Abstract
Studies on the origin of the genetic code compare measures of the degree of error minimization of the standard code with measures produced by random variant codes but do not take into account codon usage, which was probably highly biased during the origin of the code. Codon usage bias could play an important role in the minimization of the chemical distances between amino acids because the importance of errors depends also on the frequency of the different codons. Here I show that when codon usage is taken into account, the degree of error minimization of the standard code may be dramatically reduced, and shifting to alternative codes often increases the degree of error minimization. This is especially true with a high CG content, which was probably the case during the origin of the code. I also show that the frequency of codes that perform better than the standard code, in terms of relative efficiency, is much higher in the neighborhood of the standard code itself, even when not considering codon usage bias; therefore alternative codes that differ only slightly from the standard code are more likely to evolve than some previous analyses suggested. My conclusions are that the standard genetic code is far from being an optimum with respect to error minimization and must have arisen for reasons other than error minimization.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genetic code is not random. It has been proposed that the structure of the genetic code reflects the physiochemical properties of amino acids and their biosynthetic relationships. Some authors (Haig and Hurst 1991; Freeland and Hurst 1998; Knight et al. 1999; Freeland et al. 2000a) support the view that the main force that shaped the genetic code is selection for minimization of the chemical distances between amino acids, that is, error minimization at the protein level, as proposed by Woese (1965), Epstein (1966), Sonneborn (1965), and others. The main alternative view is the coevolution hypothesis, introduced by Wong (1975) and subsequently championed by Di Giulio (1989, 1997a, b, 1999, 2000a): according to this view the structure of the code reflects the biosynthetic pathway of amino acid formation but error minimization is not the main force that shaped the genetic code. The debate seems not to be resolved (Freeland et al. 2000a; Di Giulio 2000a, 2000b, 2001).
These studies on the optimization of the genetic code compare measures of error minimization of the standard code and measures produced by random variant codes. They, however, rely on different approaches: the “statistical” approach (Freeland et al. 2000a) produces a large set of random codes and observes the probability that a code with a better measure of error minimization than the standard code is observed; the “engineering” approach (named so by Freeland et al. [2000a]—referred to Di Giulio) is based on the calculation of a minimization percentage, that is, it introduces a distance function based on the physiochemical properties of amino acids and looks at the value it assumes in the genetic code with respect to both a completely randomized code and the most optimized code possible.
One critique of the statistical approach is that (Di Giulio 2000a), although the frequency of codes that perform better (in terms of relative efficiency) than the standard code is roughly 1 × 10−6 (Freeland and Hurst 1998), there are 2.4 × 1018 (=20!) possible codes in the space of permutations of the standard code, which still leaves 2.4 × 1012 possible alternative better codes. Another concern about the statistical approach is that the space of all possible permutations of the standard code contains codes that may differ very much from the standard code. Therefore most alternative codes produced by this approach are not likely to be obtained at all by mutations with small effect. A possible way to resolve this issue is to reduce the space of possible variant codes to those in the neighborhood of the standard code, that is, to generate variant codes that differ only slightly from the standard code, and, therefore, are more likely to be obtained by mutation.
All these methods, in any case, are based on the structure of the possible genetic codes, that is, on the assignment of the different amino acids to the different codons, and do not take into account at all frequency of the different codons. Yet synonymous codons are not used at random, and codon usage bias could affect the degree of error minimization because the different codons are not equal with respect to the capacity to minimize errors. This is important especially because it is supposed that life originated at high temperatures (Woese 1987; Achenbach-Ritcher et al. 1987; Di Giulio 2000c), and during the origin of the code C and G were probably more abundant than A and T, because of a more stable conformation due to the three (instead of two) hydrogen bonds. Therefore, if one allows for a bias in the CG content (for a prevalence of CG), the code should perform even better, in terms of relative efficiency, if it evolved to minimize errors.
I will measure the level of optimization of the genetic code by producing many variant codes and looking at the probability that a code that is “better” than the standard code is observed, as in the standard statistical approach, but with two main differences. First, the measure of error minimization will take into account a possible bias in codon usage. Second, the space of possible variant codes will be restricted to those codes originated by mutations of small effect, possibly taking into account the real biosynthetic pathways of amino acid formation.
Methods
Error Minimization for Single Codons: The Mean Distance
For each pair of amino acids I derive the measure DAA/AA* = ωAA/AA − ωAA/AA* from McLachlan’s (1971) matrix of chemical similarity, where ωAA/AA is the similarity of amino acid AA with itself (this value is usually the same for all amino acids, but not in all similarity matrices: in McLachlan’s it is either 8 or 9) and ωAA/AA* is the similarity of AA to the mutant amino acid AA*, obtained after an error at one of the three positions of the original codon. Hence, DAA/AA* is the distance (dissimilarity) between the original (AA) and the mutant (AA*) amino acid. Since ωAA/AA>ωAA/AA* for every amino acid, DAA/AA* is always positive, and since there are three possible mutants for each position, there are nine measures of DAA/AA* for each codon, corresponding to the nine possible mutant codons. Their mean value is taken as a measure of distance (dissimilarity) between the original codon and its possible mutants. I call this measure MD (mean distance). Since MD is a measure of dissimilarity, lower values of MD correspond to optimal codons (codons that minimize the effects of errors). For all the values of the present analysis the similarity score with the termination signal (ωAA-STOP) is set to −10 (different values ranging from 0 to −50 do not affect results significantly).
Mutation Bias
Since transitions (C↔T, A↔G) and transversions (C,T↔A,G) are not equally likely to occur, MD values are calculated with a possible transition/transversion bias for mutation and mistranslation and a possible weighting due to base position for mistranslation. The values used here are the same as used by Freeland and Hurst (1998), which coincide quite well with the empirical data and have been shown to increase the efficiency of the standard code (Freeland and Hurst 1998). The precise values are summarized in Table 1. Different values in a similar range do not change the results drastically. Moreover I consider the possibility of different mutation rates for CG and AT, because C and G, which have three hydrogen bonds, may be less error-prone than A and T, which have only two hydrogen bonds.
Rules for the Formation of Variant Genetic Codes
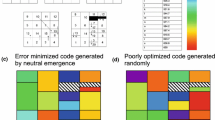
I use the following method (as in Haig and Hurst 1991; Freeland and Hurst 1998) to create random codes: the codon space (i.e., the possible 64 codons) is divided into the same 21 nonoverlapping sets of codons observed in the standard code, each set comprising all codons specifying a particular amino acid in the standard code; the three stop codons remain in the same position of the standard code for all alternative codes, while each of the 20 amino acids is assigned randomly to one of these sets to form an alternative code. In addition, in another set of random codes, I use the further constraint (not used by Haig and Hurst [1991] or by Freeland and Hurst [1998] but used by Freeland et al. [2002b] of keeping the first or the second base as in the standard code (see Fig. 1), to reduce the space of possible variant codes.
Each cell defined by three bases corresponds to a codon. Black cells correspond to stop codons. Each number (a block of cells) corresponds to an amino acid. Variant genetic codes are formed by assigning at random a position (1–20) to the 20 amino acids. Alternatively the process can be constrained at the first (a) or second (b) base position: changes are possible only among amino acids belonging (roughly—there are some exceptions for Leu, Ser, and Arg) to blocks with the same first position (a) or with the same second position (b) (same color).
Error Minimization for a Genetic Code: The Sum of the Mean Distances
The sum of MD values (SMD) is a measure of the optimization reached by the genetic code without considering codon usage bias, a measure that is similar to the “mean square” used by Haig and Hurst (1991) and Freeland and Hurst (1998), with the difference that changes to stop codons in this case are included in the calculation of MD values (but MD values for stop codons are not included in the calculation of SMD). To take into account codon usage bias, codon usage is measured on 1000 random sequences, 300 codons long, generated with a given probability of CG content and MD values are weighted by the usage frequency of each codon. If MD values are weighted according to the overall frequency of the corresponding codons, then the sum of the weighted MD values (wSMD) incorporates a bias in the importance of each amino acid, as different codon usages lead to different frequencies of amino acids. If within-amino-acid codon frequencies are used instead of overall frequencies, then the sum of the weighted MD values (zSMD) measures the optimization of the genetic code under the assumption that certain codon frequencies are used, as wSMD, but weighting all amino acids the same.
Results
Codon Usage Bias—No Constraints
To understand whether codon usage influences the level of error minimization of the code, I first evaluate the level of optimization of the standard code without codon usage bias. Of 2 million alternative codes, none is found to have a lower SMD value than the standard code, even when transition/transversion bias is taken into account and with a mutation ratio for CG/AT ranging from 2/3 to 1. This result is similar to what has been obtained by Freeland and Hurst (1998) and shows that with the similarity matrix used here the genetic code seems highly optimized when sampling in the space of all the 20! possible alternative codes. Indeed, Freeland and Hurst (1998) found one better code in 1 million, therefore with the matrix used here the idea that the standard code is the best possible code seems even more convincing.
When codon usage bias is taken into account, however, I do find codes that perform better than the standard code. For CG content below 50% no better codes are found, and nothing can be said, except that low CG contents do not seem to decrease the level of optimization of the genetic code. Even when sampling 1 million alternative codes there are no codes that perform better than the standard code, as in the case that codon usage is not taken into account. For CG content over 50%, on the contrary, it is clear (Table 2) that the probability of finding a better code increases drastically. The importance of CG content cannot be exactly measured, as we do not have a reference measure for the case of no codon bias (no better codes found with no bias of 1 million alternatives, might mean that some better codes could still be found if sampling many more alternative codes). Even if we take one in a million as a landmark, in any case, we see that there is a 100-fold increase in the probability of a better code with a 70% CG content, and even 10,000-fold with 90% CG. Of course a 90% CG content is not realistic. We are interested in the conditions that may apply to the origin of the genetic code, that is, in a CG content around 70% (Woese 1987; Achenbach-Ritcher et al. 1987; Di Giulio 2000c). Note that a 70% CG content corresponds to an effective number of codons (ENC; Wright 1990) that is about 46, which denotes quite a high codon usage bias but one that is not rarely found even in genes of extant organisms.
It must be noted, however, that wSMD values introduce a bias in the importance assigned to each amino acid, in that the frequency of each amino acid depends on the CG content. When zSMD values (that, on the other hand, weight every amino acid the same) are considered, no better code is found, whatever the CG content. The best measure is probably somewhere in between these two measures. It is difficult to know if, at the origin of the code, codon frequencies were determined mainly by temperature and CG content (in which case wSMD values are more realistic measures) or by the property of the amino acids in the protein (in which case zSMD values are more realistic values). In spite of the extensive research done on the origin of life, it is still uncertain whether the last universal common ancestor was a progenote or a cenancestor, that is, an organism in which the genotype–phenotype relationship was already well defined or not (Woese 1998).
It should also be noted that the mutation ratio CG/AT, that is, the stability of C and G (due to three hydrogen bonds instead of the two of A and T), and the transition/transversion bias do not seem to affect much the results, though higher stability of CG versus AT leads to slightly higher frequencies of better codes.
Constrained Variant Codes—No Codon Usage Bias
If we maintain the blocks of codons of the standard code, each block containing codons coding for one amino acid (see Fig. 1), and assigning one amino acid at random to each block, we obtain a space of permutations of about 2.4 × 1018 (=20!) alternative codes. Sampling in such a large space of alternative codes, to look for codes that perform better than the standard code, may be meaningless if most of this space contains variant codes that can never be obtained by small mutations of the standard code. The probability to find a better code should be measured in the neighborhood of the original (standard) code, that is, for codes that are likely to arise only by small changes in the assignment of amino acids.
One possibility is to constrain the first codon position, that is, to allow changes only among amino acids with the same first base as the standard code (see Fig. 1). This constraint reflects partially the biosynthetic pathway of amino acid formation (Freeland et al. 2000b) but reduces the possible variant codes to about 2.07 × 108 (=5!4), which is still a huge number. Moreover, it allows for variant codes with multiple position substitution. An alternative possibility is to constrain the first codon position and to allow changes only along one of the first bases (see Fig. 1). This reduces the space of possible codes to only 120 (=5!) for each of the four bases, a space that still reflects the biosynthetic pathway of amino acid formation and includes only codes that are generated by mutations of small effect, that is, codes that are in the neighborhood of the standard code. A similar procedure can be applied to constrain the second base (see Fig. 1): in this case the constraint is arbitrary; it does not necessarily reflect the biosynthetic pathway, though it does limit the space of possible alternative codes to codes that are in the neighborhood of the standard code (3.5 × 108 = 5! × 4! × 7! × 4! when multiple substitutions are allowed).
When the space of possible variant codes is reduced in these ways, the probability that a better code is found increases dramatically (Table 3). For example, about 5–20 better codes in 100,000 are found when the second base is constrained and about 1–5 when the first base is constrained (remember that with no constraints, no better codes are found in 2 million), and when changes are allowed only among one block with the same first (second) base, the percentage of better codes is up to 4% (10%). These codes differ only slightly from the standard code (see Fig. 2).
Codes with a lower SMD value than the standard code when possible changes occur only among amino acids with the same first (or second) base (no codon usage bias, no transition/transversion bias, CG/AT mutation ratio = 0.9). Only amino acid assignments that differ from the standard code are shown. The standard code is shown at the bottom right.
Constrained Variant Codes and Codon Usage Bias
When both codon usage bias and constraints for the formation of variant codes are taken into account we obtain the conditions that are most likely to have occurred during the origin of the code. In particular, we may choose to apply a CG content around 70%, as this reflects the probable CG content during the origin of the code, and to generate codes that differ from the standard code only by changing the assignment of amino acids among codons in one block with the same first (or second) base, as this reflects the probability that variant codes really arise. In this case, when both codon usage bias and mutation constraints are taken into account, the probability to find a code that performs better than the standard code is even higher.
Even when allowing multiple substitutions, that is, changes among amino acids with the same first position, for all the four possible bases, there is an up to 100-fold increase in the probability to find a better code, compared to the case of no codon usage bias. The precise values are in Table 4.
If only variant codes that differ slightly from the standard code are generated, the probability that a better code is found is even higher (see Table 4). For example, the possible variant codes with position substitutions occurring only among amino acids with T at the first position are 120 (=5!), and among them, if the CG content is 70%, between 1 and 8 codes (that is, roughly 1–7%, depending on whether amino acids are weighted all the same or not) are better than the standard code.
Different Similarity Matrices
The similarity matrix used here has been chosen because it relies on chemical similarities rather than on observed substitutions. Matrices derived from observed substitutions are probably more reliable measures of the properties of amino acids in living organisms, but they have the disadvantage to incorporate the very structure of the genetic code. That is, similarity scores between amino acids in these matrices may directly reflect the structure of the genetic code, rather than similarity between amino acids. As Di Giulio (2001) has shown, for example, the use of the PAM 74–100 matrix (Benner et al. 1994) would render tautologous an analysis of the optimization of the genetic code.
When different matrices are used, incorporating codon usage bias in the measure of error minimization, and reducing the space of possible variant codes to those originating by substitutions along single-base positions, as in the previous paragraph, the frequency of better codes is always rather high, even higher than with the matrix used throughout this paper. For example, using Woese (1973) polarity, more than 10% better codes are found among the possible variant codes with position substitutions occurring only among amino acids with T at the first position, up to 30% using the 74–100 PAM matrix (Benner et al. 1994) with C at the first position, up to 80% using other matrices based on hydrophobicity (see Table 5). Woese polarity has been used in most studies of error minimization of the genetic code because it produced better alternative codes less frequently than any other matrix (Haig and Hurst 1991). McLachlan’s (1971) chemical similarity matrix, in the study reported here, was even better than Woese’s polarity in minimizing errors. Therefore, unless McLachlan’s is the most accurate available similarity matrix to measure error minimization of the genetic code, the true frequency of better codes is probably even higher than the values discussed throughout this paper.
Discussion
Freeland and Hurst (1998) found that only one in a million possible alternative codes performs better than the standard code, in terms of relative efficiency to minimize the effects of errors. In this standard approach, codon usage was not taken into account, however, codon usage bias was probably important during the evolution of the code, as CG content was probably about 70% (Woese 1987; Achenbach-Ritcher et al. 1987; Di Giulio 2000c). Moreover, this approach considers possible alternative codes in the whole space of permutations of the standard genetic code, which allows some 2.4 × 1018 (=20!) possible variant codes. Therefore the finding of one better code in a million still leaves 2.4 × 1012 possible better codes. Freeland and Hurst (1998) concluded that the genetic code evolved to minimize errors, but the debate about this issue seems not to be resolved (Freeland et al. 2000a; Di Giulio 2000a, 2000b, 2001).
The first modification of the standard approach I used here is to take into account codon usage bias. Despite the uncertainty about the nature of the last universal common ancestor, it is probable that life originated at high temperatures (Woese 1987; Achenbach-Ritcher et al. 1987; Di Giulio 2000c) and that, during the origin of the genetic code, C and G were more abundant than A and T, because of a more stable conformation of CG-rich sequences due to the three hydrogen bonds of C and G instead of two (in A and T). In particular, CG content has been estimated for the ancestral tRNAs to be between 61% and 68%, with the latter percentage more likely (Fitch and Upper 1987; Di Giulio 2000c). Therefore, if one allows for a bias in CG content in the direction of a prevalence of CG, the code should perform even better, in terms of relative efficiency for error minimization. I have shown that, on the contrary, increasing CG content highly reduces the level of optimization of the standard code, and at a CG content around 70% the frequency of codes that perform better than the standard code is not negligible.
The second main modification of the standard approach I have used is to reduce the space of possible codes to those that are likely to arise by mutations of small effect. One could say that sampling in the space of these variant neighbor codes introduces a bias in the probability of finding better codes because in this space there exist more codes that perform better than the standard code. Indeed this is exactly the point, which is ignored by Freeland and Hurst (1998) and by the standard “statistical” approach: that natural selection for error minimization acts in the neighborhood of the original genetic code. The space of the constrained codes contains alternative codes that differ only slightly from the original (standard) code, therefore they are more likely to arise than alternative codes that differ in many positions. In other words, sampling in the space of all the possible permutations (20!) of the genetic code does not give a reliable estimate of the true probability that possible variant codes replace the standard code, simply because it is a space of codes than are unlikely to arise by small mutations of the standard code.
This concern has been considered by Freeland et al. (2000b), who take into account the biosynthetic pathway of amino acid formation to reduce the set of possible alternative codes (their set of restricted codes corresponds to my “constrained first base”) and apparently confirm the high level of error minimization of the standard code. However, as Di Giulio has shown (2001), the claim of Freeland et al. (2000b) is unsupported because their use of the PAM 74–100 matrix of amino acid similarity (which itself depends on the genetic code structure) renders their whole analysis tautologous. I have used here a similarity matrix based on chemical properties (McLachlan 1971), which does not lead to the same mistake, and I have shown that in the neighborhood of the standard code, the frequency of codes that perform better than the standard code is dramatically higher and certainly not negligible. Even when other matrices (including the PAM 74–100 matrix) are used, in any case, the results shown here do not change drastically. Indeed, with other matrices the frequency of better codes is even higher.
The conclusion of the analysis presented here is that the apparently high degree of error minimization of the genetic code is dramatically reduced when one takes into account (1) codon usage bias produced by the probable CG content occurring during the origin of the code and (2) a space of possible alternative codes that differ from the standard code only slightly. When codon usage bias and mutation constraints are taken into account, the frequency of codes that perform better than the standard code is not negligible. Therefore these results do not support the claim that the main force that shaped the genetic code is error minimization (Woese 1965; Haig and Hurst 1991; Freeland and Hurst 1998; Knight at al. 1999; Freeland et al. 2000a, b) and, though not directly supporting the coevolution theory (Wong 1975), are in favor of the view (Di Giulio 1997a, b, 1999, 1999, 2000a, 2000b; Judson and Haydon 1999) that the genetic code evolved for reasons other than the minimization of errors.
References
L Achenbach-Ritcher R Gupta KO Stetter CR Woese (1987) ArticleTitleWere the original eubacteria thermophiles? Syst Appl Microbiol 9 34–39 Occurrence Handle11542087
SA Benner MA Cohen GH Gonnet (1994) ArticleTitleAmino acid substitution during functionally constrained divergent evolution of protein sequences Protein Eng 7 IssueID11 1323–1332 Occurrence Handle1:CAS:528:DyaK2MXit1Cku7k%3D Occurrence Handle7700864
MO Dayhoff RM Schwartz BC Orcutt (1978) A model of evolutionary change in proteins MO Dayhoff (Eds) Atlas of protein sequence and structure, Vol 5, Suppl 3 National Biomedical Research Foundation Washington 352
M Di Giulio (1989) ArticleTitleThe extension reached by the minimization of the polarity distances during the evolution of the genetic code J Mol Evol 29 IssueID4 288–293 Occurrence Handle1:CAS:528:DyaL1MXmt1Ogurc%3D Occurrence Handle2514270
M Di Giulio (1997a) ArticleTitleOn the origin of the genetic code J Theor Biol 187 IssueID4 573–581 Occurrence Handle1:CAS:528:DyaK2sXmtVGlsL0%3D
M Di Giulio (1997b) ArticleTitleThe origin of the genetic code Trends Biochem Sci 22 IssueID2 49–50 Occurrence Handle1:CAS:528:DyaK2sXht1Gnu78%3D
M Di Giulio (1999) ArticleTitleThe coevolution theory of the origin of the genetic code J Mol Evol 48 IssueID3 253–255 Occurrence Handle1:CAS:528:DyaK1MXhs1ymsr0%3D Occurrence Handle10627191
M Di Giulio (2000a) ArticleTitleGenetic code origin and the strength of natural selection J Theor Biol 205 659–661 Occurrence Handle10.1006/jtbi.2000.2115 Occurrence Handle1:CAS:528:DC%2BD3cXlsVSrsrw%3D
M Di Giulio (2000b) ArticleTitleThe origin of the genetic code Trends Biochem Sci 25 IssueID2 44 Occurrence Handle10.1016/S0968-0004(99)01522-4 Occurrence Handle1:CAS:528:DC%2BD3cXhsFejsbY%3D
M Di Giulio (2000c) ArticleTitleThe universal ancestor lived in a thermophilic or hyperthermophilic environment J Theor Biol 203 203–213 Occurrence Handle1:CAS:528:DC%2BD3cXhs12hsLk%3D
M Di Giulio (2001) ArticleTitleThe origin of the genetic code cannot be studied using measurements based on the PAM matrix because this matrix reflects the code itself, making any such analyses tautologous J Theor Biol 208 141–144 Occurrence Handle10.1006/jtbi.2000.2206 Occurrence Handle1:CAS:528:DC%2BD3MXjt1yitA%3D%3D Occurrence Handle11162059
CJ Epstein (1966) ArticleTitleRole of the amino-acid “code” and of selection for conformation in the evolution of proteins Nature 210 25–28 Occurrence Handle1:CAS:528:DyaF28XpsVOksg%3D%3D Occurrence Handle5956344
WM Fitch K Upper (1987) ArticleTitleThe phylogeny of tRNA sequences provides evidence for ambiguity reduction in the origin of the genetic code Cold Spring Harbor Symp Quant Biol 52 759–767 Occurrence Handle1:CAS:528:DyaL1cXlt1Cmtrc%3D Occurrence Handle3454288
SJ Freeland LD Hurst (1998) ArticleTitleThe genetic code is one in a million J Mol Evol 47 238–248 Occurrence Handle1:CAS:528:DyaK1cXmt1ansrs%3D Occurrence Handle9732450
SJ Freeland RD Knight LF Landweber (2000a) ArticleTitleMeasuring adaptation within the genetic code Trends Biochem Sci 25 IssueID2 44–45 Occurrence Handle1:CAS:528:DC%2BD3cXhsFejtr4%3D
SJ Freeland RD Knight LF Landweber LD Hurst (2000b) ArticleTitleEarly fixation of an optimal genetic code Mol Biol Evol 17 IssueID4 511–518 Occurrence Handle1:CAS:528:DC%2BD3cXisVSgt70%3D
DG George WC Barker LT Hunt (1990) ArticleTitleMutation data matrix and its uses Meth Enzym 183 333–351 Occurrence Handle1:CAS:528:DyaK3MXivFGg Occurrence Handle2314281
D Haig LD Hurst (1991) ArticleTitleA quantitative measure of error minimization in the genetic code J Mol Evol 33 IssueID5 412–417 Occurrence Handle1:CAS:528:DyaK3MXmsleqtrs%3D Occurrence Handle1960738
OP Judson D Haydon (1999) ArticleTitleThe genetic code: What is it good for? An analysis of the effects of selection pressures on genetic codes J Mol Evol 49 IssueID5 539–550 Occurrence Handle1:CAS:528:DyaK1MXnsVajsbw%3D Occurrence Handle10552035
RD Knight SJ Freeland LF Landweber (1999) ArticleTitleSelection, history and chemistry: The three faces of the genetic code Trends Biochem Sci 24 IssueID6 241–247 Occurrence Handle10.1016/S0968-0004(99)01392-4 Occurrence Handle1:CAS:528:DyaK1MXks1ansLg%3D Occurrence Handle10366854
AD McLachlan (1971) ArticleTitleTests for comparing related amino-acid sequences. Cytochrome c and cytochrome c 551 J Mol Biol 61 409–424 Occurrence Handle1:CAS:528:DyaE38XhsF2k Occurrence Handle5167087
J Overington D Donnelly MS Johnson et al. (1992) ArticleTitleEnvironment-specific amino-acid substitution tables - tertiary templates and prediction of protein folds Protein Sci 1 IssueID2 216–226 Occurrence Handle1:CAS:528:DyaK38XksVeqtb4%3D Occurrence Handle1304904
RP Riek MD Handschumacher SS Sung M Tan MJ Glynias MD Schluchter J Novotny RM Graham (1995) ArticleTitleEvolutionary conservation of both the hydrophilic and hydrophobic nature of transmembrane residues J Theor Biol 172 IssueID3 245–258 Occurrence Handle10.1006/jtbi.1995.0021 Occurrence Handle1:CAS:528:DyaK2MXkslGmsrk%3D Occurrence Handle7715195
JL Risler MO Delorme H Delacroix A Henaut (1988) ArticleTitleAmino acid substitutions in structurally related proteins. A pattern recognition approach. Determination of a new and efficient scoring matrix J Mol Biol 204 IssueID4 1019–1029 Occurrence Handle1:CAS:528:DyaL1MXptV2jtg%3D%3D
TM Sonneborn (1965) Degeneracy of the genetic code: extent, nature and genetic implications. Academic Press, New York Academic Press New York
CR Woese (1965) ArticleTitleOn the evolution of the genetic code Proc Natl Acad Sci USA 54 1546–1552 Occurrence Handle1:CAS:528:DyaF28Xltlagsg%3D%3D Occurrence Handle5218910
CR Woese (1973) ArticleTitleEvolution of genetic code J Naturwiss 60 447–459 Occurrence Handle1:CAS:528:DyaE2cXksVOmsA%3D%3D
CR Woese (1987) ArticleTitleBacterial evolution Microbiol Rev 51 221–271
CR Woese (1998) ArticleTitleThe universal ancestor Proc Natl Acad Sci USA 95 6854–6859 Occurrence Handle1:CAS:528:DyaK1cXjslynu7w%3D Occurrence Handle9618502
JT Wong (1975) ArticleTitleA co-evolution theory of the genetic code Proc Natl Acad Sci USA 72 IssueID5 1909–1912 Occurrence Handle1:CAS:528:DyaE2MXkt1ertbg%3D Occurrence Handle1057181
F Wright (1990) ArticleTitleThe ‘effective number of codons’ used in a gene Gene 87 23–29 Occurrence Handle10.1016/0378-1119(90)90491-9 Occurrence Handle1:CAS:528:DyaK3cXktVWmsbo%3D Occurrence Handle2110097
Author information
Authors and Affiliations
Corresponding author
Additional information
[Reviewing Editor: Martin Kreitman]
Rights and permissions
About this article
Cite this article
Archetti, M. Codon Usage Bias and Mutation Constraints Reduce the Level of ErrorMinimization of the Genetic Code. J Mol Evol 59, 258–266 (2004). https://doi.org/10.1007/s00239-004-2620-0
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/s00239-004-2620-0