-1, RAIDD (supplementary 1), and procaspase 9 suggests that the structure, and therefore likely the function, of the acasp CARD in CARD/CARD interactions is similar to those in higher metazoan caspase initiation (Chou et al. 1998; Hofmann and Bucher 1997; Qin et al. 1999; Vaughn et al. 1999). This indicates that elements of complexity of the protein/protein interactions that comprise the apoptosis cascade are ancient and conserved.
Based on the placement of A. pallida acasp, a CARD-containing putative executioner, at the base of the executioner clade, we hypothesize that the absence of CARD domains from executioner caspases from higher invertebrates and vertebrates is a derived loss (see block arrow; Fig. 2). This hypothesis suggests that caspase progenitors contained CARD domains that have been retained in the initiator and cytokine activator caspases but were lost early in the evolution of executioners. Acasp would therefore resemble such a progenitor. Additional caspase sequences from other lower metazoans will provide a test of this hypothesis.
The Relationship of acasp with Other Invertebrate Caspases
The phylogenetic analyses of acasp and other caspase homologues (Fig. 2) is the first extensive analysis of caspases to include cnidarian sequences. They showed both consistencies and inconsistencies with other investigations. Acasp grouped closely with caspase 3B from fellow cnidarian Hydra vulgaris within the executioner clade. The Cikala et al. (1999) study characterized both H. vulgaris 3A and 3B sequences as “3-like executioner caspases.” Our analyses agreed with this designation for 3B but not for 3A, which formed a single lineage outside of any subfamily. Therefore H. vulgaris caspases 3A and 3B could represent ancient paralogues. Although both H. vulgaris caspases were reported to have large prodomains, sequences were not published or included in the phylogenetic analysis (Cikala et al. 1999). It is possible that acasp and H. vulgaris 3B share not only homology to the executioner caspases in their large and small subunit domains, but also a large prodomain that may include a CARD. It would be interesting to compare the prodomains of these two cnidarian caspases to look for common, potentially ancestral qualities.
Other invertebrate caspases occurred at several locations across the tree. For example a caspase from the sponge Subertes domincula grouped with initiator caspases 8 and 10, one from the sponge Geodia cydonium formed a monophyletic group outside of any clade, ced3 from C. elegans grouped with the initiator caspase 2s, and all four D. melanogaster sequences fell into the executioner clade with acasp and H. vulgaris 3B. This surprising grouping of ced3 from C. elegans with inflammatory caspase 2s, despite its function as an executioner caspase, agrees with the Lamkanfi et al. (2002) study. The breadth of occurrence of invertebrate homologues suggests that the evolution of caspases and apoptosis occurred very early in metazoan evolution and that apoptotic pathways in lower invertebrates are likely to include a complexity of interacting genes that warrant further study.
Abhp Shares Characteristics with Some Anti-Apoptotic Bcl-2 Members
The comparative analysis of A. pallida abhp with other Bcl-2 family members is more difficult and less clear than the caspase comparisons because the Bcl-2 family encompasses a broad variety of genes whose structures and functions are still not completely understood. Abhp is a Bcl-2 family homologue that contains a BH1 and BH2 domain, and based on the presence of only these two domains, it most closely resembles the anti-apoptotic A1 (Fig. 4). The most significant blast hits and alignments of the highly conserved BH1 and BH2 domains also associate abhp with anti-apoptotic Bcl-2 family members including Bcl-2L10 from R. norvegicus, A1, and Mcl-1. The Bayesian analysis grouped abhp with Bcl-2L10 but with weak support (Fig. 6), and this relationship was not observed in the WP analysis. However, although Bcl-2L10 has BH1 and BH2 domains like abhp, it is the BH4 and transmembrane domains, both absent from abhp, which are key to the Bcl-2L10 function (Zhang et al. 2001). Therefore, although the sequence identity between Bcl-2L10 and abhp is the highest of any Bcl-2 homologue and they group together in one phylogenetic analysis, the protein architecture of the two is different.
Although all of the above-mentioned vertebrate Bcl-2 homologues are considered anti-apoptotic, their specific roles in the cell death cascade are highly diverse and often not completely understood. A1 is expressed specifically in hemopoietic macrophages, T-helper cells, and neutrophils (Lin et al. 1993) and, in mice, has been shown to accumulate in response to bacterial lipopolysaccharide. A1 homologues in humans, GRS (Kenny et al. 1997) and Bfl-1(Choi et al. 1997; Ko et al. 2003), are anti-apoptotic targets of NF-κB in response to pro-inflammatory TNFα stimulation (Zong et al. 1999). Mcl-1 is associated with differentiating cell lines and viability of differentiated cells (Kozopas et al. 1993; Leo et al. 1999). Bcl-2L10 blocks the intrinsic apoptotic pathway by preventing release of cyt c from mitochondria and is redundant with the extrinsically mediated pathway (Zhang et al. 2001). The Bcl-2L10 homologues, Diva and Boo, regulate cell death by forming a complex with Apaf-1 and caspase-9 (Inohara et al. 1998; Song et al. 1999).
The complexity of the Bcl-2 gene family and the lack of sequence data for the diverse set of homologues across taxa resulted in phylogenetic trees with several strongly supported terminal clades whose interrelationships, i.e., more basal nodes, are weakly supported (Fig. 6). Abhp grouped with anti-apoptotic Bcl-2L10, A1, and Mcl-1 in a weakly supported clade. More sequence data from across taxa are needed before the relationships within this group can be confidently resolved. In addition, further investigations into the function of abhp are required to determine whether it shares functional anti-apoptotic properties with other members of its clade.
There are very few invertebrate Bcl-2 homologues that shared enough sequence identity with abhp to be included in phylogenetic analyses. However, as with the invertebrate caspases, the broad occurrence of these invertebrate homologues across the trees suggests that the Bcl-2 family arose very early in metazoan evolution.
Conclusion
In summary, in this study we describe two novel components of the apoptotic pathway in the sea anemone Aiptasia pallida, a caspase, acasp, and a Bcl-2 family member, abhp. Both are highly similar to the corresponding vertebrate homologues in amino acid sequence as well as predicted protein structure. This corroborates early data from other lower metazoan groups that suggest that apoptosis genes and cellular pathways are ancient and highly conserved through metazoan evolution (Cikala et al. 1999; Wiens et al. 2003). The phylogenetic and structural analyses of these components are consistent with predicted protein structure and provide new insight into cnidarian apoptosis previously observed by Dunn et al. (2002, 2004) and highlight the value of using lower invertebrate models to understand the evolution of genetic and cellular processes in higher metazoans.
References
Adams JM, Cory S (1998) The Bcl-2 protein family: arbiters of cell survival. Science 281:1322–1326
Ashkenazi A, Dixit VM (1998) Death receptors: signaling and modulation. Science 281:1305–1308
Cepero E, Johnson BW, Boise LH (2001) Cloning and analysis of Bcl-2 family genes. In: Schwartz LM, Ashwell JD (eds) Apoptosis. Academic Press, San Diego, pp 29–47
Choi SS, Park SH, Kim UJ, Shin HS (1997) Bfl-1, a Bcl-2-related gene, is the human homolog of the murine A1, and maps to chromosome 15q24.3. Mamm Genet 8:781–782
Chou JJ, Matsou H, Duan H, Wagner G (1998) Solution structure of the RAIDD CARD and model for CARD/CARD interaction in caspase-2 and caspase-9 recruitment. Cell 94:171–180
Cikala M, Wilm B, Hobmayer E, Böttger A, David CN (1999) Identification of caspases and apoptosis in the simple metazoan Hydra. Curr Biol 9:959–962
Donepudi M, Grütter MG (2002) Structure and zymogen activation of caspases. Biophys Chem 101–102:145–153
Droin NM, Green DR (2004) Role of Bcl-2 family members in immunity and disease. Biochim Biophys Acta 1644:179–188
Dunn SR, Thomason JC, Le Tissier MDA, Bythell JC (2002) Detection of cell death activity during bleaching of the symbiotic sea anemone Aiptasia sp. In: Kasim Moosa MK, Soemodihardjo S, Nontji A, Soegiarto A, Romimohtarto K, Sukarno S (eds) Proceedings of the Ninth International Coral Reef Symposium. Indonesian Institute of Sciences in cooperation with the State Ministry for Environment, Bali, Indonesia, pp 145–155
Dunn SR, Thomason JC, Le Tissier MDA, Bythell JC (2004) Heat stress induces different forms of cell death in sea anemones and their endosymbiotic algae depending on temperature and duration. Cell Death Diff 11:1213–1222
2llis RE, Yuan J, Horvitz HR (1991) Mechanisms and functions of cell death. Annu Rev Cell Biol 7:663–698
Fuentes-Prior P, Salvesen GS (2004) The protein structures that shape caspase-activity, specificity, activation and inhibition. Biochem J 384:201–232
Green DR, Ferguson TA (2001) The role of Fas ligand in immune privilege. Nature Rev Mol Cell Biol 2:917–924
Hall TA (1999) Bioedit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hengartner MO, Bryant JA (2000) Apoptotic cell death: from worms to wombats...but what about the weeds? In: Bryant JA, Hughes SG, Garland JM (eds) Programmed cell death in animals and plants. BIOS Scientific, Oxford, pp 1–9
Hofmann K, Bucher P (1997) The CARD domain: a new apoptotic signaling motif. Trends Biol Sci 22:155–156
Huang Q, Petros AM, Virgin HW, Fesik E, Olejniczak T (2002) Solution structure of a Bcl-2 homolog from Kaposi’s sarcoma virus. Proc Nat Acad Sci USA 99:3428–3433
Huelsenbeck JP (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755
Hulsenbeck JP, Rannala B (2004) Frequentist properties of Bayesian posterior probabilities of phylogenetic trees under simple and complex substitution models. Syst Biol 53:904–913
Humke EW, Shriver SK, Starovasnik MA, Fairbrother WJ, Dixit VM (2000) Iceberg; a novel inhibitor of interleukin-1beta generation. Cell 103:99–111
Inohara N, Gourley TS, Carrio R, Muniz M, Merino J, Garcia I, Koseki T, Hu Y, Chen S, Nenez G (1998) Diva, a Bcl-2 homologue that binds directly to Apaf-1 and induces BH3-independent cell death. J Biol Chem 273:32479–32486
Itoh T, Itoh A, Pleasure D (2003) Bcl-2 related protein family gene expression during oligodendroglial differentiation. J Neurochem 85:1500–1512
Jones DT (1999) PSIPRED secondary structure prediction based on position-specific scoring matrices. J Mol Biol 292:195–202
Kenny JJ, Knobloch TJ, Augustus M, Carter KC, Rosen CA, Lang JC (1997) GRS, a novel member of the Bcl-2 gene family, is highly expressed in multiple cancer cell lines and in normal leukocytes. Oncogene 14:997–1001
Ko JK, Lee MJ, Cho SH, Cho JA, Lee BY, Koh JS, Lee SS, Shim YH, Kim CW (2003) Bfl-1, a novel alternative splice variant of Bfl-1, localises in the nucleus via its C-terminus and prevents cell death. Oncogene 22:2457–2465
Kortschak DR, Samuel G, Saint R, Miller DJ (2003) EST analysis of the cnidarian Acropora millepora reveals extensive gene loss and rapid sequence divergence in the model invertebrates. Curr Biol 13:2190–2195
Kozopas KM, Yang T, Buchan HL, Zhou P, Craig RW (1993) MCL1, a gene expressed in programmed myeloid cell differentiation, has sequence similarity to Bcl2. Proc Natl Acad Sci USA 90:3516–3520
Kuo J, Chen M-C, Lin C-H, Fang LS (2004) Comparative gene expression in the symbiotic and aposymbiotic Aiptasia pulchella by expressed sequence tag analysis. Biochem Biophys Res Commun 318:176–186
Kusserow A, Pang K, Sturm C, Hrouda M, Lentfer J, Schmidt HA, Technau Y, Haeseler Av, Hobmayer B, Martindale MQ, Holstein TW (2005) Unexpected complexity of the Wnt gene family in a sea anemone. Nature 433:156–160
Kuznetsov SG, Anton-Erxleben F, Bosch TCG (2002) Epithelial interactions in Hydra: apoptosis in interspecies grafts is induced by detachment from the extracellular matrix. J Exp Biol 205:3809–3817
Lamkanfi M, Declercq W, Kalai M, Saelens X, Vandenabeele P (2002) Alice in caspase land. A phylogenetic analysis of caspases from worm to man. Cell Death Differ 9:358–361
Leo CP, Hsu SY, Chun SY, Bae HW, Hsueh AJ (1999) Characterisation of the anti-apoptotic Bcl-2 family member myeloid cell leukemia-1 (Mcl-1) and the stimulation of its message by gonadotropins in the rat ovary. Endocrinology 140:5469–5477
Li X, Darzynkiewicz Z (2000) Cleavage of Poly(ADP-ribose) polymerase measured in situ in individual cells: relationship to DNA fragmentation and cell cycle position during apoptosis. Exp Cell Res 255:125–132
Lin EY, Orlofsky A, Berger MS, Prystowsky MB (1993) Characterization of A1, a novel hemopoietic-specific early response gene with sequence similarity to bcl-2. J Immunology 151:1979–1988
MacDonald G, Shi LF, Velde CV, Lieberman J, Greenberg AH (1999) Mitochondria-dependent and independent regulation of granzyme B-induced apoptosis. J Exp Med 189:131–143
McGuffin LJ, Jones DT (2003) Improvement of the GenTHREADER method for genomic fold recognition for genomic sequences. Bioinformatics 19:874–881
McGuffin LJ, Bryson K, Jones DT (2000) The PSIPRED protein structure prediction server. Bioinformatics 16:404–405
Mire P, Venable S (1999) Programmed cell death during longitudinal fission in a sea anemone. Invert Biol 118:319–331
Muchmore SW, Sattler M, Liang H, Meadows RP, Harlan JE, Yoon HS, Nettlesheim D, Chang BS, Thompson CB, Wong SL, Ng SL, Fesik SW (1996) X-Ray and NMR structure of human Bcl-xL, an inhibitor of programmed cell death. Nature 381:335–341
Nicholson DW (1999) Caspase structure, proteolytic substrates, anf function during apoptotic cell death. Cell Death Differ 6:1028–1042
Nicholson DW, Thornberry NA (1997) Caspases: killer proteases. Trends Biol Sci 22:299–306
Okamoto Y, Anan H, Nakai E, Morihira K, Kurihara H, Sakashita H, Terai Y, Takeuchi M, Shibanuma T, Isomura Y (1999) Peptide based interleukin-1beta converting enzyme (ICE) inhibitors: synthesis, structure, activity relationship and crystallographic study of the inhibitor complex. Chem Pharm Bull 47:11–21
Oltvai ZN, Milliman CL, Korsmeyer SJ (1993) Bcl-2 heterodmerizes in-vivo with a conserved homolog, Bax that accelerates programmed cell death. Cell 74:609–619
Piana S, Sulpizi M, Rothlisberger U (2003) Structure-based thermodynamic analysis of caspases reveals key residues for dimerization and activity. Biochemistry 42:8720–8728
Qin H, Srinivasula MS, Wu G, Fernandes-Alnemri T, Alnemri ES, Shi Y (1999) Structural basis of pro-caspase-9 recruitment by apoptotic protease-activating factor 1. Nature 399:549–557
Reed JC (1998) Bcl-2 family proteins. Oncogene 17:3225–3236
Rotonda J, Nicholson DW, Fazil KM, Gallant M, Labelle M, Peterson EP, Rasper DM, Ruel R, Vaillancourt JP, Thornberry NA, Becker JW (1996) The three dimensional structure of apopain/CPP32, a key mediator of apoptosis. Nat Struct Biol 3:619–625
Sander C, Schneider R (1991) Database of homology-derived protein structures and the structural meaning of sequence alignment. Proteins Struct Funct Genet 9:56–68
Schweizer A, Briand C, Grutter MG (2003) Crystal structure of caspase-2, apical initiator of the intrinsic apoptotic pathway. J Biol Chem 278:42441–42447
Song Q, Kuang Y, Dixit VM, Vincenz C (1999) Boo, a novel negative regulator of cell death interacts with Apaf-1. EMBO J 18:167–178
Srinivasula MS, Saleh A, Ahmad M, Fernandes-Alnemri T, Alnemri ES (2001) Isolation and assay of caspases. In: Schwartz LM, Ashwell JD (eds) Apoptosis. Academic Press, San Diego
Suryaprasad AG, Prindiville T (2003) The biology of the TNF blockade. Autoimmun Rev 2:346–357
Suzuki M, Youle RJ, Tjandra N (2000) Structure of Bax: coregulation of dimer formation and intracellular localization. Cell 103:645–654
Swofford D (2004) PAUP: phylogenetic analysis using parsimony. Sinauer Associates, Sunderland, MA
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24:4876–4882
Uren AG, O’Rourke K, Aravind L, Pisabarro MT, Seshagirl S, Koonin EV, Dixit VM (2000) Identification of paracaspases and metacaspases: two ancient families of caspase-like proteins, one of which plays a key role in MALT Lymphoma. Mol Cell 6:961–967
Vaughn DE, Rodriguez J, Lazebnik Y, Joshua-Tor L (1999) Crystal structure of the Apaf-1 caspase recruitment domain: an alpha-helical greek key fold for apoptosis signaling. J Mol Biol 293:439–447
Wiens M, Krasko A, Müller CI, Müller WEG (2000) Molecular evolution f apoptotic pathways: Cloning of key domains from sponges (Bcl-2 homology domains and death domains) and their phylogenetic relationships. J Mol Evol 50:520–531
Wiens M, Krasko A, Perovic S, Müller WEG (2003) Caspase-mediated apoptosis in sponges: cloning and function of the phylogenetic oldest apoptotic proteases from Metazoa. Biochim Biophys Acta 1593:179–189
Wilkins MR, Lindskog I, Gasteiger E, Bairoch A, Sanchez J-C, Hochstrasser DF, Appel RD (1997) Detailed peptide characterisation using PEPTIDEMASS—a World-Wide Web accessible tool. Electrophoresis 18:403–408
Woo EJ, Kim YG, Kim MS, Han WD, Shin S, Robinson H, Park SY, Oh BH (2004) Structural mechanism for inactivation and activation of CAD/DFF40 in the apoptotic pathway. Mol Cell 14:531–539
Wyllie AH, Kerr JFK, Currie AR (1980) Cell death: the significance of apoptosis. Int Rev Cytol 68:251–306
Yan N, Gu L, Kodel D, Chai J, Li W, Han A, Chen L, Xue D, Shi Y (2004) Structural, biochemical, and functional analysis of Ced-9 recognition by the proapoptotic proteins Egl-1 and Ced-4. Mol Cell 15:999–1006
Yuan J, Shaham S, Ledoux S, Ellis HM, Horvitz HR (1993) The C. elegans cell death gene ced-3 encodes a protein similar to mammalian interlukin-1b-converting enzyme. Cell 75:641–652
Zhang H, Holzgreve W, De Geyter C (2001) Bcl2-L-10, a novel anti-apoptotic member of the Bcl-2 family, blocks apoptosis in the mitochondrial death pathway but not in the death receptor pathway. Hum Mol Genet 10:2329–2339
Zong WX, Edelstein LC, Chen C, Bash J, Gelinas C (1999) The prosurvival Bcl-2 homolog Bfl-1/A1 is a direct transcriptional target of NF-kappa B that blocks TNFalpha induced apoptosis. Genes Dev 13:382–387
Acknowledgments
We gratefully thank members of the Weis lab for their comments on the manuscript. This work was supported by a grant from the National Science Foundation (02372230-MCB) to V.W. and D.G.
Author information
Authors and Affiliations
Corresponding author
Additional information
[Reviewing Editor: Dr. Stuart Newfeld]
Electronic Supplementary Material
Rights and permissions
About this article
Cite this article
Dunn, S.R., Phillips, W.S., Spatafora, J.W. et al. Highly Conserved Caspase and Bcl-2 Homologues from the Sea Anemone Aiptasia pallida: Lower Metazoans as Models for the Study of Apoptosis Evolution. J Mol Evol 63, 95–107 (2006). https://doi.org/10.1007/s00239-005-0236-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-005-0236-7


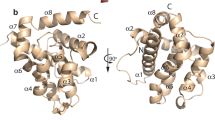




 , Highly conserved BH domains;
, Highly conserved BH domains; 

