Abstract
Introduction
This study aims to develop an automatic segmentation framework on the basis of extreme value distribution (EVD) for the detection and volumetric quantification of white matter hyperintensities (WMHs) on fluid-attenuated inversion recovery (FLAIR) images.
Methods
Two EVD-based segmentation methods, namely the Gumbel and Fréchet segmentation, were developed to detect WMHs on FLAIR (slice thickness = 5 mm; TR/TE/TI = 11,000/120/2,800 ms; flip angle = 90°) images. Another automatic segmentation method using a trimmed likelihood estimator (TLE) was implemented for comparison with our proposed segmentation framework. The performances of the three automatic segmentation methods were evaluated by comparing with the manual segmentation method.
Results
The Dice similarity coefficients (DSCs) of the two EVD-based segmentation methods were larger than those of the TLE-based segmentation method (Gumbel, 0.823 ± 0.063; Fréchet, 0.843 ± 0.057; TLE, 0.817 ± 0.068), demonstrating that the EVD-based segmentation outperformed the TLE-based segmentation. The Fréchet segmentation obtained larger DSCs on patients with moderate to severe lesion loads and a comparable performance on patients with mild lesion loads, indicating that the Fréchet segmentation was superior to the Gumbel segmentation. The Gumbel segmentation underestimated the lesion volumes of all patients, whereas the Fréchet and TLE-based segmentation methods obtained overestimated lesion volumes (Manual, 13.71 ± 14.02 cc; Gumbel, 12.73 ± 13.21 cc; Fréchet, 13.88 ± 13.96 cc; TLE, 13.54 ± 12.27 cc). Moreover, the EVD-based segmentation was demonstrated to be comparable to other state-of-the-art methods on a publicly available dataset.
Conclusion
The proposed EVD-based segmentation framework is a promising, effective, and convenient tool for volumetric quantification and further study of WMHs in aging and dementia.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
White matter hyperintensities (WMHs), also known as white matter lesions or leukoaraiosis [1, 2], are focal or diffused lesions of high signals that are visualized on T2-weighted and fluid-attenuated inversion recovery (FLAIR) magnetic resonance imaging scans [3]. WMHs are commonly detected on elderly patients and are accompanied by other symptoms, such as cognition impairment [1, 4], gait dysfunction [5], dementia [6], and depression [7]. The exact pathological mechanism of WMHs remains unclear, but WMHs are reportedly associated with myelin or axonal loss, diminished pallor or rarefaction, and mild gliosis [1, 8].
Volumetric quantification data of WMHs are valuable for disease tracking, therapeutic evaluation and prognosis, and neuroscience research [2, 9]. WMHs are possible independent risk factors for symptomatic intracerebral hemorrhage in patients treated with thrombolysis for acute stroke [10, 11]. The relative risk of stroke increases significantly as the WMH burden increases. Furthermore, the study of the relationship between the severity of WMHs and cognitive impairment in demented patients [6, 12, 13] has attracted considerable increasing attention from researchers worldwide.
Accurate segmentation of WMHs on MR images is required before volumetric quantification. Automatic segmentation methods can be categorized into two groups [9]: lesion-based and tissue-based methods. Lesion-based methods [14–16] consider the WMH as a separate class and use empirical rules to detect specific types of lesions directly. By contrast, tissue-based segmentation [17–20] avoids the need to model the intensity of the heterogeneous lesions directly. Instead, the tissue-based methods model the intensity distribution of normal brain tissues (e.g., gray matter (GM), white matter (WM), and cerebral spinal fluid (CSF)) using the Gaussian mixture model (GMM) [21] and detect the WMHs as tissue outliers. For example, some methods [18–20] employed a trimmed likelihood estimator (TLE) proposed by Neykov et al. [22] to identify a fraction of the abnormal voxels from normal brain tissues. For all the aforementioned techniques, two important issues should be considered. First, magnetic resonance imaging (MRI) sequences should be chosen properly. The FLAIR sequence is more advantageous in suppressing the CSF signal and enhancing the contrast between the WMHs and other normal tissues [3]. Thus, FLAIR images are widely employed in WMH segmentation [23–25]. The second important issue to consider is the WMH lesion load, which can vary greatly in different patients [2].
In this study, we propose a new segmentation framework that combines unilateral normal distributions (UND) and extreme value distributions (EVD) [26–28] for the automatic segmentation and volumetric quantification of WMHs on FLAIR images. The EVD-based segmentation is divided into two subtasks, given that the two subtypes of the EVDs (namely, the Gumbel and Fréchet distributions) are employed to detect the WMHs. Different similarity measures are used to assess the volumetric agreement between the EVD-based segmentation and manual segmentation on 60 patients with different lesion loads. Furthermore, the proposed automatic segmentation framework is compared with the aforementioned method, which employs TLE to detect the WMHs as outliers to the normal brain tissue model.
Material and methods
Patient selection and image acquisition
This study was approved by the institutional review board, and written informed consent was obtained from all patients. The FLAIR images of 60 elderly patients scanned as part of normal patient care were enrolled in this study. The patients were between 61 and 86 years old (mean age, 68.2 years) and suffered from various degrees of vascular white matter abnormalities. All the patients participated in a mini-mental state examination to screen for cognitive impairment. A score was awarded to indicate whether a person has normal cognition or different levels of cognitive impairment. The patients were classified into three classes according to the locations of WMHs. The demographic characteristics of the patient cohort are shown in Table 1.
Brain MR images were acquired through a 3 T clinical MR system (Inter Achieva SMI-2.1, Philip Medical System). All the patients were scanned by using the same MR protocol of the brain with a T1-weighted fast-field echo sequence (slice thickness = 5 mm; TR/TE = 250/2.3 ms; flip angle = 70°; FOV = 280 mm; scan matrix = 640 × 640; and voxel size = 5 × 0.4375 × 0.4375 mm3), a T2-weighted turbo-spin echo sequence (slice thickness = 5 mm; TR/TE = 3,508.4/80 ms; flip angle = 90°; FOV = 280 mm; scan matrix = 640 × 640; and voxel size = 5 × 0.4375 × 0.4375 mm3), and FLAIR sequence (slice thickness = 5 mm; TR/TE/TI = 11,000/120/2,800 ms; flip angle = 90°; FOV = 280 mm; scan matrix = 640 × 640; and voxel size = 5 × 0.4375 × 0.4375 mm3). The magnetic resonance (MR) images of these three sequences were acquired in axial direction. Besides the clinical MR images mentioned before, a subset of the Medical Image Computing and Computer-Assisted Intervention (MICCAI) data (http://www.ia.unc.edu/MSseg/index.html) was also employed to assess our segmentation methods. The FLAIR images of ten cases from the training dataset provided by Children’s Hospital of Boston (CHB) were used to detect multiple sclerosis (MS) lesions. The manual segmentation of the training cases was performed by an expert from CHB, and the manual segmentation results were publicly available.
Preprocessing with already available software
Two steps of image preprocessing were used before WMH segmentation: intensity inhomogeneity (IIH) correction and skull stripping. The images were loaded and displayed using the default view in the MIPAV software (http://mipav.cit.nih.gov/). During this process, a robust range scaling method was automatically performed to transform the raw images into new images with intensity values between 0 and 255. Two parameters, namely the window and level, defined a specific range of voxel intensities to be displayed. A window/level function was applied to make sure all the voxel intensities falling inside the specified range. The N3 inhomogeneity correction module [29] in the MIPAV software was employed to correct the MR inhomogeneity on the FLAIR images. The results were exported as regular bmp-images. Non-brain tissues, such as the skull and the scalp, which might cause misclassification of the WMHs, were eliminated by using the brain extraction tool (BET) [30] in MRIcro software (http://www.mccauslandcenter.sc.edu/mricro/mricro/). Image binarization was performed to get a binary brain template. The brain region was extracted from the FLAIR images by combining the bmp-images processed by N3 inhomogeneity correction and the binary brain template obtained before.
EVD-based WMH segmentation
Before WMH segmentation, Otsu’s threshold method [31] was employed to divide the FLAIR image into two parts: the foreground and background regions. The foreground region was composed of the WM, GM, and WMHs, whereas the background region consisted of the CSF and the region outside the brain. The intensities and locations for all the voxels in the foreground region were recorded so that the following WMH segmentation would only occur at the foreground area.
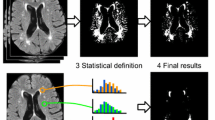
For some outlier detection issues, the EVD concerns the data distribution of the abnormally low or high values in the tails of some data-generating distributions. Notably, the histogram of the FLAIR image (Fig. 1) contains a single central peak, which represents normal brain tissues. The left-hand tail indicates the CSF region and the dark background, whereas the right-hand tail corresponds to the WMHs. Thus, the EVD can be employed to describe the intensity distribution of WMHs on FLAIR images. Given that we mainly focus on the right-hand tail of the histogram, the normal brain tissues (WM and GM) in the foreground region are modeled by using a UND.
Example of the WMHs on the FLAIR images. The WMHs are located at the periventricular WM in a single slice of FLAIR image (a). A pseudo-color FLAIR image (b) and the pseudo-color histogram of the colored FLAIR image (c) are provided to better understand the positional correspondence of specific tissues on the two colored graphics
The WMH segmentation is conducted by comparing the probability density functions (PDFs) of the UND and the EVD. Two different kinds of EVDs, namely, the Gumbel and Fréchet distributions [26, 27, 32], are suitable for detecting abnormally high signals in FLAIR images. The PDFs of the Gumbel and Fréchet distributions are given by
where x represents the intensity of each voxel in the foreground region. The PDF of the Gumbel distribution was evaluated through Eq. 1, using the mean (μ) and standard variation (σ) of the foreground region. The parameters α, m, and s in Eq. 2 correspond to the shape, scale, and location parameters, respectively, of the Fréchet distribution. UND was employed to model the intensity distribution property of the normal tissues (WM and GM), and the PDF is given by [28]
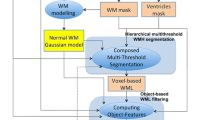
where μ and σ are the mean and standard variations of the foreground region. The PDFs between the UND and the EVD were compared to understand the WMH segmentation better (Fig. 2). Notably, the EVD-based segmentation was divided into two subtypes according to the different EVDs enrolled in our WMH segmentation. These subtypes are the Gumbel and Fréchet segmentations. The complete segmentation scheme of the proposed approach (Fig. 3) is composed of the following steps:
Comparison of the Gaussian distribution and the two EVDs. a The PDFs of the Gaussian distribution and Gumbel distribution, where the center of the Gaussian curve corresponds to the normal brain tissue (WM and GM) and the right tail of the Gumbel curve indicate the WMHs. b The PDFs of the Gaussian and Fréchet distributions
-
Step 1:
Calculation of the mean value μ and standard variation σ, with regard to the foreground region. The PDFs of Fréchet distributions using different combination of parameters (α, m, and s) were compared and the recommended empirical values of the parameters employed in the Fréchet segmentation should be α = 2, m = μ, and s = 3 σ.
-
Step 2:
Calculation of the PDFs, with respect to the UND and the two EVDs for each voxel in the foreground region.
-
Step 3:
Classification of the normal appearing brain tissues and the abnormal WMHs, by comparing the PDFs of the UND and the EVD. For Gumbel segmentation, the voxel would be considered as a possible WMH candidate if P UND < P Gumbel; otherwise, the voxel would be classified as the normal appearing brain tissue candidate if P UND ≥ P Gumbel. Likewise, Fréchet segmentation was performed by comparing the PDFs of the UND (P UND) and the Fréchet distribution (P Fréchet) for each voxel in the foreground region.
-
Step 4:
Performing the false-positive (FP) minimization. After the segmentation, there were some FP signals, like the bony artifacts and the flow artifacts in the WMH candidates. These FP signals mainly occurred around the interface of the CSF and cortical GM as well as the juxtacortical lesions. Thus, two morphological operations, including a single dilation and erosion, were performed on the binary WMH segmentation results to reduce the FP signals. A 3 × 3 convolution kernel was used for the morphological operations.
The two EVD-based WMH segmentation methods (the Gumbel and Fréchet segmentations) were implemented by using Matlab R2012b (The MathWorks Inc, Natick, MA, USA).
TLE-based WMH segmentation
TLE was proposed by Neykov et al. [22] to avoid outliers in the maximum likelihood estimation of the robust GMM. The TLE has been used to detect MS lesions in multidimensional MR sequences (T1w, T2w, and proton density weighted images) [18–20]. In our study, we implemented TLE-based segmentation to detect the WMHs on the FLAIR images. The required steps for the TLE-based segmentation are as follows:
-
Step 1:
Classification of the foreground and background regions using the EM algorithm. A two-component GMM was employed to model the intensity distribution of the FLAIR images that were not preprocessed via skull stripping. The foreground region consisted of the WM, GM, WMHs, and non-brain tissues (e.g., skull and scalp). The background region was composed of the CSF and the background outside the brain. The PDFs of all the voxels were computed with respect to the GMM.
-
Step 2:
Detection of abnormally high signals. With the computed PDFs for all the voxels in the FLAIR images, the TLE was employed to detect abnormally high signals (WMHs and non-brain tissues). The proportion of abnormally high signals in the FLAIR images has to be estimated before WMH segmentation because this estimation is a required parameter in TLE. An anatomy image atlas was constructed by using the FLAIR images of 20 patients with different WMH volumes, varying from small to large. The tissues, namely, the WMHs, skull, and scalp, that presented abnormally high signals were delineated or detected to estimate the proportion of the abnormally high signals on the FLAIR images. The proportion of the abnormally high signals was denoted by h, which indicates that approximately h × 100 % voxels in the FLAIR images can be considered as abnormally high signals. Thus, TLE detected the abnormally high signal by sorting the PDFs of all voxels in descending order and by considering the last h × 100 % voxels as abnormally high signals. When performing the TLE-based segmentation, we chose h = 0.035 to detect the patients with different lesion loads.
-
Step 3:
Refinement step. The abnormally high signals detected may include both WMHs and non-brain tissues. A binary brain template was generated through skull stripping with the use of BET in MRIcro. Non-brain tissues, namely, abnormally high signals that did not overlap with the binary brain template, were removed through morphology processing. Moreover, the morphology operations, including erosion and dilation, were performed on the binary segmentation results to eliminate the FP signals, such as the bony artifacts and flow artifacts.
The TLE-based WMH, segmentation on the FLAIR images was also implemented by using Matlab R2012b (The MathWorks Inc., Natick, MA, USA).
Statistical analysis
Three different automatic segmentation methods were developed to detect WMHs on FLAIR images: Gumbel, Fréchet, and TLE-based segmentations. Notably, the Gumbel and Fréchet segmentations are two subtypes of EVD-based segmentation. Manual segmentation was performed by an experienced neurologist and an experienced radiologist to delineate the WMHs on the FLAIR images, referring to the corresponding T1-w and T2-w images. The bmp-images were loaded in Adobe Photoshop CS5 [33], and the boundaries of the WMHs were delineated by the neurologist and radiologist separately. Two groups of the results were obtained after manual segmentation. The final results of manual segmentation were assessed in a consensus meeting. For each patient, the better manual segmentation result of the two groups was considered the ground truth and used to assess the performance of automatic segmentation methods. According to the WMH volumes obtained through manual segmentation, we categorized 60 patients into three groups [34, 35]: severe (WMH volume >18 cc; 14 patients), moderate (4 cc < WMH volume <18 cc; 31 patients), and mild (WMH volume <4 cc; 15 patients).
The performances of these automatic segmentation methods were assessed by using three different similarity measures: Dice similarity coefficient (DSC) [36], false-positive ratio (FPR), and false-negative ratio (FNR) [24, 37]. These similarity measures were defined as follows:
where A and M represent the WMH areas detected by automatic and manual segmentation, respectively. Similarly, Ā and \( \overline{\mathrm{M}} \) denote the background areas in the FLAIR images derived from the automatic and manual segmentation. M ∩ A represents the true positive (TP), which is the area of the WMHs that was correctly detected through automatic segmentation. The volume of \( \overline{\mathrm{M}}\cap \mathrm{A} \) corresponds to the FP, which is the area of the background that is falsely classified as WMHs. M ∩ Ā denotes the volume of the FN, which is the area of the WMHs that is falsely classified as the background. The DSC measures the percentage of the correctly detected WMH area relative to the total area of the WMHs derived from the manual and automatic segmentations. The value of the DSC varies between 0 and 1. Practically, a DSC value of 0.7 or higher indicates good agreement between automatic and manual segmentations [2, 37]. FPR and FNR respectively measure the FP (\( \overline{\mathrm{M}}\cap \mathrm{A} \)) and FN (M ∩ Ā) classifications of the WMHs in the area of the WMHs detected via manual segmentation. In fact, the evaluation of the three similarity measures mentioned above is limited, since these similarity metrics were sensitive to volumes. Thus, we also computed the outlier error rate (OER) [38], detection error rate (DER) [39], and SIestimate [38, 39] to evaluate the automatic segmentation methods. Moreover, a volumetric comparison between automatic and manual segmentations was performed using linear regression and Bland–Altman analysis [40, 41]. The intraclass correlation coefficient (ICC) and the Pearson’s correlation coefficient (PCC) were computed to evaluate volumetric agreement between manual and automated segmentation. Of note, the ICC values were obtained using two-way mixed model with absolute agreement definition. SPSS for Windows (version 19.0; SPSS, Chicago, IL, USA) was used for the statistical computations.
Results
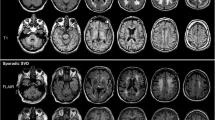
EVD- and TLE-based WMH segmentations were performed on 60 patients with different lesion loads. The three different automatic segmentation methods, namely, Gumbel, Fréchet, and TLE segmentations, were employed to detect the WMHs on the FLAIR images. The WMHs detected by the automatic and manual segmentations were illustrated on the images of three patients with a mild, moderate, and severe lesion loads (Fig. 4). For a clear evaluation, the regions of the TP, FP, and FN were detected using the binary results of automatic and manual segmentations. An illustrative example is provided to facilitate understanding of the real TP, FP, and FN signals in the segmentation results, which were obtained on a sample slice of a patient with a moderate lesion load (Fig. 5). The result of the WMH segmentation on the sample slices demonstrated the minimum FP signal for the Gumbel segmentation among the three automatic methods employed. Interestingly, Gumbel segmentation also produced the maximum FN signal.
The results of applying the different WMH segmentation methods on the FLAIR images of three patients with different lesion loads. From top to bottom: sample slices of three patients with mild (a–e), moderate (f–j), and severe (k–o) lesion loads. From left to right, the segmented WMHs derived from the manual (b, g, l), Gumbel (c, h, m), Fréchet (d, i, n), and TLE-based (e, j, o) segmentations are illustrated, respectively
Sample images of an axial slice for assessing the segmentation performance on a patient with moderate lesion load. From top to bottom: the TP (a–c), FP (d–f), and TN (g–i) signals derived from the automatic segmentation. From left to right: the results of different automatic segmentation methods, namely the Gumbel (a, d, g), Fréchet (b, e, h), and TLE-based (c, f, i) segmentations
Similarity measures
To measure the spatial accuracy of the three automatic segmentation methods, we computed and compared the DSCs, FPRs, and FNRs for all patients with different lesion loads (Table 2 and Fig. 6). The mean DSCs with respect to the Gumbel, Fréchet, and TLE segmentations for all patients were 0.823 ± 0.063, 0.843 ± 0.057, and 0.817 ± 0.068 (Table 2), respectively. These values indicate that the EVD-based (Gumbel and Fréchet) segmentations were more accurate than the TLE-based WMH segmentation (Fig. 6a). The comparison of these three segmentation methods revealed that Fréchet segmentation outperformed the other two segmentation methods on the patients with different lesion loads, considering all the similarity measures (e.g., DSC, FPR, and FNR). Gumbel segmentation performed slightly better than the TLE segmentation.
Similarity measures between the automatic segmentation and the ground truth derived from the manual segmentation as a function of lesion loads. The DSCs (a), FPRs (b), and FNRs (c) of the Gumbel, Fréchet, and TLE-based segmentation are obtained to assess the performance of the different automatic segmentation methods
The DSCs for all of the three automatic segmentation methods increased as the WMH volumes increased (Table 2 and Fig. 6a). In particular, for patients with mild lesion loads, Gumbel segmentation obtained higher DSC and lower FPR than the Fréchet and TLE-based segmentations. For patients with moderate and severe lesion loads, Fréchet segmentation outperformed the other two automatic methods. TLE-based segmentation performed well on patients with moderate and severe lesion loads. Nevertheless, the TLE-based segmentation remains limited, given that this method is prone to generate more FP signals than the EVD-based segmentation methods (Fig. 6b), especially on patients with mild lesion loads (FPR = 0.363 ± 0.175). The results of the Gumbel segmentation were more susceptible to the effect of the FN signals than those of the Fréchet and TLE-based segmentations (Fig. 6c).
The DER, OER, and SIestimate were listed in the Table 3. Similar to SI (Table 2), the SIestimate indicates the Fréchet segmentation has the best performance among the three automatic segmentation methods. The Fréchet segmentation obtained the minimum mean DER and mean OER for patients with mild and severe lesion loads. In general, the DERs and OERs decreased as the lesion volume increased.
Volumetric comparison
Linear regression and Bland–Altman analysis were performed to compare the volumetric agreement between the automatic and manual segmentations (Figs. 7 and 8). In the Bland–Altman plot, the difference between the two measurements (manual vs. automatic), which is referred to as the bias, is plotted against the average of the two measurements (Fig. 8).
Linear regression analysis. Manual delineated WMH volumes are plotted over automatically segmented WMH volumes. a Gumbel versus manual volume measurement. b Fréchet versus manual volume measurement. c TLE-based versus manual volume measurement. The solid lines represent the regression lines, while the dotted lines correspond to the equality line (y = x)
The regression analysis (Fig. 7a and Table 4) shows excellent agreement (ICC = 0.989; p < 0.01) between Gumbel and manual segmentations. The slope of the regression line of 0.936 did not differ significantly from the equality (y = x), and the intercept of −0.014 was close to 0. The Bland–Altman plot (Fig. 8a) indicates a system bias of 0.98 cc and a standard deviation (SD) of 1.76 cc over a range of 1.47 to 62.35 cc. The ICC value between Fréchet and manual segmentations (ICC = 0.993; p < 0.01) is a little larger than that between the Gumbel and manual segmentations (ICC = 0.989; p < 0.01). The slope and intercept for the regression line of Fréchet versus manual segmentation (Fig. 7b) were 0.989 and 0.318, respectively. The Bland–Altman plot (Fig. 8b) shows the absence of a systematic bias of −0.17 cc and a narrow 95 % limit of agreement (1.92 cc, 64.57 cc) with SD of 1.62 cc, which demonstrates a general overestimation of the WMH volume for Fréchet segmentation compared with manual segmentation.
The ICC value between TLE-based and manual segmentations (ICC = 0.981; p < 0.01) was lower than that between EVD-based (Gumbel and Fréchet) and manual segmentations. The slope of the regression line was 0.866, and the intercept was 1.669. The Bland–Altman analysis between the two measurements (manual versus TLE-based) (Fig. 8c) illustrates a system bias of 0.19 cc and SD of 2.58 cc over a range of 2.49 cc to 56.78 cc.
The average volumes of the WMHs quantified by these three automatic methods for patients with different lesion loads were summarized and compared with those derived from the manual segmentation (Table 5). Compared with manual segmentation, Gumbel segmentation underestimated the WMH volumes for patients with all lesion loads as a whole. An overestimation was observed for the WMH volumes derived by the Fréchet and TLE-based segmentations on patients with mild and moderate lesions. The WMH volumes for patients with severe lesion loads were prone to underestimation by all three automatic segmentation methods.
Validation on public datasets
The automatic segmentation methods were also validated on the MICCAI data. FLAIR images of ten cases from the training dataset provided by CHB were used to detect MS lesions. The manual segmentation results of the training cases were used to evaluate our segmentation methods. For the purpose of comparison, two similarity metrics, namely the true positive rate (TPR) and the positive predictive value (PPV) were employed to compare our methods with other two state-of-art segmentation methods proposed by Geremia et al. [42] and Abdullah et al. [43], respectively. The TPR was defined as TPR=TP/(TP+FN), and the PPV was computed by PPV=TP/(TP+FP) [43]. A larger value of TPR or PPV indicates less FN or FP signals in the segmented MS lesions. The results of the comparison between our automatic methods and other state-of-art methods were illustrated in Table 6. Given that the TLE-based segmentation obtained the minimum PPV, we considered that TLE-based segmentation produced more FP signals than the other two automatic methods (the Gumbel and Fréchet segmentation). The performance of the EVD-based segmentation was comparable to those of the other proposed methods. Of note, the automatic segmentation methods performed better on our own clinical images than the MICCAI dataset. In fact, the quality of the images in the MICCAI dataset is poorer given that various artifacts were found in most of the images [2].
Discussion
In this study, we proposed an EVD-based segmentation framework using the Gumbel and Fréchet distributions to detect and quantify WMHs on FLAIR images. A TLE-based segmentation method was implemented for comparison with the proposed EVD-based segmentation. The results of the three automatic segmentation methods (Gumbel, Fréchet, and TLE segmentations) were validated against the manually drawn WMHs on FLAIR images of 60 patients with different lesion loads. The comparison between our EVD-based segmentation and the TLE-based method yields higher DSCs and lower FPRs, indicating that the EVD-based segmentation (the Gumbel and Fréchet segmentation) outperformed TLE-based segmentation. Moreover, the Fréchet segmentation performed better and was more robust than the Gumbel segmentation on patients with different lesion loads.
EVD-based segmentation combined outlier detection theory with the adaptive threshold technique to detect WMHs. In EVD-based segmentation, the normal brain tissue (WM and GM) in the foreground region was modeled by using the UND. The WMHs were considered as outliers in the normal brain tissue model and were illustrated by the EVD. After comparing the PDFs of the UND and the EVD, the voxels whose probability density values of EVD were equal or greater than those with respect to UND were considered as the possible WMH candidates. The minimum intensity of these voxels corresponds to the threshold that distinguishes the WMHs from the normal brain tissue. Thus, the process of determining the thresholding process is completely automated and self-adaptive. The TLE-based segmentation employed the two-component GMM to model the foreground and background of the FLAIR images, and used the TLE to detect the WMHs as outliers of the GMM. The performance of the TLE-based segmentation depended on the parameter h, which indicated the possible ratio between the abnormal WMHs and the normal brain tissue in the FLAIR images. In fact, the TLE-based segmentation resembles a proximity-based thresholding technique [44], where a voxel is considered the possible WMH candidate if threshold distance from the normal brain tissue model (GMM) is exceeded. By contrast, the EVD-based segmentation is more principled than the TLE-based segmentation, given that that the EVD-based segmentation method uses an adaptive thresholding technique for WMH detection.
Compared with other state-of-the-art segmentation methods, the EVD-based segmentation is more automated and has better robustness on MR images collected, using different imaging sequence parameters or imaging devices. For example, supervised segmentation methods (e.g., k-nearest neighbors (k-NN), artificial neural networks (ANNs), or support vector machine (SVM)) [45–47] are commonly employed for various lesion segmentation tasks. These methods involve a training stage that uses a training database to learn the feature of the lesions to discriminate the WMHs from the normal brain tissues. However, the construction of a well-performed training database is difficult because the training database should cover all possible cases, and all the lesions should be previously segmented using the labor-intensive and time-consuming manual segmentation [2, 9]. This way, the EVD-based segmentation is more automated and does not need manual intervention compared with the supervised segmentation. The EVD-based segmentation is independent of the acquisition protocol and should not be affected by variability in lesion size, shape, and location. We employed a subset of the MICCAI data to evaluate our segmentation methods. The comparison of our methods and other two state-of-art supervised segmentation methods [42, 43] demonstrated that the EVD-based segmentation can be generalized to other scanners and to other types of white matter damage, e.g., MS lesions, which are less confluent and can be more diffuse. Furthermore, the EVD-based segmentation achieved good segmentation performance when dealing with different lesion types (e.g., periventricular, deep white matter, juxtacortical, or infratentorial lesions). The EVD-based segmentation is also more advantageous than the other unsupervised methods. For instance, the lesion-based segmentation using empirical rules performs well for a particular type of lesion. However, the segmentation performance of this method can be negatively affected by the image intensity non-standardness [48], which implies that intensities do not have a tissue-specific numeric meaning for images collected from different patients. Likewise, the method is negatively influenced when distinct imaging parameters are used. Given that the lesion-based segmentation often directly uses the intensity of the MR images to model and detect WMHs, then intensity standardization [49] should be performed on all of the MR images requiring segmentation. Intensity standardization is complicated and can have noticeable effects on the results of the lesion-based segmentation. By contrast, based on the outlier detection theory and adaptive threshold technique, the EVD-based WMH segmentation is flexible with no restrictions on the MRI imaging parameters or image intensities.
Lesion load is an important issue that should be considered during the evaluation of a segmentation method, as the segmentation performance can differ depending on whether the lesion load is mild or severe. Generally, in our study, the EVD-based segmentation performed better against the TLE-based segmentation regardless of the lesion load. For the patients with mild lesion loads, the Gumbel segmentation outperformed the Fréchet and TLE-based segmentations because the Gumbel segmentation method has a maximum DSC and a minimum FPR. Among the three automatic segmentation methods employed in our study, the Fréchet segmentation performed best on the patients with moderate and severe lesion loads. Although the TLE-based segmentation performed well on the patients with moderate and severe lesion loads, this segmentation method produced an excessive amount of FP signals compared with the two EVD-based segmentation methods, especially on the patients with mild lesion loads. Hence, the TLE-based segmentation is inappropriate, considering the effect of the FP signals on the final results. The FNR was computed to measure the percentage of the missed WMH voxels. A comparison of the two EVD-based segmentation methods yields that the FNR for the Gumbel segmentation is higher than that for the Fréchet segmentation. The probabilistic characteristics of the Gumbel and Fréchet distributions are well explained. The Gumbel distribution is actually more specifically suited for outlier detection, with respect to data mixed with minimal abnormal data samples. Hence, the Gumbel segmentation most probably underestimated the WMHs for the patients with moderate and severe lesion loads. By contrast, the Fréchet distribution is flexible enough to handle different outlier detection tasks, regardless of the amount of the abnormal data samples. Thus, the Fréchet segmentation should conform more to the demands of the WMH segmentation, considering the varying lesion loads for different patients.
The primary sources of the FP signals in the results of the EVD-based segmentation are directly linked to some bony and CSF flow artifacts. To eliminate the FP signals, some studies [50] proposed to segment the WM region on the T1-w images first. Subsequently, a morphological operation is performed on the binary segmentation results and the binary WM mask, to eliminate the FP signals outside the WM. However, the difficulty of detecting the complete region of the WM is an issue. Given that the signal of the WMHs in the T1-w images is lower than that of the WM, different-sized “black holes” will probably be detected in the WM region. Thus, a morphological closing operation is required to fill these “black holes” and obtain a contiguous WM region. Nevertheless, obtaining a satisfactory WM region is made difficult by the different-sized “black holes.” The binary mathematical morphological operations of dilation and erosion were performed in our study to eliminate the FP signals that present either threadiness or isolated voxels. Of note, the EVD-based segmentation behaved well in the presence of hyperintense choroid plexus whereas the TLE-based segmentation method may classified little hyperintense choroid plexus as possible lesions. Thus, as far as the FPR is concerned, the EVD-based segmentation is likely to maintain the FPR signals in a relatively lower level compared with the other methods (for example, the TLE-based segmentation).
Conversely, our method is limited because different normal tissues (WM, GM, and CSF) were undetected along with the WMHs, given that only the FLAIR images were employed in our work. Moreover, the FLAIR may overestimate the lesion loads in the lateral ventricle or supratentorial region when compared with the T2-w or proton density-weighted images [2]. To solve this problem, some methods were proposed to detect the WMHs, combing information from different MRI sequences, particularly for the supervised segmentation methods. These methods are advantageous in segmenting the WM, GM, and CSF along with the WMHs. However, using multimodal MR images presents some disadvantages. Before the segmentation, the different MR sequences of the same patients should be registered into the same space through a proper image registration algorithm, which is a labor-intensive and time-consuming task. Moreover, the MR images collected from the elderly patients are often accompanied by motion artifacts and bony artifacts, which can negatively influence the outcome of the registration and make the segmentation results unconvincing. In this study, we solely focus on the automatic segmentation and volumetric quantification of the WMHs in the WM areas. Actually, the FLAIR images are more advantageous over the other conventional MRI techniques when detecting WMHs because of the high contrasts between the WMHs and the other normal brain tissues (e.g., WM, GM, and CSF). Using the FLAIR images provides a fast and efficient WMH segmentation method to meet the required accuracy (DSC > 0.7).
In conclusion, we presented an EVD-based segmentation framework to detect WMHs on FLAIR images. The EVD-based segmentation framework has been applied to detect and quantify the WMH volume of patients with different lesion loads. The evaluation of the EVD-based segmentation on the MICCAI data indicated that the performance of this segmentation is comparable to other state-of-art segmentation methods. This EVD-based segmentation outperformed the TLE-based segmentation method. Notably, two different EVDs (Gumbel and Fréchet distributions) were employed in the EVD segmentation framework. The Fréchet segmentation generally performed better than the Gumbel segmentation. Thus, the EVD-based segmentation using the Fréchet distribution can be the best choice, by weighing various factors comprehensively. In the future, we will release the EVD-based segmentation framework as a user-friendly software for larger cohorts to employ this segmentation framework for the study of WMHs in aging and dementia.
Abbreviations
- WMHs:
-
White matter hyperintensities
- FLAIR:
-
Fluid-attenuated inversion recovery
- MRI:
-
Magnetic resonance imaging
- TLE:
-
Trimmed likelihood estimator
- EVD:
-
Extreme value distribution
- UND:
-
Unilateral normal distribution
- PDF:
-
Probability density function
- DSC:
-
Dice similarity coefficient
- FPR:
-
False-positive rate
- FNR:
-
False-negative rate
References
Debette S, Markus H (2010) The clinical importance of white matter hyperintensities on brain magnetic resonance imaging: systematic review and meta-analysis. BMJ 341:c3666
García-Lorenzo D, Francis S, Narayanan S, Arnold DL, Collins DL (2013) Review of automatic segmentation methods of multiple sclerosis white matter lesions on conventional magnetic resonance imaging. Med Image Anal 17:1–18
Barkhof F, Scheltens P (2002) Imaging of white matter lesions. Cerebrovasc Dis 13:21–30
Au R, Massaro JM, Wolf PA et al (2006) Association of white matter hyperintensity volume with decreased cognitive functioning: the Framingham Heart Study. Arch Neurol 63:246–250
Whitman G, Tang T, Lin A, Baloh R (2001) A prospective study of cerebral white matter abnormalities in older people with gait dysfunction. Neurology 57:990–994
Smith CD, Snowdon DA, Wang H, Markesbery WR (2000) White matter volumes and periventricular white matter hyperintensities in aging and dementia. Neurology 54:838–842
Herrmann LL, Le Masurier M, Ebmeier KP (2008) White matter hyperintensities in late life depression: a systematic review. J Neurol Neurosurg Psychiatry 79:619–624
Pantoni L, Garcia JH (1997) Pathogenesis of leukoaraiosis a review. Stroke 28:652–659
Lladó X, Oliver A, Cabezas M et al (2012) Segmentation of multiple sclerosis lesions in brain MRI: a review of automated approaches. Inf Sci 186:164–185
Neumann Haefelin T (2006) Leukoaraiosis is a risk factor for symptomatic intracerebral hemorrhage after thrombolysis for acute stroke. Stroke 37:2463–2466
Palumbo V (2007) Leukoaraiosis and intracerebral hemorrhage after thrombolysis in acute stroke. Neurology 68:1020–1024
Brickman AM, Siedlecki KL, Muraskin J et al (2011) White matter hyperintensities and cognition: testing the reserve hypothesis. Neurobiol Aging 32:1588–1598
Prins ND, van Dijk EJ, den Heijer T et al (2004) Cerebral white matter lesions and the risk of dementia. Arch Neurol 61:1531–1534
Bedell BJ, Narayana PA (1998) Automatic segmentation of gadolinium enhanced multiple sclerosis lesions. Magn Reson Med 39:935–940
Boudraa A-O, Dehak SMR, Zhu Y-M, Pachai C, Bao Y-G, Grimaud J (2000) Automated segmentation of multiple sclerosis lesions in multispectral MR imaging using fuzzy clustering. Comput Biol Med 30:23–40
Datta S, Sajja BR, He R, Gupta RK, Wolinsky JS, Narayana PA (2007) Segmentation of gadolinium-enhanced lesions on MRI in multiple sclerosis. J Magn Reson Imaging 25:932–937
Van Leemput K, Maes F, Vandermeulen D, Colchester A, Suetens P (2001) Automated segmentation of multiple sclerosis lesions by model outlier detection. IEEE Trans Med Imaging 20:677–688
Aït-Ali LS, Prima S, Hellier P, Carsin B, Edan G, Barillot C (2005) STREM: a robust multidimensional parametric method to segment MS lesions in MRI. Medical Image Computing and Computer-Assisted Intervention–MICCAI 2005. Springer, pp 409–416
Bricq S, Collet C, Armspach J-P (2008) Lesions detection on 3D brain MRI using trimmed likelihood estimator and probabilistic atlas. Biomedical Imaging: From Nano to Macro, 2008 ISBI 2008 5th IEEE International Symposium on. IEEE, pp 93–96
García-Lorenzo D, Prima S, Arnold DL, Collins DL, Barillot C (2011) Trimmed-likelihood estimation for focal lesions and tissue segmentation in multisequence MRI for multiple sclerosis. IEEE Trans Med Imaging 30:1455–1467
Zhang Y, Brady M, Smith S (2001) Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans Med Imaging 20:45–57
Neykov N, Filzmoser P, Dimova R, Neytchev P (2007) Robust fitting of mixtures using the trimmed likelihood estimator. Comput Stat Data Anal 52:299–308
Simões R, Mönninghoff C, Dlugaj M et al (2013) Automatic segmentation of cerebral white matter hyperintensities using only 3D FLAIR images. Magn Reson Imaging 31:1182–1189
Gibson E, Gao F, Black SE, Lobaugh NJ (2010) Automatic segmentation of white matter hyperintensities in the elderly using FLAIR images at 3T. J Magn Reson Imaging 31:1311–1322
Khayati R, Vafadust M, Towhidkhah F, Nabavi M (2008) Fully automatic segmentation of multiple sclerosis lesions in brain MR FLAIR images using adaptive mixtures method and Markov random field model. Comput Biol Med 38:379–390
de Haan L, Ferreira A (2006) Extreme value theory: an introduction. Springer, New York
Kotz S, Nadarajah S (2000) Extreme value distributions. Imperial College Press, London
Roberts SJ (2000) Extreme value statistics for novelty detection in biomedical data processing. Science, Measurement and Technology, IEE Proceedings-. IET, pp 363–367
Sled JG, Zijdenbos AP, Evans AC (1998) A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imaging 17:87–97
Smith SM (2002) Fast robust automated brain extraction. Hum Brain Mapp 17:143–155
Otsu N (1979) A threshold selection method from gray-level histograms. IEEE Trans Syst Man Cybern 9:62–66
MacDonald A, Scarrott CJ, Lee D, Darlow B, Reale M, Russell G (2011) A flexible extreme value mixture model. Comput Stat Data Anal 55:2137–2157
Park JS, Chung MS, Hwang SB, Lee YS, Har D-H (2005) Technical report on semiautomatic segmentation using the Adobe Photoshop. J Digit Imaging 18:333–343
Admiraal-Behloul F, Van Den Heuvel D, Olofsen H et al (2005) Fully automatic segmentation of white matter hyperintensities in MR images of the elderly. Neuroimage 28:607–617
Anbeek P, Vincken KL, van Osch MJ, Bisschops RH, van der Grond J (2004) Probabilistic segmentation of white matter lesions in MR imaging. Neuroimage 21:1037–1044
Dice LR (1945) Measures of the amount of ecologic association between species. Ecology 26:297–302
Shiee N, Bazin P-L, Ozturk A, Reich DS, Calabresi PA, Pham DL (2010) A topology-preserving approach to the segmentation of brain images with multiple sclerosis lesions. Neuroimage 49:1524–1535
Wack DS, Dwyer MG, Bergsland N et al (2012) Improved assessment of multiple sclerosis lesion segmentation agreement via detection and outline error estimates. BMC Med Imaging 12:17
Steenwijk MD, Pouwels PJ, Daams M et al (2013) Accurate white matter lesion segmentation by k nearest neighbor classification with tissue type priors (kNN-TTPs). Neuroimage Clin 3:462–469
Altman DG, Bland JM (1983) Measurement in medicine: the analysis of method comparison studies. Stat 32:307–317
Bland JM, Altman DG (1999) Measuring agreement in method comparison studies. Stat Methods Med Res 8:135–160
Geremia E (2011) Spatial decision forests for MS lesion segmentation in multi-channel magnetic resonance images. Neuroimage 57:378–390
Abdullah BA, Younis AA, John NM (2012) Multi-sectional views textural based SVM for MS lesion segmentation in multi-channels MRIs. Open Biomed Eng J 6:56
Hodge VJ, Austin J (2004) A survey of outlier detection methodologies. Artif Intell Rev 22:85–126
Wu Y, Warfield SK, Tan IL et al (2006) Automated segmentation of multiple sclerosis lesion subtypes with multichannel MRI. Neuroimage 32:1205–1215
Cerasa A, Bilotta E, Augimeri A et al (2012) A cellular neural network methodology for the automated segmentation of multiple sclerosis lesions. J Neurosci Methods 203:193–199
Yamamoto D, Arimura H, Kakeda S et al (2010) Computer-aided detection of multiple sclerosis lesions in brain magnetic resonance images: false positive reduction scheme consisted of rule-based, level set method, and support vector machine. Comput Med Imaging Graph 34:404–413
Madabhushi A, Udupa JK (2005) Interplay between intensity standardization and inhomogeneity correction in MR image processing. IEEE Trans Med Imaging 24:561–576
Zhuge Y, Udupa JK (2009) Intensity standardization simplifies brain MR image segmentation. Comput Vis Image Underst 113:1095–1103
Ong KH, Ramachandram D, Mandava R, Shuaib IL (2012) Automatic white matter lesion segmentation using an adaptive outlier detection method. Magn Reson Imaging 30:807–823
Acknowledgments
This study has received funding by National Basic Research Program of China (973 Program, No. 2010CB732506), National Natural Science Foundation of China (No. 81301213), National Natural Science Foundation of China (No. 81000609), National Natural Science Foundation of China (No. 60972110), and Major Program of Social Science Foundation of China (No. 11&ZD174).
Ethical standards and patient consent
We declare that all human studies have been approved by the Institutional Review Board of Shanghai Jiao Tong University Sixth Affiliated People’s Hospital and have therefore been performed in accordance with the ethical standards laid down in the 1964 Declaration of Helsinki and its later amendments. We declare that all patients gave informed consent prior to inclusion in this study.
Conflict of interest
We declare that we have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wang, R., Li, C., Wang, J. et al. Automatic segmentation and volumetric quantification of white matter hyperintensities on fluid-attenuated inversion recovery images using the extreme value distribution. Neuroradiology 57, 307–320 (2015). https://doi.org/10.1007/s00234-014-1466-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00234-014-1466-4