Abstract
The toxoglossate mollusks are a large group of venomous animals (>10,000 species) conventionally divided into three groups, the cone snails, the auger snails, and the turrid snails; turrids account for >90% of the biodiversity of toxoglossans. Only the venoms of cone snails have been intensively investigated, with little work focused on turrids. We report the first broad characterization of genes expressed in venom ducts of any turrid species. Twenty-three different cDNA clones encoding putative toxins were characterized from the venom duct of the turrine species Lophiotoma olangoensis Olivera 2002 and belong to 16 different gene families. Of the 16 different Lophiotoma olangoensis gene families that encode putative toxins, for only 1 was there clear evidence of sequence similarity with any conotoxin gene family. The I-like gene family of Lophiotoma olangoensis was found to be related to the K channel-targeted I2 conotoxin superfamily. Most putative Lophiotoma toxins are cysteine-rich polypeptides, with a significant fraction much larger (>80 amino acids) than the toxins from cone snails. A small number were not cysteine-rich but had hydrophobic amino acid clusters interspersed with arginine residues. This is only 1 of >10,000 different turrid venoms that needs to be characterized. From this study, a common origin with Conus for one family of putative turrid toxins is indicated.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Most people do not immediately associate snails with venomous predators—however, the biodiversity of venomous snails is in fact very impressive. It is likely that >10,000 different species of marine snails use venom as the primary weapon for prey capture. Most well-known are the cone snails (Conus), comprising 500–700 species (Röckel et al. 1995). Two other large groups of gastropod mollusks are venomous—the auger snails (family Terebridae), which are specialized for foraging in sandy environments, and the turrid snails (family Turridae, broadly defined). The latter group probably comprises >90% of the biodiversity of venomous snails (Bouchet et al. 2002; Powell 1964; Powell 1966). Although the venoms of cone snails have been intensively investigated in the past few decades (Olivera 2002a), little is known about the venoms of turrids. For this study, we carried out the first broad molecular characterization of genes expressed in the venom duct of any turrid species and compared these genes to those in Conus venom ducts.
A number of criteria can be used to identify genes that encode putative toxins (the term toxin is broadly defined to include all pharmacologically active components of a venom that contribute to prey capture or to defense against predators or to competitor deterrence). All venomous animals have glandular epithelia that produce the polypeptidic components of their venoms; in all venomous mollusks, this is a well-defined, easily recognized structure, the venom duct (or venom gland) (Taylor et al. 1993). Thus, toxin genes would be expressed in the venom duct. Furthermore, since venom injected into a targeted animal is an extracellular fluid produced by the duct, all functional toxin genes must be translated and then secreted. Therefore, a signal sequence should be present in the open reading frame of any gene encoding a putative toxin.
Several characteristic features of toxins help to confirm that a gene is toxin-encoding. Since most toxins need to circulate or disperse within the targeted animal, they tend to be smaller, highly stable polypeptides—in general, disulfide-rich polypeptides under 100 amino acids (AA) in size. Toxin genes from cone snails have additional special features: invariably, these genes have a canonical organization—at the N-terminus is a signal sequence, followed by an intervening “pro” region, and at the C-terminus of the open reading frame, the mature toxin region, always in single copy and generally rich in Cys residues (Olivera et al. 1999; Woodward et al. 1990). Within the mature toxin region, only a few characteristic arrangements of cysteine residues occur; these are referred to as the conotoxin “Cys patterns.” Each individual Cys pattern specifies a particular disulfide framework, the dominant structural determinant which stabilizes these unusually small (typically 12–30 AA) peptide toxins. A most remarkable feature of Conus toxin precursors is that all toxins with a particular Cys pattern also share a highly conserved signal sequence; this combination of a characteristic consensus signal sequence and a specific Cys pattern defines members of a Conus toxin gene superfamily (Terlau and Olivera 2004).
Cone snails, auger snails, and turrids are conventionally placed in the same superfamily or suborder (Conoidea Rafinesque 1815, also referred to as either Toxoglossa or Conacea). Our goal in the molecular characterization of a turrid venom duct was both to provide an overview of the genes expressed in a single turrid snail species and to explore which of the general characteristics of Conus venoms are also to be found in the turrids. A hallmark of Conus venoms is the diversity of toxins found; we address whether a similar degree of genetic and biochemical complexity occurs in the mRNA transcripts found in the venom duct of a turrid. We have characterized the genes expressed in the venom duct of a single turrid species, Lophiotoma olangoensis Olivera 2002, which belongs to the subfamily Turrinae.
In particular, we assessed whether in this turrid species, there was any overlap with the toxin gene superfamilies that have been characterized from cone snails. The major conotoxin gene superfamilies are ubiquitously found in all species of Conus; any toxin family also found in a turrid implies that toxins belonging to that family were present in the last common ancestor of these now divergent taxa of venomous snails. A systematic investigation of such gene families within the >10,000 toxoglossan species could be useful for understanding the biochemical/pharmacological diversity and evolutionary history of the venomous mollusks, even where the detailed biology is mostly unknown. The extent of overlap in putative toxin gene families provides an initial assessment of the degree of similarity or divergence between turrid and Conus toxinology.
Materials and Methods
Specimen Collection; RNA Isolation
The turrid species analyzed was initially identified as Xenuroturris cingulifera. After the analysis described in this paper was initiated, it became apparent that the form being examined (see Fig. 1) was distinct from that species and, therefore, was reclassified as a new species, Lophiotoma olangoensis Olivera 2002 (Olivera 2002b). The status of Xenuroturris as a putative genus/subgenus was evaluated, and it was suggested that all forms previously included in Xenuroturris be incorporated into Lophiotoma.
The shells of animals in different toxoglossan groups. Top left: the shell of Lophiotoma olangoensis, the turrid species whose venom is characterized in this report. Top right: examples of all of the major groups in the subfamily Turrinae. A Two examples of the genus Turris: left, Turris ruthae, South Africa, and right, Turris normandavidsoni, Philippines. B Two examples of Lophiotoma: left, Lophiotoma notata, Japan, and right, Lophiotoma friedrichbonhoefferi, Philippines. C Two specimens of Gemmula: left, Gemmula sp. (unnamed), Philippines, and right, Gemmula periscelida, off East Mexico. D Two speciamens of Polystira: left, Polystira albida, Caribbean and Eastern Atlantic, and right, Polystira nobilis, West Mexico. Bottom E Specimens of Conidae. F Specimens of Terebridae. G Specimens in the turrid subfamily Borsoniinae; in one proposal (Taylor et al. 1993), this was one of several turrid subfamilies transferred from the Turridae to the Conidae.
Specimens of Lophiotoma olangoensis were collected between 20 and 50 m off Olango Island, Cebu, Philippines; specimens were dissected and venom ducts immediately frozen. A Lophiotoma olangoensis venom duct was homogenized in 2 ml TRIzol reagent, using a disposable microtissue grinder. The RNA was isolated by phase separation and precipitation, according to the manufacturer’s standard protocol (TRIzol Total RNA Isolation; Life Technologies/Gibco BRL, Grand Island, NY).
A similar, less extensive analysis was carried out on several other Lophiotoma species collected from the same locality (Lophiotoma acuta, Lophiotoma albina, and Lophiotoma cingulifera).
Preparation and Sequencing of cDNA Clones
First-strand cDNA was prepared from 2 μg RNA, followed by second-strand synthesis and amplification, which were performed by long-distance polymerase chain reaction (PCR; Clonetech SMART PCR cDNA Synthesis; Clontech Laboratories, Palo Alto, CA) using an MJ Research PTC-200 Peltier Thermal Cycler. The resulting PCR products were purified using the High Pure PCR Product Purification Kit (Roche Diagnostics; Indianapolis, IN) following the manufacturer’s suggested protocol. A second amplification, using tailed PCR primers, was performed on the PCR product. The resulting PCR products were gel-purified, and material >400 bp in length was recovered from agarose. Eluted DNA was annealed to pAMP1 vector and the resulting products transformed into competent DH5a cells, using the CloneAmp pAMP System for Rapid Cloning of Amplification Products (Life Technologies/Gibco BRL; Grand Island, NY). The nucleic acid sequences of the resulting clones were determined using an ABI automated sequencer.
Results
DNA Clones from Lophiotoma
cDNA clones from a Lophiotoma olangoensis venom duct library were analyzed as described under Materials and Methods; from this library, a total of 190 cDNAs were sequenced. Using the criteria normally applied for predicting toxin genes in Conus (as described in the Introduction), these generally had clearly defined signal sequences, and most were small disulfide-rich peptides. Twenty different putative toxin precursors were identified. A smaller group of seven predicted polypeptides had some toxin-like properties, but there were reservations with including these in the putative toxin group (lack of a definitive signal sequence, etc.). There were 31 encoded polypeptides identified that were clearly not toxins; of these, 18 had significant sequence similarity to known gene products, 9 to components involved in translation (ribosomal proteins, etc.), and 5 to tyrosinase/peroxidase/metalloproteinases. The latter two groups are not discussed further.
The cDNA clones that encoded putative toxins can be grouped into three classes. The first class encodes polypeptides that after processing are predicted to generate peptides with cysteine patterns resembling those of one of the standard conotoxin families. A second class comprises cysteine-rich peptides that deviate in their cysteine patterns from standard Cys patterns found in the major conotoxin families. Finally, there is a small group of putative toxins that lack Cys residues. We discuss these groups sequentially in the sections that follow.
Venom Peptides with Conotoxin-like Cysteine Patterns
Among the clones characterized from the Lophiotoma olangoensis cDNA library, eight had cysteine patterns similar to those characteristic of standard conotoxin gene superfamilies; these were of particular interest since they seemed most likely to reveal any potential links between turrid and Conus toxins. Two of the peptides shown in Fig. 2 encoded by clones OL67 and OL105 have an I-conotoxin-like cysteine pattern (–C–C–CC–CC–C–C–); these predicted mature gene products share considerable sequence identity between them (69%), suggesting that they belong to the same Lophiotoma olangoensis gene family. We refer to the putative peptide family to which these belong as the “I-like” Lophiotoma gene family. Similarly, two peptides have a cysteine framework seen in the peptides of the O-conotoxin superfamily (–C–C–CC–C–C–). These two peptides appear to be related (but somewhat less so than the I-like peptides); we will refer to an “O-like” gene family. We adopt a modified conotoxin nomenclature for these peptides: the clones encoding the two peptides that have an O-conotoxin-like cysteine pattern are designated Lo1 6.1 and Lo1 6.2 (since they clearly have a Conus class 6 cysteine pattern), while the two peptides with the I-superfamily-like cysteine pattern are designated Lo1 11.1 and Lol 11.2 (since they exhibit a Conus class 11 cysteine pattern; see the recent review of conotoxins by Terlau and Olivera [2004]). The initials “Lol” indicate that the predicted peptides are from Lophiotoma olangoensis.
Four peptides have a P-superfamily-type cysteine pattern (–C–C–C–C–C–C–), but these appear to be a heterogeneous group; the predicted sequences of the mature P-type peptides are also shown in Fig. 2. The spacing between cysteine residues varies considerably between the four peptides, unlike the pairs of Lophiotoma O-like and I-like peptides, where the size of the “loops” between cysteine residues is generally conserved. It is likely that these do not belong to a single gene family; this is discussed at greater length under Discussion, below.
cDNA Clones Encoding Putative Lophiotoma olangoensis Toxins with Cysteine Patterns Different from Standard Conotoxin Families
A larger number of cDNA clones that had strong signal sequences were predicted to encode cysteine-rich mature gene products that differed from the standard conotoxin cysteine patterns; these are summarized in Fig. 3. Some of the predicted mature peptides have 10–16 cysteine residues, and are considerably larger than Conus peptides, in the size range of 80–115 AA. The two peptides with 10 cysteine residues share considerable sequence identity and are clearly related to each other. However, other peptides have cysteine patterns different from each other and appear to belong to different toxin gene families (except for OL22 and OL78 that probably encode polymorphic variants).
Comparison of Precursors
The peptides shown in Figs. 2 and 3 vary considerably in the size of the propeptide regions (Table 1 summarizes the length of the putative signal sequence, propeptide, and mature toxin regions). Although for some Lophiotoma olangoensis peptide precursors (OL38, OL142, OL108) a conotoxin-like distribution of pre, pro, and mature toxin is observed, for most of the peptide precursors, the predicted propeptide region is shorter than in comparable conotoxin precursors, and in some cases, there is no propeptide region at all. In all cases except OL172, which is discussed below, the predicted mature peptide is found in single copy, as is characteristic of the conotoxins.
Secreted Peptides That Lack Cys Residues
Of all the clones analyzed that encoded putative toxins, only two (shown in Fig. 4) had open reading frames predicted to yield mature peptides without disulfide cross-links. The open reading frame of clone OL179 encodes a polypeptide with a strong signal sequence of 21 AA. Although the precise cleavage site to yield the mature toxin is somewhat ambiguous, one likely alternative is shown in Fig. 4A; proteolysis at this site would yield a propeptide region of 11 AA and a mature toxin region of 38 AA. There are no cysteine residues predicted; instead, hydrophobic or aromatic amino acids are present approximately every sixth position in the polypeptide chain, with arginine residues in between.
The processing of two peptide precursors deduced from open reading frames of two cDNA clones sequenced from the venom duct of Lophiotoma olangoensis. a This Lophiotoma peptide precursor yields a single predicted mature peptide sequence, very similar to the turritoxins isolated from Gemmula periscelida and Polystira albida (López-Vera et al. 2004). b This Lophiotoma peptide precursor is predicted to be processed to yield multiple small peptides without Cys residues and one conotoxin-like peptide. The latter type of processing has never been observed for Conus toxins.
There is a second clone that was isolated twice which is also predicted to yield peptides without disulfide cross-links. The predicted processing (see Fig. 4B) is quite different from that predicted for the precursor encoded by clone OL179, which had a typical conotoxin-like prepropeptide organization in which the putative mature toxin region is found at the C-terminus in single copy. The open reading frame of the second clone, OL172, comprises 127 AA, and if standard processing is carried out with cleavage after the strong signal sequence of 20 AA and the RK sequence before the first cysteine residue, it would yield an unusually large pro region (86 AA) and a Cys-rich, conotoxin-like C-terminal region. However, it seems highly likely that further proteolytic processing occurs: there are two RKQ sequences which are potential proteolytic cleavage sites (at AA 74 and 105); the larger of the resulting peptides is likely to be further processed as indicated in Fig. 4. Thus, multiple mature peptides are probably generated from the initial translation product encoded by this one clone, with one conotoxin-like peptide and three other processed mature peptides without disulfides. The latter show considerable sequence identity in their C-terminal sequences. The conotoxin-like peptide would have two disulfide bonds; presumably, the glutamine at the N-terminus would be cyclized to pyroglutamate, making the peptide blocked at the N-terminus. The generation of multiple peptides from a single precursor has not been reported in any Conus clone, and this is the only example characterized in this study from Lophiotoma olangoensis strongly predicted to yield multiple mature peptide products. As is the case for many neuropeptide precursors, three of the four peptides generated after posttranslational proteolytic processing of the initial translation product share considerable sequence similarity.
The two clones shown in Fig. 4 were the only ones encoding peptides that lack disulfide bonds; all other open reading frames are predicted to encode cysteine-rich peptides in single copy.
Discussion
The characterization of Conus venoms in the last 20 years has provided powerful neuropharmacological tools, as well as general insights into the biology and evolution of Conus. It has even led to the development of a commercial drug for pain, Prialt. The recent pioneering field work by Bouchet and coworkers (2002) has established that there is a much greater biodiversity of venomous turrids than of cone snails; although Conus is a large group (500–700 species), the number of turrid species is more than 10-fold greater. In New Caledonia alone, the extensive fieldwork carried out by the Bouchet team revealed the presence of ∼1800 different turrids and only 51 Conus species; the great majority of turrids have not even been described (Bouchet et al. 2002). Thus, not surprisingly, the study of their venoms has been largely neglected. Turrid venoms are unexplored—an obvious question to address is how divergent these venoms are from those of Conus. One initial issue to clarify is to what extent turrid toxins are encoded by genes homologous to conotoxin families expressed in cone snail venom ducts vs. gene families without homology to conotoxins.
A molecular cloning strategy was employed for a broad overview of the genes expressed in the venom duct of the turrid species Lophiotoma olangoensis. The immediate aim of our study was to identify genes likely to encode toxins expressed in the venom duct and to compare these to the conotoxins, the well-characterized peptide toxins from Conus venoms. As far as we know, this is the first comprehensive molecular characterization carried out for any turrid venom.
The majority of the cDNA clones identified from Lophiotoma olangoensis venom ducts that encoded polypeptides (18/20) were cysteine-rich and biochemically generally similar to the conotoxins. These, like conotoxin precursors, encoded a single copy of a predicted mature disulfide-rich peptide. A significant group of Lophiotoma olangoensis peptides (8/20) was conotoxin-like in the arrangement of cysteine residues in the primary sequence. Two peptides with a cysteine pattern similar to conotoxins of the O-superfamily (–C–C–CC–C–C–) had sufficient sequence similarity to conclude that they belong to the same gene family. Although the cysteine patterns suggest an O-like toxin family, the degree of sequence similarity to the Conus O-conotoxin superfamily was insufficient to establish that these were related gene families (see Fig. 5). The lack of apparent sequence similarity in the N-terminal region of the precursors has two alternative explanations: (1) independent origins for the corresponding Lophiotoma and Conus families or (2) a common origin and subsequent sequence divergence, possibly accentuated by a potentially more relaxed conservation of signal sequences in the turrid gene families (M.Watkins, unpublished results). The accumulation of a more extensive database of toxin gene sequences from turrid venoms may help to distinguish between these alternatives, particularly if these gene families occur in turrid groups believed to be more closely related to Conus than is Lophiotoma olangoensis. If a wider data set of sequences from turrid toxin gene families were available, those gene families that share a common origin with conotoxin genes might provide more convincing sequence similarities.
Comparison between Lophiotoma peptides and conotoxin families with the same Cys pattern. *Glu residues are known to be posttranslationally carboxylated. Boxes with solid lines denote sequence identities between Lophiotoma and Conus signal sequences. Clone Em11.10 from Conus emaciatus encodes an I-conotoxin that is almost identical to a peptide from Conus virgo, VrTx, that targets voltage-gated K channels (Kauferstein et al. 2003); there are two conservative substitutions in the toxin region. The BeTx sequence shown is almost identical to that published by Fan et al. (2003) with one substitution in the signal sequence region.
Four putative toxins from Lophiotoma olangoensis venom (OL9, 11, 135, and 142) had an arrangement of cysteine residues similar to the P-superfamily (–C–C–C–C–C–C–). These appear to be a heterogeneous group, exhibiting no apparent sequence similarity to each other nor to the Conus spasmodic peptides, the defining members of the P-conotoxin superfamily (see Fig. 5). The four Lophiotoma olangoensis peptides with a P-type Cys pattern differ from each other in the number of amino acids between cysteine residues, as well as in their N-terminal sequences.
In addition to the cysteine-rich peptides that have O- and P-conotoxin superfamily-like patterns, an even greater fraction of venom-expressed gene products identified from Lophiotoma olangoensis had a different number or arrangement of cysteine residues compared to established conotoxin families. This is an extremely diverse set of cysteine-rich polypeptides, with only two pairs that seem to belong to the same family of toxins. These Lophiotoma olangoensis venom peptides are apparently unrelated to any standard conotoxin gene family identified to date.
Of the 20 different open reading frames identified that are putative toxin precursors, 2 encode polymorphic variants, and three pairs of peptides are probably encoded by different genes belonging to the same gene families. The majority (12 of the mature peptides) each belong in a different gene family, sharing no apparent sequence similarity. Thus, it appears that our analysis has identified 16 different Lophiotoma gene families from a single venom that encode putative toxins. There are likely significantly more such gene families in this species, since the analysis carried out, though representing a fair sampling of expressed gene products, was far from saturation.
Of the 16 different putative toxin gene families transcribed in Lophiotoma olangoensis venom ducts, for only 1 is there a convincing case for significant similarity to a toxin gene family from Conus. This is the Lophiotoma I-like gene family with eight cysteine residues (see Figs. 2 and 5). The I-like peptide gene family from Lophiotoma olangoensis appears to be related to one of the conotoxin superfamilies with a class 11 Cys pattern (–C–C–CC–CC–C–C–) (Fan et al. 2003; Jimenez et al. 2003); the same Cys pattern is found in the Lophiotoma I-like toxins. As a group, conotoxins with this Cys pattern have a number of unusual features compared to other Conus peptides: the predicted toxins are larger than usual, and this group of conotoxins has branches that diverge more than other groups of conotoxins sharing the same Cys pattern. We recently found that these Conus peptides can be divided into the I1 and the I2 conotoxin superfamilies; the I1 conotoxins all exhibit the usual canonical organization of conotoxin precursors, e.g., an N-terminal signal sequence, followed by an intervening “pro” region with the C-terminal region encoding the mature toxin in single copy; many of these peptides apparently have a D-amino acid (Buczek et al. 2004). The I2 peptides have an unrelated signal sequence (Fan et al. 2003; M. Watkins, J. Garrett, and B. Olivera, unpublished results) and, unique among conotoxins, have no intervening “pro” regions: the mature toxin region immediately follows the signal sequence. A more detailed and systematic characterization of the I1- and I2-conotoxin superfamilies is being prepared for publication elsewhere (O. Buczek, E. Jimenez, M. Watkins, and B. Olivera, in preparation).
Not only do the I-like Lophiotoma olangoensis peptides have the same Cys pattern (C–C–CC–CC–C–C–) seen in the I2-superfamily (Fig. 5), but in addition, when the signal sequences of the I-like Lophiotoma toxin precursors and the I2 conotoxins are compared, there is considerable sequence similarity; 40% of the amino acids in the signal sequence of the BeTx conotoxin precursor are conserved in the signal sequences of the Lophiotoma I-like clones OL105 and OL67 (see Fig. 5). This indicates a common origin for the two groups of peptides, a suggestion further strengthened by the fact that uniquely among all known conotoxin families, and like only a small minority of the putative Lophiotoma olangoensis toxins (see Fig. 5), both the I2 conotoxins and the I-like Lophiotoma family lack a propeptide region. Thus, the similarity found in their signal sequences, the shared Cys patterns of their mature toxin regions, and the unusual absence of an intervening propeptide region in the precursors of both groups of peptides make a compelling case for common ancestry between the I-like Lophiotoma gene family and the I2 conotoxins. The physiological targets of the I2 conotoxins that have been identified are K channels (Fan et al. 2003; Kauferstein et al. 2003). This provides an initial hypothesis for a potential mechanism-of-action of the Lophiotoma olangoensis I-like peptides: that they may target K channels.
This is the first turrid from which multiple secreted gene products from a venom duct have been identified; general similarities and differences between Lophiotoma olangoensis and Conus venoms can be gleaned. All molluscan venoms (Conus, Terebra, Lophiotoma) are similar in that the majority of secreted gene products are cysteine-rich (and presumably multiply disulfide-bonded) polypeptides; a small minority are not cysteine-rich, similar to those previously characterized from Gemmula and Polystira (see Fig. 2) (López-Vera et al. 2004).
The expressed Lophiotoma genes encoding putative toxins have open reading frames similar in their organization to Conus venom peptide precursors. In most cases, the Conus paradigm for proteolytic processing of toxin precursors can be applied directly: a signal peptidase cleaving off the N-terminal signal sequence and a protease recognizing an –XR– sequence to cleave between the propeptide and the mature peptide region. In 19 of 20 toxin-encoding clones, a single copy of the mature peptide is predicted to be generated from each gene, as is found in Conus. However, one clone (OL172) encodes a precursor predicted to yield four peptides after processing, one that is Cys-rich and three that are devoid of Cys residues. Although processing of precursors to yield multiple mature neuropeptides is the norm in nervous and endocrine systems, this situation has never been found in Conus venom ducts. It is clear, however, that Conus-type processing is the dominant pattern in Lophiotoma olangoensis, with the one exception.
One general biochemical difference between the predicted mature Cys-rich Lophiotoma toxins and those found in Conus venoms is that while most major Conus venom peptides are in the size range of 12–30 AA, in Lophiotoma venom, the putative toxins are larger. A major fraction (11/20) are 30–60 AA, with a small but significant number (6/20) being even greater in size; these are >80 AA in length. While it is conceivable that the latter are not toxins, their expression in venom duct and the high frequency of Cys residues all make it likely that these are pharmacologically active venom constituents injected into prey, predator, or competitor.
The direct demonstration that the predicted gene products encoded by the cDNA clones characterized in this study are toxins would require biochemical purification of the individual toxins from Lophiotoma olangoensis venom. For turrids in general, this is a much more challenging problem than for other venomous animals; this is largely a deep-water group, most species found at depths >100 m. Those found in shallower waters tend to be smaller species that do not occur in large, concentrated populations. Furthermore, their venom ducts are extremely fine; thus, to obtain sufficient venom to do a biochemical characterization as well as functional assays to demonstrate toxic mechanisms is a formidable challenge. It is likely that, ultimately, toxins will need to be characterized after either chemical synthesis or expression of the cloned sequences. Polypeptides with multiple disulfide cross-links are problematic to synthesize or express in a biologically active form, particularly if the native, biologically active polypeptide isolated from the natural source with the native disulfide cross-linking pattern is unavailable. Even if sufficient venom were made available, a potential problem in demonstrating toxicity of the gene products is the paucity of information regarding the biology of turrids; there is essentially no information in the literature regarding prey, predators, and competitors of any species in the genus Lophiotoma. Lophiotoma toxins could very well be nontoxic to mice, though extremely toxic to their natural prey.
However, there is one general evolutionary prediction for bona fide toxin genes that can be tested. Different species in the same genus would be expected to occupy different ecological niches, and therefore the spectrum of prey, predators, and competitors for one species would differ from that of any other. Genes that encoded either a housekeeping or an endogenous signaling gene product (not targeted for injection into another animal) would not be under strong selective pressure for rapid interspecific divergence. A hallmark of toxins, however, is that because speciation results in a different set of biotic interactions, the genes are under diversifying selection and exhibit accelerated evolution. Thus, one prediction that can be tested is that if the Lophiotoma olangoensis venom duct genes that we characterized do indeed encode toxins, rapid divergence should be observed if interspecific comparisons are made. In contrast, if these were housekeeping genes, amino acid sequences should be highly conserved.
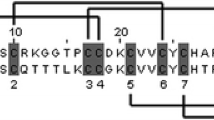
We have tested this prediction by using cDNA libraries; we carried out a preliminary analysis of genes expressed in the venom ducts of three other species of the genus Lophiotoma (i.e., Lophiotoma acuta, Lophiotoma albina, and Lophiotoma cingulifera). Polypeptides were identified that belong to the same gene families as had been identified in Lophiotoma olangoensis venom ducts. A comparison for two of the families is shown in Fig. 6. The cysteine patterns for the two families align very well, suggesting that the gene products have the same pattern of disulfide cross-links and, therefore, the same general structural framework. However, the sequence divergence observed is very high; in all cases, the gene family members identified for other Lophiotoma species differ by >30% from the corresponding Lophiotoma olangoensis peptide. The sequence divergence observed is comparable to the interspecific sequence divergence observed for conotoxins. This rate of interspecific divergence is far higher than would be expected for a housekeeping gene, or one used for endogenous signaling purposes. This result supports the conclusion that gene products from the OL55 and OL11 families are toxins, with their targets being in the different prey, predators, and competitors of these Lophiotoma species.
Interspecific comparison of two Lophiotoma gene families. Amino acid sequences for two venom gene families deduced from cDNA clones from the venom ducts of different Lophiotoma species. The amino acids that are identical to the predicted Lophiotoma olangoensis peptides are shaded. Pairwise comparisons of the different predicted sequences show 30–50% AA sequence nonidentity.
Of the diverse Lophiotoma gene families identified in this study, only for the I-like peptide family is there substantial evidence for a common origin with the conotoxins. The I-like Lophiotoma peptides and the I2 conotoxin group appear to be a venom gene family that has persisted in both turrid and cone snails since at least the early Cenozoic, and potentially even earlier. We therefore expect that peptides belonging to this family will be widely distributed in toxoglossan venoms, and genes belonging to the I-like/I2-conotoxin toxin family should be useful markers for understanding patterns of toxoglossan evolution.
References
Bouchet P, Lozouet P, Maestrat P, Heros V (2002) Assessing the magnitude of species richness in tropical marine environments: high numbers of mollusks at a New Caledonia site. Biol J Linnean Soc 75:421–436
Buczek O, Yoshikami D, Bulaj G, Jimenez EC, Olivera BM (2005) Posttranslational amino acid isomerization: a functionally important D-amino acid in an excitatory peptide. J Biol Chem 280:4247–4253
Fan C-X, Chen X-K, Zhang C, Wang L-X, Duan KL, He LL, Cao Y, Liu S-Y, Zhong M-N, Ulens C, Tytgat J, Chen J-S, Chi C-W, Zhou Z (2003) A novel conotoxin from Conus betulinus, k-BtX, unique in cysteine pattern and in function is a specific BK channel modulator. J Biol Chem 278:12624–12633
Jimenez EC, Shetty RP, Lirazan M, Rivier J, Walker C, Abogadie FC, Yoshikami D, Cruz LJ, Olivera BM (2003) Novel excitatory Conus peptides define a new conotoxin superfamily. J Neurochem 85:610–621
Kauferstein S, Huys I, Lamthanh H, Stöcklin R, Sotto F, Menez A, Tytgat J, Mebs D (2003) A novel conotoxin inhibiting vertebrate voltage-sensitive potassium channels. Toxicon 42:43–52
López-Vera E, Heimer de la Cotera EP, Maillo M, Riesgo-Escovar JR, Olivera BM, Aguilar MB (2004) A novel structural class of toxins: the methionine-rich peptides from the venoms of turrid marine snails (Mollusca, Conoidea). Toxicon 43:365–374
Olivera BM (2002a) Conus venom peptides: reflections from the biology of clades and species. Annu Rev Ecol Syst 33:25–42
Olivera BM, (2002b) The gastropod genus Xenuroturris (Iredale, 1929) evaluated and a new turrid, Lophiotoma olangoensis, described from the Central Philippines. Sci Diliman 14:39–49
Olivera BM, Walker C, Cartier GE, Hooper D, Santos AD, Schoenfeld R, Shetty R, Watkins M, Bandyopadhyay P, Hillyard DR (1999) Speciation of cone snails and interspecific hyperdivergence of their venom peptides. Potential evolutionary significance of introns. Ann NY Acad Sci 870:223–237
Powell AWB (1964) The family Turridae in the Indo-Pacific. Part 1. The subfamily Turrinae Indo-Pacific. Mollusca 1:227–346
Powell AWB (1966) The molluskan families Speightiidae and Turridae. Bull Auckland Inst Mus 5:1–184
Röckel D, Korn W, Kohn AJ (1995) Manual of the living Conidae. Verlag Christa Hemmen, Wiesbaden, Germany
Taylor JD, Kantor YI, Sysoev AV (1993) Foregut anatomy, feeding mechanisms, relationships and classification of the Conoidea (=Toxoglossa) (Gastropoda). Bull Nat Hist Mus Lond (Zool) 59:125–170
Terlau H, Olivera BM (2004) Conus venoms: a rich source of novel ion channel-targeted peptides. Physiol Rev 84:41–68
Woodward SR, Cruz LJ, Olivera BM, Hillyard DR (1990) Constant and hypervariable regions in conotoxin propeptides EMBO J 1:1015–1020
Acknowledgments
This work was supported in part by NIH Grant GM 48677 and in part by funds from the University of Utah. We would like to thank Noel Saguil and Victor Pagobo for their help in collecting specimens of Lophiotoma olangoensis.
Author information
Authors and Affiliations
Corresponding author
Additional information
[Reviewing Editor: Dr. Rafael Zardoya]
Rights and permissions
About this article
Cite this article
Watkins, M., Hillyard, D.R. & Olivera, B.M. Genes Expressed in a Turrid Venom Duct: Divergence and Similarity to Conotoxins. J Mol Evol 62, 247–256 (2006). https://doi.org/10.1007/s00239-005-0010-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-005-0010-x