Abstract
At the neuromuscular junction (NMJ), the nicotinic acetylcholine receptor (nAChR) self-associates to give rise to rapid muscle movement. While lipid domains have maintained nAChR aggregates in vitro, their specific roles in nAChR clustering are currently unknown. In the present study, we carried out coarse-grained molecular dynamics simulations (CG-MD) of 1–4 nAChR molecules in two membrane environments: one mixture containing domain-forming, homoacidic lipids, and a second mixture consisting of heteroacidic lipids. Spontaneous dimerization of nAChRs was up to ten times more likely in domain-forming membranes; however, the effect was not significant in four-protein systems, suggesting that lipid domains are less critical to nAChR oligomerization when protein concentration is higher. With regard to lipid preferences, nAChRs consistently partitioned into liquid-disordered domains occupied by the omega-3 (\(\omega\)-3) fatty acid, docosahexaenoic acid (DHA); enrichment of DHA boundary lipids increased with protein concentration, particularly in homoacidic membranes. This result suggests dimer formation blocks access of saturated chains and cholesterol, but not polyunsaturated chains, to boundary lipid sites.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The muscle-derived nicotinic acetylcholine receptor (nAChR) (PDB 2BG9) (Unwin 2005) is the most abundant neurotransmitter receptor at the neuromuscular junction (NMJ) in most vertebrates, including humans (Albuquerque et al. 2009). Within the postsynaptic membrane, nAChRs cluster in high densities (\(10,000\,\text {per}\,\upmu \text {m}^{2}\)) to properly activate the skeletal muscle (Ramarao and Cohen 1998; Breckenridge et al. 1972). As a major transmembrane protein, nAChR depends upon a highly specific lipid environment to maintain functionality. Lipids influence nAChR activity by affecting both function and organization. It is essential to understand how changes in lipid environment impact nAChR’s structure and activity, given that lipids can change in response to aging and disease (Yadav and Tiwari 2014), and also vary across tissue and organism.
Over the past few decades, considerable progress has been made in uncovering lipid sensitivities associated with nAChR (Criado et al. 1982). In a majority of experiments, researchers have prioritized studying cholesterol over other membrane lipids. Early studies revealed that, when reconstituted into membrane mixtures, nAChR failed to conduct cations across the lipid bilayer unless cholesterol was present (Fong and McNamee 1986; Sunshine and McNamee 1992; Butler et al. 1993; Fong and McNamee 1987; daCosta et al. 2001). More recently, researchers have examined the effects of membrane dynamics on nAChR organization (Baenziger et al. 1848; Brusés et al. 2001; Marchand et al. 2002; Oshikawa et al. 2003; Pato et al. 2008; Zhu et al. 2006; Baenziger et al. 2017; Barrantes 2007; Barrantes et al. 2000; Bermudez et al. 2010; Barrantes et al. 2010; Perillo et al. 2016; Wenz and Barrantes 2005; Borroni et al. 2016; Unwin 2017). In-vitro studies (Barrantes 2007; Barrantes et al. 2010) indicate that nAChRs form larger aggregates upon cholesterol depletion. Experimental evidence suggest that cholesterol-rich lipid domains, known as lipid rafts, facilitate clustering of nAChRs (Campagna and Fallon 2006; Marchand et al. 2002; Pato et al. 2008). More specifically, after disrupting lipid raft formation, Zhu et al. observed a significant loss of nAChR clusters in vitro (Zhu et al. 2006). In the mature neuromuscular membrane, nAChRs are linked by the intracellular anchoring protein, rapsyn, which bridges receptors together at their bases (Zuber and Unwin 2013). According to fluorescent studies (Marchand et al. 2002), lipid rafts mediate the association between rapsyn and neighboring nAChR molecules. In the mid-2000s, Willmann et al. and Stetzkowski-Marden et al. proposed that lipid rafts can stabilize receptor networks and may even provide a localized environment for nAChR (Willmann et al. 2006; Stetzkowski-Marden et al. 2006).
Domain formation occurs when a membrane is comprised of at least three lipid types: cholesterol, unsaturated fatty acids, and a molecule that interacts closely with cholesterol such as saturated fatty acids or sphingomyelin (Feller 2008; Yeagle 2016). The membrane separates into at least two domains, a liquid-ordered or “raft” phase containing cholesterol and saturated lipids/sphingomyelin, and a liquid-disordered phase containing unsaturated lipids. Polyunsaturated phospholipids make these domains more well defined (Levental et al. 2016). Some results from atomistic simulations (Sodt et al. 2014; Iyer et al. 2018) indicate that cholesterol may preferentially partition to the boundary between liquid-ordered phases and liquid-disordered phases composed of monounsaturated lipids. In neuromuscular membranes, intrinsic domain formation is dependent upon several lipid species, including the widely influential omega-3 (\(\omega\)-3) fatty acids. One \(\omega\)-3 in particular, Docosahexaenoic acid (DHA), is prevalent in the native neuromuscular membrane and is strongly associated with flexible and well-defined domains (Turk and Chapkin 2013; Shaikh et al. 2004). Additionally, DHA is a major contributor to brain functioning, motor activity, and cardiac health; however, its specific effects on neuromuscular health are poorly understood (Lavandera et al. 2017; Wassall and Stillwell 2008; Georgieva et al. 2015).
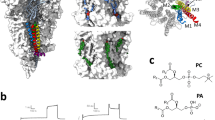
Nicotinic acetylcholine receptor (nAChR) structure. a Atomistic and coarse-grained representations of Unwin et al’s cryo-EM structure, PDB 2BG9. The nAChR is colored by subunit (\(\alpha :green,\beta :purple,\delta :gray,\gamma :cyan\)) and labeled by structure. The extracellular domain (ECD) is located above the bilayer and is critical for ligand binding. The transmembrane domain (TMD) is positioned within the lipid bilayer, and the intracellular domain (ICD) is located in the cytoplasm. The coarse-grained model omits the ICD since it is poorly resolved and not necessary for this study. b TMD from the extracellular perspective, with its four alpha-helices labeled in a single \(\alpha\) subunit. The outermost M4 helices closely interact with surrounding membrane lipids, while the M2 helices outline the ion pore; the M1 and M3 helices make up the body of the transmembrane domain (Unwin 2005) (Color figure online)
Characterizing such complex lipid–protein interactions requires detailed information on each molecular structure. The nAChR is a member of a family of ion channels, known as pentameric ligand-gated ion channels (pLGICs), which has a number of recent structures (Laverty et al. 2019, 2017; Masiulis et al. 2019; Althoff et al. 2014; Hibbs and Gouaux 2011; Morales-Perez et al. 2016a; Baenziger and Corringer 2011; Corringer et al. 2012; Nemecz et al. 2016; Prevost et al. 2012; Sauguet et al. 2014); however, only single structures have been determined, rather than dimers. pLGICs are composed of five subunits; each subunit is composed of an extracellular domain for ligand binding, a transmembrane domain (TMD), and an intracellular domain (ICD) (Fig. 1a). The TMD is the embedded portion of pLGICs that interacts most often with surrounding membrane lipids; this region of pLGICs is composed of four alpha-helices, M1–M4, with the innermost, M2 helices forming the ion pore. The ICD, or cytoplasmic domain, is highly disordered, making it challenging to obtain information on its structure. Currently, there are two structures of nAChR pentamers resolved from X-ray crystallography (neuronal type) and cryo-electron microscopy (muscle-type) (Morales-Perez et al. 2016b; Unwin 2005).
Other closely related channels have been resolved with bound lipids. In 2017, the inhibitory neurotransmitter receptor, the gamma-aminobutyric acid (\(\text {GABA}_A\)) receptor, was crystallized with a cholesterol molecule protruding between its M1 and M3 helices (Laverty et al. 2017), similar to our previous prediction (Hénin et al. 2014). Interestingly, X-ray structures of the glutamate-gated chloride channel (GluCl) revealed embedded POPC phospholipids in the same location (Althoff et al. 2014). Together, these data provide evidence for lipid-based modulation of pLGICs, which can potentially be applied to the gating of nAChR.
Structural information alone, however, is insufficient for answering questions about native cell membranes. For one, most structures are obtained under artificial conditions, using detergents or nanodisks with non-native lipid composition. Additionally, current structures do not represent oligomers, particularly in a liquid state. Fully atomistic molecular dynamics (MD) simulations (Brannigan et al. 2008; Cheng et al. 2009; Hénin et al. 2014; Carswell et al. 2015) have complemented experimental investigations of pLGICs interacting with lipids, but they are limited in their ability to capture domain formation since atomistic lipids cannot diffuse over typical simulation microsecond time scales (Ingólfsson et al. 2014; Bond and Sansom 2006; Parton et al. 2013; Goose and Sansom 2013; Scott et al. 2008). Furthermore, it is unfeasible to incorporate multiple nAChRs in simulations with atomistic resolution. Coarse-grained MD (CG-MD) simulations are run over longer length and time scales, making them suitable for exploring complex model membranes. CG-MD is widely applied in simulations of both lipid–protein binding and domain formation (Bond and Sansom 2006; Scott et al. 2008; Parton et al. 2013; Goose and Sansom 2013). Additionally, CG-MD can capture large-scale membrane phenomenon such as protein self-assembly and lipid-mediated oligomerization (Gahbauer and Böckmann 2016; Baaden and Marrink 2013).
Recently, we (Sharp et al. 2019) conducted CG-MD simulations of a single nAChR from the electric ray Torpedo in mixed membranes. Contrary to expectations, nAChR consistently preferred a local lipid environment rich in PUFAs rather than cholesterol, especially long-chained \(\omega\)-3s, such as DHA. While cholesterol occupied the transmembrane gaps of nAChR, PUFAs were even more likely to be embedded, regardless of their phospholipid headgroup.
The present study adopts a similar approach, with a particular focus on nAChR-associated clustering. Through molecular dynamics simulations, we investigate nAChR lipid preferences and clustering behavior in membranes with and without domains. For this study, we tested three major hypotheses: (1) Membrane organization affects nAChR boundary lipid specificity: when PUFA chains are prevented from forming PUFA-rich domains, their prevalence among nAChR boundary lipids will be significantly reduced. (2) Domain formation will indirectly facilitate the clustering of nAChRs, by inducing partitioning preferences and restricting diffusion within the membrane. (3) Within a dimer, we will observe sequence preferences in facing subunits.
Methods
System Setup
In order to isolate the role of lipid domain formation on nAChR-based lipid sorting and clustering, our simulations compared two different membranes with distinct phospholipid topology. Each membrane had 30% cholesterol and 70% phospholipids. The phospholipids all had phosphatidylcholine (PC) headgroups, and half the total number of acyl chains were saturated (16:0) chains, while the other half were polyunsaturated (22:6) chains. For homoacid membranes, all phospholipids had either two polyunsaturated acyl chains (didocosahexaenoylphosphatidylcholine or dDHA-PC) or two saturated chains (dipalmitoylphosphatidylcholine or DPPC), facilitating domain formation. In the heteroacid membranes, all phospholipids were hybrid lipids with one polyunsaturated acyl chain and one saturated chain (1-palmitoyl-2-docosahexaenoyl-phosphatidylcholine or PDPC), which are topologically unable to separate into PUFA-rich and PUFA-poor domains.
Lipids and proteins were modeled using the coarse-grained (CG) Martini 2.2 force field (Marrink et al. 2007). Systems included between 1–4 nAChR molecules, derived from the Torpedo electric organ (Unwin 2005) (PDB 2BG9). This structure is the only pLGIC structure obtained in a native membrane.
We converted protein structures into CG models using the Martini script “martinize.py,” mapping four non-hydrogen atoms to one CG interaction. We constructed and assembled our protein-bilayer systems using the Martini script, “insane.py,” using a box sizes of \(29\times 29\times 21\,\text {nm}^{3}\) for one-to-two proteins, \(40\times 40\times 20\,\text {nm}^{3}\) for three proteins, and \(44\times 44\times 22\,\text {nm}^{3}\) for four-protein systems, respectively (Marrink et al. 2007). Initially, the proteins were in a circle of about 13 nm. The receptors were evenly spaced, and their \(\delta\) subunits were facing the same direction. Once simulations started, nAChRs shifted from their initial orientations. We ran 24 CG-MD simulations containing 1–4 nAChRs (3 replicas per system).
Simulation Details
Simulations were run using the Martini 2.2 force field parameters and the Gromacs 5.1.2 simulation package (Marrink et al. 2007; Pronk et al. 2013) as in our previous work (Sharp et al. 2019). Each simulation consisted of two steps: energy minimization and molecular dynamics. For each system, we ran two consecutive energy minimizations for 10,000 steps. Harmonic restraints between backbone atoms were imposed to preserve nAChR conformation. More specifically, we applied an elastic force constant of 750 kJ/mol and set lower and upper bounds on the bond with using a bond length of 0.7 nm (Marrink et al. 2007; Pronk et al. 2013). The molecular dynamics simulations ran for 10–20 \(\upmu \text {s}\) at a 0.025 ps time-step. Simulation temperature and pressure were kept constant at values of 323 K and a reference pressure of 1 bar. The isotropic pressure coupling compressibility constant was maintained at \(3.0 \times {10^{-5}}\,\text {bar}^{-1}\).
Analysis
For a given nAChR, \(n_{\mathrm{emb}}^{\alpha }\) is the total number of embedded lipids of lipid species \(\alpha\), where embedded lipids satisfy the following criteria: the headgroup (in the case of cholesterol) or the terminal bead on the acyl chains (in the case of phospholipids) is within 10 Å of the M2 helices. Visual inspection indicated any lipids outside of this range were also outside of the TMD bundle.
Similarly, \(n_{\mathrm{ann}}^{\alpha }\) is the total number of annular lipids of lipid species \(\alpha\), where annular lipids satisfy the following criteria: headgroup or the terminal bead on the acyl chains is between 10 and 35 Å of the M2 helices. This range of lipids corresponded to the third lipidation shell around nAChR.
(For phospholipids, each acyl chain was counted separately as a half-lipid, so it was possible for, e.g., the sn-1 chain to be embedded and the sn-2 chain to be annular.)
Boundary lipid fractions for a given species \(\alpha\) are defined as
where \(n_{\mathrm{emb}}\) and \(n_{\mathrm{ann}}\) are the total number of embedded and annular lipids, respectively.
Two-dimensional density distributions of boundary acyl chain species (B), \(\tilde{\rho }_{\text {B}}(r_i,\theta _j),\) were calculated, as a function of radius, \(r_i\), and angle \(\theta _j\) projected onto the membrane.
where \(r_i = i \varDelta r\) is the distance from the origin to the center of bin i, \(\varDelta r\) is the radial bin width, and \(\varDelta \theta\) is the angular bin width. \(\left\langle n_{\text {B}}(r_i,\theta _j) \right\rangle\) is the time-averaged number of acyl chain beads of species B within bin i.
To quantify enrichment or depletion of a given acyl chain with respect to random distribution, the normalized density, \(\tilde{\rho }_{\text {B}}(r_i,\theta _j)\), was calculated.
where \(s_{\text {B}}\) is the number of beads of lipid species B, \(N_L\) is total number of lipids in a system, and \(\langle L^{2}\rangle\) is the average projected box area, and \(x_\text {B}\) is lipid B concentration. The expression does not take into consideration the protein footprint or undulations present within the system, and as such is an approximation.
The radial distribution function g(r) of multiple nAChRs was calculated using the three-dimensional distance r between the centers of mass of the receptors for each of the 8000 frames, evenly distributed through the simulation. It was derived from the distribution of pairwise separations, P(r), by dividing by the expected separations in a random distribution: \(g(r) = \frac{P(r)\,\text {d}r}{r\,\text {d}r}\times \int _{0}^{R} r \text {d}r\). The height of the g(r) peak corresponds to the amount of enrichment in probability for a given distance; for example, if \(\text {g}(10\,\text {nm}) = 50\), it is 50 times more probable that two monomers will be 10 nm apart than expected in a random distribution.
The total number of observed dimers \(n_{d}\) is given by the number of pairs (across all analyzed proteins) where \(r<100\) Å. For each observed dimer, the closest subunit pair was determined, and the enrichment calculated as \(F_{x,y}\):
where \(n_{x,y}\) represents the number of dimers in which subunits x and y form the closest pair, and 1/25 is the expectation for a random distribution.
We visualized and imaged all simulation results using visual molecular dynamics (VMD) (Humphrey et al. 1996).
Results
nAChR Boundary Lipid Preferences
In order to determine the significance of domain formation on enrichment of polyunsaturated acyl chains among boundary lipids, we calculated the fraction of embedded and annular boundary lipids in two types of membranes with equal amounts of cholesterol and saturated and polyunsaturated acyl chains. In the domain-forming membranes, all phospholipids had either two polyunsaturated acyl chains or two saturated chains (homoacids), while in the non-domain-forming membranes, all phospholipids had one polyunsaturated acyl chain and one saturated chain (heteroacids).
Distributions for \(f_\text {emb}\) and \(f_\text {ann}\) are shown in Fig. 2 in heteroacids compositions without domains and in Fig. 3 for homoacids compositions with domains, while mean values are given in Table 1. For heteroacids, PUFA chains are slightly enriched among embedded chains and slightly depleted among annular chains, while the reverse trend is observed for saturated lipids. Saturated chains are significantly depleted from the embedded lipids, but due to the topology of these lipids, a fully embedded polyunsaturated acyl chain must also contribute a saturated chain to the protein annulus. Distributions were remarkably consistent across simulations containing 1, 2, 3, or 4 proteins.
Figure 3 shows the results of the same analysis for homoacidic membranes. Replacing heteroacids with homoacids substantially increases the fraction of polyunsaturated chains among both embedded and annular lipids, with the peak of the distribution shifting further to the right as more proteins are included (Fig. 3).
Together, these results are consistent with our previous (Sharp et al. 2019) observation that polyunsaturated chains can displace cholesterol from embedded sites. For heteroacids, embedding a PUFA constrained the linked saturated chain to the nAChR annulus. In this case, cholesterol (which introduces no such constraint) was enriched among embedded lipids. For homoacids, embedded PUFAs constrained a corresponding PUFA chain to the nAChR annulus, and here cholesterol was not enriched among embedded lipids.
The fraction of embedded chains that were polyunsaturated increased as more proteins were added to the homoacidic membrane. This could be consistent with each nAChR monomer in an oligomer blocking access to embedded lipid binding sites in the other monomers: flexible PUFA chains have multiple routes to access an embedded site, and rigid lipids have only one or two.
Distribution of nAChR boundary cholesterol or acyl chain fractions in mixed membranes containing heteroacidic phospholipids. Probability density distribution of fraction of embedded (\(f_{\text {emb}}\)) or annular lipids (\(f_{\text {ann}}\)) as defined in Eq. 2 are shown for 1 to 4 proteins. Dashed lines represent expected boundary ratios for a randomly mixed membrane, based on bulk lipid composition (\(x_{\text {CH}} = 0.3\), \(x_{\text {SAT}}=x_{\text {PUFA}}= 0.35\))
Distribution of nAChR boundary cholesterol or acyl chain fractions in mixed membranes containing homoacidic phospholipids. Probability density distribution of fraction of embedded (\(f_{\text {emb}}\)) or annular lipids (\(f_{\text {ann}}\)) as defined in Eq. 2 are shown for 1 to 4 proteins. Dashed lines represent expected boundary ratios for a randomly mixed membrane, based on bulk lipid composition (\(x_{\text {CH}} = 0.3\), \(x_{\text {SAT}}=x_{\text {PUFA}}= 0.35\))
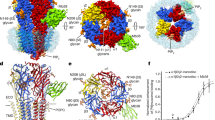
Enrichment or depletion of lipid species surrounding a single nAChR. Heatmaps represent normalized lipid densities (\(\tilde{\rho }_{\text {B}}(r_i,\theta _j)\)) as defined in Eq. 4, on a scale of − 3 (blue, indicating depletion) to 1 (red, indicating enrichment). Densities were averaged over the last \(8\,\upmu s\) of the \(10\,\upmu s\) simulations and across three replicas (Color figure online)
The two-dimensional density distribution of each lipid species from the center of a single nAChR is shown in Fig. 4. For all three lipids, there was a fivefold symmetry of densities surrounding nAChR, with lipid preferences determined by transmembrane helix, rather than by subunit. In heteroacid mixtures, cholesterol is enriched near the subunit interfaces, and even buried more deeply within the \(\gamma /\alpha\) interface, with saturated acyl chains packed just outside cholesterol. The outermost M4 helices, however, were packed with PUFAs; PUFA chains also diffused throughout the TMD bundle. In homoacid membranes, saturated lipids and cholesterol were depleted among embedded lipids, but they maintained the same helix association observed in heteroacid membranes. PUFAs were especially enriched in domain-forming membranes, with high densities around all four transmembrane helices.
nAChR Clustering in the Presence and Absence of Lipid Domains
Here, we repeatedly observed spontaneous formation of receptor dimers in the simulations containing multiple proteins (Fig. 5). To investigate the role of lipid domain formation in driving nAChR oligomerization, we calculated the radial distribution function for the pairwise distances between centers of mass, as shown in Fig. 6. For both heteroacid and homoacid membranes, we observed a peak at \(r\sim 7.5\,\text {nm}\), corresponding to dimerization. The differences in profiles between domain and non-domain-forming membranes were primarily quantitative: most significantly, the peak corresponding to dimers was substantially higher for 2 and 3 proteins in homoacids than in heteroacids. For two proteins, the distribution for homoacids was shifted to the left, relative to the distribution for heteroacids, indicating that the peak for dimerization was at a shorter distance in domain-forming membranes. This difference is consistent with results indicating a role for domain formation in aggregation and clustering of nAChRs (Barrantes 2007; Oshikawa et al. 2003; Pato et al. 2008). The simplest explanation of this difference is the higher effective concentration of proteins when domains are present: all nAChRs are corralled in a single liquid-disordered domain, with approximately half the area of the overall membrane.
Protein clustering in domain-forming (top row) and hybrid (bottom row) membranes. View of the membrane from the extracellular region at the final frame of 10 \(\upmu\)s simulations. nAChRs were colored by subunit (\(\alpha :green,\beta :purple,\delta :gray,\gamma :cyan\)), and lipids by acyl chain (DHA: white, saturated: blue, and cholesterol: red). Clustering of nAChRs on the borders of lipid rafts is visible in the domain-forming membranes (Color figure online)
Closest Subunits Across Dimerizing Proteins
In order to determine whether nAChR dimers were more likely to form with specific subunit interactions, we determined the closest interacting subunit pair for nAChR dimers within each frame. To ensure that dimers, rather than larger oligomers, were being analyzed, we excluded trimers and tetramers from the analysis and only considered two protein systems, with increased sampling. Figure 7 shows the amount of enrichment for each possible subunit pairing, relative to an expected random distribution. Results were very sensitive to the use of domain-forming compositions. In heteroacidic membranes, the \(\alpha _{\delta }\) subunit formed a monomer–monomer interaction with the \(\beta\) subunit, while the \(\alpha _{\gamma }\) most favorably interacted with the \(\delta\) subunit. In domain-forming membranes, the \(\delta{-}\alpha _{\delta }{-}\gamma\) interface was two to five times as likely to pair with a \(\alpha _{\delta }\) subunit compared to a random distribution, but substantially more dispersion was detected than in non-domain-forming membranes. This difference suggests that nAChRs have preferred dimerization orientations, but membrane organization can reorient receptors. Although \(\delta\) subunits are linked by a disulfide bond at the NMJ (Chang and Bock 1977), our simulations suggest that without this link, \(\delta\) subunits do not form the closest pair among dimerizing proteins.
Most probable subunit pairs among dimerizing proteins. Heatmap showing the likelihood that two subunits were closest together among dimerizing proteins (100 Å threshold). Distinct subunits are each expected to form the closest pair 8% of the time, while identical subunits are each expected to dimerize only 4% of the time. Each color on the heatmap represents multiplicative depletion or enrichment relative to the pair’s expectation value, ranging from complete depletion (red) to no enrichment (white) to fivefold enrichment (blue) (Color figure online)
Discussion
The nAChR is one of the most well-studied, fundamental pLGICs for understanding human cognition, memory, and muscle contraction (Gotti et al. 1997). As an integral membrane protein, the function and organization of nAChR is strongly dictated by its surrounding lipid environment. DHA is an \(\omega\)-3 polyunsaturated fatty acid abundant in synaptic membranes and the Torpedo electric organs, which are both native nAChR membranes. With its six double bonds, DHA is considered highly disordered and can induce domain formation in membranes (Wassall and Stillwell 2008). We previously observed (Sharp et al. 2019) favorable interactions between a single nAChR and DHA-rich, cholesterol-poor domains using coarse-grained simulations. Here, we have extended this approach to investigate the role of lipid topology and domain formation on boundary lipids of individual nAChRs. We have also conducted the first simulations containing multiple nAChRs, which has allowed us to observe spontaneous dimerization.
While DHA is implicated in human health and disease, (Lavandera et al. 2017) experimental studies considering its interactions with nAChR have exclusively considered its free-fatty acid form (FFA), (Antollini and Barrantes 2016) rather than as an acyl chain component of a phospholipid. Application of \(\omega\)-3 FFAs causes a significant reduction in open times observed through single-channel recordings (Bouzat and Barrantes 1993). Here, we observe a substantial effect of lipid topology on both embedded and annular lipids: DHA chains in homoacidic phospholipids are far more likely to be found as either annular or embedded boundary lipids. We previously observed only quantitative effects of swapping PE with PC on partitioning, but the headgroup does serve to anchor the lipid at the membrane/protein interface (Sharp et al. 2019). DHA in its FFA form (without a headgroup) may diffuse into an open nAChR pore, blocking the channel.
We find that, consistent with coarse-grained MD simulations using one nAChR (Sharp et al. 2019), multiple nAChRs continue to prefer the liquid-disordered phase containing long-chain \(\omega\)-3 fatty acids. While the number of nAChRs in the system did not affect the partitioning profile in these simulations, it did affect the composition of embedded lipids. Our results are consistent with each nAChR monomer in an oligomer blocking access to embedded lipid binding sites in the other monomers, suggesting an intriguing coupling between specific binding and membrane organization.
Interestingly, upon removing membrane organization, embedded lipids cluster around specific transmembrane helices in a fivefold symmetry around nAChR. This finding suggests that intrinsic lipid preferences are primarily helix dependent, rather than subunit dependent. In homoacid membranes, a lipid preference for DHA was observed across all transmembrane helices, with shells of PUFAs found even at the border of the liquid-ordered phase. Although saturated acyl chains and cholesterol were generally depleted around nAChR, the highest densities for both lipids were found around the M1 and M3 helices, as seen in heteroacid membranes.
In native membranes, nAChR dimers can be stabilized by a disulfide bond between \(\delta\) subunits (Chang and Bock 1977). An early controversy (Anholt et al. 1980; Rüchel et al. 1981; Zingsheim et al. 1982; Schindler et al. 1984) concerned whether the disulfide bond was necessary for dimer formation. There is no mechanism for covalent bonds between monomers in these coarse-grained simulations. All dimers were stabilized by non-covalent interactions, consistent with the results of Rüchel et al. (1981) and Schindler et al. (1984). We observed far more stable dimers in the homoacid mixtures than the heteroacid mixtures, which would be consistent with high sensitivity to experimental conditions. We did not observe a consistent \(\delta -\delta\) preference for interfacing subunits in either homoacid or heteroacid mixtures. Schindler et al. (1984) observed an apparent gain-of-function for single-channels within dimers relative to monomers, regardless of disulfide linking (Schindler et al. 1984), suggesting that lipid modulation of oligomerization could also provide a pathway for modulating single-channel function.
References
Albuquerque EX, Pereira EFR, Alkondon M, Rogers SW (2009) Mammalian nicotinic acetylcholine receptors: from structure to function. Physiol Rev 89(1):73–120. https://doi.org/10.1152/physrev.00015.2008
Althoff T, Hibbs RE, Banerjee S, Gouaux E (2014) X-ray structures of glucl in apo states reveal a gating mechanism of cys-loop receptors. Nature 512(7514):333–337. https://doi.org/10.1038/nature13669
Anholt R, Lindstrom J, Montal M (1980) Functional equivalence of monomeric and dimeric forms of purified acetylcholine receptors from torpedo californica in reconstituted lipid vesicles. Eur J Biochem 109:481–487
Antollini SS, Barrantes FJ (2016) Fatty acid regulation of voltage- and ligand-gated ion channel function. Front Physiol 7:573. https://doi.org/10.3389/fphys.2016.00573
Baaden M, Marrink SJ (2013) Coarse-grain modelling of protein–protein interactions. Curr Opin Struct Biol 23(6):878–886. https://doi.org/10.1016/j.sbi.2013.09.004
Baenziger JE, Corringer PJ (2011) 3D structure and allosteric modulation of the transmembrane domain of pentameric ligand-gated ion channels. Neuropharmacology 60(1):116–125. https://doi.org/10.1016/j.neuropharm.2010.08.007
Baenziger JE, Hénault CM, Therien JPD, Sun J (2015) Nicotinic acetylcholine receptor-lipid interactions: mechanistic insight and biological function. Biochimica Biophysica Acta 1848(9):1806–1817. https://doi.org/10.1016/j.bbamem.2015.03.010
Baenziger JE, Domville JA, Therien JPD (2017) The role of cholesterol in the activation of nicotinic acetylcholine receptors. Curr Topics Memb 80:95–137. https://doi.org/10.1016/bs.ctm.2017.05.002
Barrantes FJ (2007) Cholesterol effects on nicotinic acetylcholine receptor. J Neurochem 103(s1):72–80
Barrantes FJ, Antollini SS, Blanton MP, Prieto M (2000) Topography of nicotinic acetylcholine receptor membrane-embedded domains. J Biol Chem 275(48):37333–37339
Barrantes FJ, Bermudez V, Borroni MV, Antollini SS, Pediconi MF, Baier JC, Bonini I, Gallegos C, Roccamo AM, Valles AS, Ayala V, Kamerbeek C (2010) Boundary lipids in the nicotinic acetylcholine receptor microenvironment. J Mol Neurosci 40:87–90. https://doi.org/10.1007/s12031-009-9262-z
Bermudez V, Antollini SS, Nievas GAF, AveldaÒo MI, Barrantes FJ (2010) Partition profile of the nicotinic acetylcholine receptor in lipid domains upon reconstitution. J Lipid Res 51(9):2629–2641
Bond PJ, Sansom MSP (2006) Insertion and assembly of membrane proteins via simulation. J Am Chem Soc 128(8):2697–2704. https://doi.org/10.1021/ja0569104
Borroni MV, Vallés AS, Barrantes FJ (2016) The lipid habitats of neurotransmitter receptors in brain. Biochimica Biophysica Acta 1858:2662–2670. https://doi.org/10.1016/j.bbamem.2016.07.005
Bouzat CB, Barrantes FJ (1993) Effects of long-chain fatty acids on the channel activity of the nicotinic acetylcholine receptor. Recept Channels 1:251–258
Brannigan G, Hénin J, Law R, Eckenhoff R, Klein ML (2008) Embedded cholesterol in the nicotinic acetylcholine receptor. Proc Natl Acad Sci 105(38):14418–14423
Breckenridge W, Gombos G, Morgan I (1972) The lipid composition of adult rat brain synaptosomal plasma membranes. Biochimica Biophysica Acta (BBA) 266(3):695–707. https://doi.org/10.1016/0005-2736(72)90365-3
Brusés JL, Chauvet N, Rutishauser U (2001) Membrane lipid rafts are necessary for the maintenance of the (alpha)7 nicotinic acetylcholine receptor in somatic spines of ciliary neurons. J Neurosci 21(2):504–512
Butler DH, McNamee MG (1993) FTIR analysis of nicotinic acetylcholine receptor secondary structure in reconstituted membranes. Biochimica Biophysica Acta (BBA) 1150(1):17–24. https://doi.org/10.1016/0005-2736(93)90116-h
Campagna J, Fallon J (2006) Lipid rafts are involved in c95 (4, 8) agrin fragment-induced acetylcholine receptor clustering. Neuroscience 138(1):123–132
Carswell CL, Hénault CM, Murlidaran S, Therien J, Juranka PF, Surujballi JA, Brannigan G, Baenziger JE (2015) Role of the fourth transmembrane helix in the allosteric modulation of pentameric Ligand-Gated ion channels. Structure 23(9):1655–64. https://doi.org/10.1016/j.str.2015.06.020
Chang HW, Bock E (1977) Molecular forms of acetylcholine receptor: effects of calcium ions and a sulfhydryl reagent on the occurrence of oligomers. Biochemistry 16:4513–4520
Cheng MH, Xu Y, Tang P (2009) Anionic lipid and cholesterol interactions with \(\alpha 4 \beta 2\) nachr: insights from md simulations. J Phys Chem B 113(19):6964–6970
Corringer PJ, Poitevin F, Prevost MS, Sauguet L, Delarue M, Changeux JP (2012) Structure and pharmacology of pentameric receptor channels: from bacteria to brain. Structure 20(6):941–956. https://doi.org/10.1016/j.str.2012.05.003
Criado M, Eibl H, Barrantes FJ (1982) Effects of lipids on acetylcholine receptor: essential need of cholesterol for maintenance of agonist-induced state transitions in lipid vesicles. Biochemistry 21(15):3622–3629. https://doi.org/10.1021/bi00258a015
daCosta CJB, Ogrel AA, McCardy EA, Blanton MP, Baenziger JE (2001) Lipid–protein interactions at the nicotinic acetylcholine receptor. J Biol Chem 277(1):201–208. https://doi.org/10.1074/jbc.m108341200
Feller SE (2008) Acyl chain conformations in phospholipid bilayers: a comparative study of docosahexaenoic acid and saturated fatty acids. Chem Phys Lipids 153(1):76–80. https://doi.org/10.1016/j.chemphyslip.2008.02.013
Fong T, McNamee M (1986) Correlation between acetylcholine receptor function and structural properties of membranes. Biochemistry 25(4):830–840
Fong T, McNamee M (1987) Stabilization of acetylcholine receptor secondary structure by cholesterol and negatively charged phospholipids in membranes. Biochemistry. https://doi.org/10.1021/bi00387a020
Gahbauer S, Böckmann RA (2016) Membrane-mediated oligomerization of g protein coupled receptors and its implications for gpcr function. Front Physiol 7:494. https://doi.org/10.3389/fphys.2016.00494
Georgieva R, Chachaty C, Hazarosova R, Tessier C, Nuss P, Momchilova A, Staneva G (2015) Docosahexaenoic acid promotes micron scale liquid-ordered domains: a comparison study of docosahexaenoic versus oleic acid containing phosphatidylcholine in raft-like mixtures. Biochimica Biophysica Acta (BBA) 1848(6):1424–1435. https://doi.org/10.1016/j.bbamem.2015.02.027
Goose JE, Sansom MS (2013) Reduced lateral mobility of lipids and proteins in crowded membranes. PLoS Comput Biol 9(4):1003033. https://doi.org/10.1371/journal.pcbi.1003033
Gotti C, Fornasari D, Clementi F (1997) Human neuronal nicotinic receptors. Prog Neurobiol 53(2):199–237
Hénin J, Salari R, Murlidaran S, Brannigan G (2014) A predicted binding site for cholesterol on the GABAA receptor. Biophys J 106(9):1938–1949. https://doi.org/10.1016/j.bpj.2014.03.024
Hibbs RE, Gouaux E (2011) Principles of activation and permeation in an anion-selective cys-loop receptor. Nature 474(7349):54–60
Humphrey W, Dalke A, Schulten K (1996) VMD—visual molecular dynamics. J Mol Graph 14:33–38
Ingólfsson HI, Melo MN, Van Eerden FJ, Arnarez C, Lopez CA, Wassenaar TA, Periole X, De Vries AH, Tieleman DP, Marrink SJ (2014) Lipid organization of the plasma membrane. J Am Chem Soc 136(41):14554–14559. https://doi.org/10.1021/ja507832e
Iyer SS, Tripathy M, Srivastava A (2018) Fluid phase coexistence in biological membrane: insights from local nonaffine deformation of lipids. Biophys J 115(1):117–128. https://doi.org/10.1016/j.bpj.2018.05.021
Lavandera JV, Saín J, Fariña AC, Bernal CA, González MA (2017) N-3 fatty acids reduced trans fatty acids retention and increased docosahexaenoic acid levels in the brain. Nutr Neurosci 20(7):424–435
Laverty D, Thomas P, Field M, Andersen OJ, Gold MG, Biggin PC, Gielen M, Smart TG (2017) Crystal structures of a GABAA-receptor chimera reveal new endogenous neurosteroid-binding sites. Nat Struct Mol Biol. https://doi.org/10.1038/nsmb.3477
Laverty D, Desai R, Uchański T, Masiulis S, Stec WJ, Malinauskas T, Zivanov J, Pardon E, Steyaert J, Miller KW, Aricescu AR (2019) Cryo-em structure of the human 1 3 2 gaba, javax.xml.bind.jaxbelement@18520d8a, receptor in a lipid bilayer. Nature 565:516–520. https://doi.org/10.1038/s41586-018-0833-4
Levental K, Lorent J, Lin X, Skinkle A, Surma M (2016) Polyunsaturated lipids regulate membrane domain stability by tuning membrane order. Biophys J. https://doi.org/10.1016/j.bpj.2016.03.012
Marchand S, Devillers-Thiéry A, Pons S, Changeux JP, Cartaud J (2002) Rapsyn escorts the nicotinic acetylcholine receptor along the exocytic pathway via association with lipid rafts. J Neurosci 22(20):8891–8901
Marrink SJ, Risselada HJ, Yefimov S, Tieleman DP, de Vries AH (2007) The martini force field: coarse grained model for biomolecular simulations. J Phys Chem B 111(27):7812–7824. https://doi.org/10.1021/jp071097f
Masiulis S, Desai R, Uchañski T, Serna Martin I, Laverty D, Karia D, Malinauskas T, Zivanov J, Pardon E, Kotecha A, Steyaert J, Miller KW, Aricescu AR (2019) Gabaa receptor signalling mechanisms revealed by structural pharmacology. Nature 565:454–459. https://doi.org/10.1038/s41586-018-0832-5
Morales-Perez CL, Noviello CM, Hibbs RE (2016a) X-ray structure of the human \(\alpha 4 \beta 2\) nicotinic receptor. Nature 538(7625):411–415. https://doi.org/10.1038/nature19785
Morales-Perez CL, Noviello CM, Hibbs RE (2016b) X-ray structure of the human [alpha]4[beta]2 nicotinic receptor. Nature 538(7625):411–415
Nemecz A, Prevost MS, Menny A, Corringer PJ (2016) Review: emerging molecular mechanisms of signal transduction in pentameric ligand-gated ion channels. Neuron 90:452–470
Oshikawa J, Toya Y, Fujita T, Egawa M, Kawabe J, Umemura S, Ishikawa Y (2003) Nicotinic acetylcholine receptor alpha 7 regulates cAMP signal within lipid rafts. Am J Physiol Cell Physiol 285(3):C567–74. https://doi.org/10.1152/ajpcell.00422.2002
Parton D, Tek A, Baaden M, Sansom M (2013) Formation of raft-like assemblies within clusters of influenza hemagglutinin observed by md simulations. PLoS Comput Biol 9(4):e1003034
Pato C, Stetzkowski-Marden F, Gaus K, Recouvreur M, Cartaud A, Cartaud J (2008) Role of lipid rafts in agrin-elicited acetylcholine receptor clustering. Chemico-Biol Interactions 175(1–3):64–67. https://doi.org/10.1016/j.cbi.2008.03.020
Perillo VL, Peñalva DA, Vitale AJ, Barrantes FJ, Antollini SS (2016) Transbilayer asymmetry and sphingomyelin composition modulate the preferential membrane partitioning of the nicotinic acetylcholine receptor in Lo domains. Arch Biochem Biophys 591:76–86. https://doi.org/10.1016/j.abb.2015.12.003
Prevost MS, Sauguet L, Nury H, Van Renterghem C, Huon C, Poitevin F, Baaden M, Delarue M, Corringer PJ (2012) A locally closed conformation of a bacterial pentameric proton-gated ion channel. Nat Struct Mol Biol 19(6):642–649
Pronk S, Páll S, Schulz R, Larsson P, Bjelkmar P et al (2013) Gromacs 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. https://doi.org/10.1093/bioinformatics/btt055
Ramarao MK, Cohen JB (1998) Mechanism of nicotinic acetylcholine receptor cluster formation by rapsyn. Proc Natl Acad Sci USA 95(7):4007–4012. https://doi.org/10.1073/pnas.95.7.4007
Rüchel R, Watters D, Maelicke A (1981) Molecular forms and hydrodynamic properties of acetylcholine receptor from electric tissue. Eur J Biochem 119:215–223
Sauguet L, Shahsavar A, Poitevin F, Huon C, Menny A, Nemecz A, Haouz A, Changeux JP, Corringer PJ, Delarue M (2014) Crystal structures of a pentameric ligand-gated ion channel provide a mechanism for activation. Proc Natl Acad Sci 111(3):966–971
Schindler H, Spillecke F, Neumann E (1984) Different channel properties of torpedo acetylcholine receptor monomers and dimers reconstituted in planar membranes. Proc Natl Acad Sci USA 81:6222–6226
Scott KA, Bond PJ, Ivetac A, Chetwynd AP, Khalid S, Sansom MSP (2008) Coarse-grained MD simulations of membrane protein-bilayer self-assembly. Structure 16(4):621–630. https://doi.org/10.1016/j.str.2008.01.014
Shaikh SR, Dumaual AC, Castillo A, Locascio D, Siddiqui RA, Stillwell W, Wassall SR (2004) Oleic and docosahexaenoic acid differentially phase separate from lipid raft molecules: a comparative nmr, dsc, afm, and detergent extraction study. Biophys J 87(3):1752–1766. https://doi.org/10.1529/biophysj.104.044552
Sharp L, Salari R, Brannigan G (2019) Boundary lipids of the nicotinic acetylcholine receptor: spontaneous partitioning via coarse-grained molecular dynamics simulation. Biochimica Biophysica. https://doi.org/10.1016/j.bbamem.2019.01.005
Sodt AJ, Sandar ML, Gawrisch K, Pastor RW, Lyman E (2014) The molecular structure of the liquid-ordered phase of lipid bilayers. J Am Chem Soc 136(2):725–732. https://doi.org/10.1021/ja4105667
Stetzkowski-Marden F, Gaus K, Recouvreur M, Cartaud A, Cartaud J (2006) Agrin elicits membrane lipid condensation at sites of acetylcholine receptor clusters in c2c12 myotubes. J Lipid Res 47(10):2121–2133
Sunshine C, McNamee MG (1992) Lipid modulation of nicotinic acetylcholine receptor function: the role of neutral and negatively charged lipids. Biochim Biophys Acta 1108(2):240–246. https://doi.org/10.1016/0005-2736(92)90031-G
Turk HF, Chapkin RS (2013) Membrane lipid raft organization is uniquely modified by n-3 polyunsaturated fatty acids. Prostaglandins Leukotrienes Essential Fatty Acids. https://doi.org/10.1016/j.plefa.2012.03.008
Unwin N (2005) Refined structure of the nicotinic acetylcholine receptor at 4 Å resolution. J Mol Biol 346(4):967–989. https://doi.org/10.1016/j.jmb.2004.12.031
Unwin N (2017) Segregation of lipids near acetylcholine-receptor channels imaged by cryo-em. IUCrJ 4:393–399. https://doi.org/10.1107/S2052252517005243
Wassall SR, Stillwell W (2008) Docosahexaenoic acid domains: the ultimate non-raft membrane domain. Chem Phys Lipids 153:57–63
Wenz JJ, Barrantes FJ (2005) Nicotinic acetylcholine receptor induces lateral segregation of phosphatidic acid and phosphatidylcholine in reconstituted membranes. Biochemistry 44(1):398–410
Willmann R, Pun S, Stallmach L, Sadasivam G, Santos AF et al (2006) Cholesterol and lipid microdomains stabilize the postsynapse at the neuromuscular junction. EMBO J 25(17):4050–4060
Yadav RS, Tiwari NK (2014) Lipid integration in neurodegeneration: an overview of Alzheimer’s Disease. Mol Neurobiol 50:168–76
Yeagle PL (2016) Chapter 7-structures of lipid assemblies. pp 115–154
Zhu D, Xiong WC, Mei L (2006) Lipid rafts serve as a signaling platform for nicotinic acetylcholine receptor clustering. J Neurosci 26(18):4841–4851. https://doi.org/10.1523/JNEUROSCI.2807-05.2006
Zingsheim HP, Neugebauer DC, Frank J, Hänicke W, Barrantes FJ (1982) Dimeric arrangement and structure of the membrane-bound acetylcholine receptor studied by electron microscopy. EMBO J 1:541–547
Zuber B, Unwin N (2013) Structure and superorganization of acetylcholine receptor-rapsyn complexes. Proc Natl Acad Sci USA 110(26):10622–7. https://doi.org/10.1073/pnas.1301277110
Acknowledgements
GB was supported by research Grants NSF MCB1330728 and NIH P01GM55876. GB and LM were also supported through a Grant from the Research Corporation for Scientific Advancement. This project was supported with computational resources from the National Science Foundation XSEDE program through allocation NSF-MCB110149, a local cluster funded by NSF-DBI1126052, the Rutgers University Office of Advanced Research Computing (OARC) and the Rutgers Discovery Informatics Institute (RDI2), which is supported by Rutgers and the State of New Jersey. We are grateful to Dr. Jérôme Hénin for his helpful suggestions throughout this study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest. This research was supported in part by the National Science Foundation, the National Institutes of Health, and the Research Corporation for Scientific Advancement.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Woods, K., Sharp, L. & Brannigan, G. Untangling Direct and Domain-Mediated Interactions Between Nicotinic Acetylcholine Receptors in DHA-Rich Membranes. J Membrane Biol 252, 385–396 (2019). https://doi.org/10.1007/s00232-019-00079-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00232-019-00079-0