Abstract
The effect of glycosylation following limited enzymatic hydrolysis on conformational and functional properties of black bean protein isolate (BBPI) was investigated in this study. The black bean protein hydrolysate (HBBPI) was prepared using alcalase at pH 9.0 and then glycosylated with glucose (G) at 80 °C for different incubation times from 1 to 6 h. Grafted HBBPI with glucose (HBBPI-G conjugates) had a higher molecular weight than HBBPI by SDS-PAGE analysis. HBBPI-G conjugates had a higher content of β-turn and random coil, but a lower content of α-helix and β-sheet structure than BBPI. HBBPI-G conjugates had lower fluorescence intensity and exhibited bathochromic shift compared with BBPI. Subsequently, the functional properties were also evaluated. Results indicated that the emulsifying activity and solubility were obviously improved (P < 0.05) by HBBPI-G conjugates incubated for 4 h with 6.5% degree of hydrolysis compared to BBPI. Additionally, the glycosylation had positive effects on the reducing power and hydroxyl radical scavenging rate. Therefore, the combination of limited hydrolysis and glycation can be used as an effective method for BBPI modification to obtain enhanced functional properties.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Black beans (Phaseolus vulgaris L.) are a prime source of high-quality proteins and contain a favorable balance of amino acids, and they are also nutritionally rich in isoflavones, carotenoids, saponins, anthocyanins, and polyphenols [1,2,3]. The protein content of black beans is higher than that of soy bean and even that of milk or eggs, which makes black beans an attractive source of proteins for extraction and modification [4, 5]. Owing to the high contents of hydrophobic amino acids, black bean protein isolate appears excellent emulsifying property and antioxidant activity, and become an important emulsifying agent in food industry [3]. Moreover, these functional properties can be improved via various modification methods, such as enzymolysis, ultrasound, glycosylation and so on, according to the previous studies [5,6,7].
Now, the enzymatic modification is a widely accepted and safe method to produce the high physiological activity protein hydrolysate with difference degrees of hydrolysis. For instance, limited hydrolysis with pepsin and pancreatin was sufficient to improve emulsifying and foaming properties of hard-to-cook bean protein concentrate [8]. Chen et al. [9] found that the significant improvement of emulsifying capability of soy protein hydrolysates was related to the increased solubility and decreased molecular weight. Enhanced antioxidant activity of zein was observed after hydrolysis with alcalase or papain [10].
The Maillard reaction, a chemical reaction between reducing sugars and available amino groups of proteins, also has proven to be extremely important in protein modification [11]. There are two main ways to cause the Maillard reaction: dry heating method and wet-heating method. The dry-heating Maillard reaction is not applicable for mass production in the food factory due to the long time requirement (up to a few days). The wet-heating Maillard reaction takes less time, usually 1–10 h. Zhu et al. [6] developed a wet-heating method, which shortened the reaction time to only several hours thereby limiting the Maillard reaction to the initial stage of Schiff base formation and produced protein–polysaccharide conjugates. In addition, Zhang et al. [12] found that the emulsifying properties and antioxidant activities of soybean protein isolate were increased by the wet-heating Maillard reaction and controlled enzyme hydrolysis. The similar results were obtained when using the same method to improve the emulsifying stability of rice proteins [13]. However, it is unclear whether functional properties of black bean protein isolate (BBPI) could be improved by the wet-heating Maillard reaction and limited enzyme hydrolysis method.
In this paper, we prepared BBPI-modified products by limited enzymatic hydrolysis using alcalase, and glycosylation with glucose and characterized their electrophoretic property, secondary structure, tertiary structure, functional properties (surface hydrophobicity, solubility, emulsifying property), as well as antioxidant activity. Our results show functionality changes of BBPI after limited enzymatic hydrolysis and wet-heating Maillard reaction and suggest an efficient way for modification of other proteins.
Materials and methods
Materials
Black beans were obtained from Hei Long Jiang Agriculture Company Limited (Hei Long Jiang, China). Glucose was from Baolingbao Biology Company (Shandong, China). Alcalase was purchased from Aoboxing biotechnology Co. (Beijing, China). Soya oil of first grade of gold arowana was obtained from Yihai Kerry foodstuffs marketing company (Jiangsu, China). Sodium dodecyl sulphate (SDS) and Ferrozine were purchased from Sigma-Aldrich Co. (St. Louis, MO, USA). All other chemicals were of analytical grade.
Preparation of black bean protein isolate
BBPI was prepared according to the method described by Jiang et al. [7] with some modifications. The defatted black bean meal powder was suspended in tenfold water and adjusted to pH 9 with 2 M NaOH. After stirring for 1 h, the solution was centrifuged at 10,000g for 30 min to collect the supernatant. Then the supernatant was subjected to isoelectric precipitation by adjusting pH to 4.5 with 2 M HCl and centrifuged at 10,000g for 30 min to obtain the precipitate. The precipitate was dialyzed against deionized water for 48 h at 4 °C, neutralized to pH 7.0 using 2 M NaOH, and then freeze-dried. The final BBPI product had a protein content of 83.71 ± 1.53%, as determined using the Kjeldahl method (N × 5.8).
Preparation of black bean protein isolate hydrolysate
The 5 mg/mL aqueous solution of BBPI was completely hydrated at room temperature, followed by heating treatment at 80 °C for 10 min. The BBPI solution was then cooled down to 50 °C and adjusted to pH 9.0. Alcalase was added (enzyme/substrate, E/S, 0.3%, w/w) and the pH value was kept at 9.0 with pH–stat method [14] using the 719S Titrino (Metrohm, Switzerland). The pH of the dispersion was kept constant by addition of 0.5 M NaOH. The enzyme reaction was stopped by heating the dispersion at 80 °C for 10 min. The dispersion was then cooled and stored at 4 °C prior to analysis. Degree of hydrolysis (DH) was determined according to the following equation [15]:
where B is the base consumption (mL), N is the concentration of base (0.5 M NaOH), α is the degree of dissociation of the α-amino groups, M p is the mass of protein (g), h is the hydrolysis equivalents, and for black bean protein, htot is the total number of peptide bonds in the protein substrate (mequiv/g of protein). Values taken for 1/α and htot are 2.27 and 7.75.
Preparation of Maillard reaction products
The hydrolysates of black bean protein isolate (HBBPI) were dissolved in phosphate buffer (0.1 M, pH 7.0) and then stirred with a magnetic stirrer at room temperature for 2 h. The completely hydrated hydrolysate solution was incubated with glucose at the HBBPI/glucose ratio of 2/1 (w/w) at 80 °C, pH 7.0 for 1, 2, 3, 4, 5, 6 h, respectively. These samples were lyophilized and referred as HBBPI-G conjugates.
Degree of glycation (DG)
Free amino groups were measured by the o-phthaldialdehyde (OPA) assay with some modifications [16]. 40 mg OPA was dissolved in 1 mL methanol and mixed with 25 mL of 10 mM sodium tetraborate, 2.5 mL of 20% (w/v) SDS, and 100 µL of β-mercaptoethanol. The solution was diluted to a final volume of 50 mL with distilled water to make the OPA reagent. Aliquots of 200 µL of the samples (5 mg/mL) were directly added into 4 mL of OPA reagent in the tubes, and a blank was prepared with 200 µL of deionized water. The solutions were mixed and incubated for 2 min at 35 °C, and the absorbance was measured at 340 nm. The free amino groups content was calculated from the standard curve according to the value of absorbance. The standard curve was constructed using 0.25–2 mM l-lysine. The degree of glycation (DG) was calculated using the equation:
where A0 is the absorbance of free amino groups of the HBBPI, and A t is the absorbance of free amino groups of the sample after Maillard reaction for t h.
Browning value
The browning value was measured according to the method described by Li et al. [13] with a slight modification: the samples were diluted with 0.1% (w/v) SDS to a protein concentration of 0.2% (w/v) and the blank sample was the 0.1% (w/v) SDS solution with no protein sample. The browning value was recorded as the absorbance at 420 nm.
Sodium dodecylsulfate polyacrylamide gel electrophoresis (SDS-PAGE)
SDS-PAGE was performed using a discontinuous-buffer system [17] with a 12% (v/v) separating gel and a 5% (v/v) stacking gel. The samples were dissolved in the buffer containing 50 mM Tris–HCl, 10% (v/v) glycerin, 2% (w/v) SDS, 5% (v/v) 2-mercaptoethanol, and 0.1% (w/v) bromophenol blue, and heated for 5 min at 100 °C. 10 µL of sample was loaded onto the gel. After running, the gel was stained with Coomassie Blue.
Fourier transform infrared (FTIR) spectroscopy
The protein sample was analyzed by Fourier transform infrared (FTIR) spectroscopy by a Bruker Vertex 70 FTIR spectrometer (Bruker Optics, Ettlingen, Germany). The spectroscopic measurements were performed using 1 mg of each sample mixed with 100 mg of dried KBr. The FTIR spectra were recorded with 64 scans at 4 cm− 1 resolution from 4000 to 400 cm− 1 at 25 °C. The software “Peakfit Version 4.12” was used to analyze the secondary structure of protein and “Gaussian peak fitting” was used as the algorithm [18].
Fluorescence spectroscopy
The fluorescence spectra of the protein samples were measured by the F-4500 Fluor photometer (Hitachi, Tokyo, Japan) according to the method described by Liu et al. [19] with some modifications. The sample solution (0.2 mg/mL) was prepared in 10 mM phosphate buffer (pH 7.0). The excitation wavelength was 295 nm, and the emission spectra were recorded from 300 to 440 nm at a constant slit of 5 nm for both excitation and emission. Each scan was repeated ten times.
Surface hydrophobicity (H 0)
Surface hydrophobicity (H0) was determined according to the method described by Haskard et al. [20] with some modifications. Surface hydrophobicity (H0) values of proteins were determined using 1,8-anilinonaphthalene sulfonate (ANS) as the fluorescence probe. The protein solution was diluted by phosphate buffer (10 mM, pH 7.0) to the concentration ranging from 0.05 to 0.5 mg/mL, and then 40 µL of ANS (8.0 mM) was added to 4 mL of each diluted sample solution. Fluorescence intensity (FI) was measured at wavelengths 390 nm (excitation) and 470 nm (emission) using the F-4500 fluorescence spectrophotometer (Hitachi Ltd, Tokyo, Japan) with a slit width of 5 nm. The initial slope of FI versus protein concentration plot was used as the index of H0.
Solubility
Protein solubility was determined according to the method of Cattaneo et al. [21] with modifications. 100 mg Coomassie G-250 was dissolved in 50 mL ethyl alcohol (95%, v/v) and 100 mL phosphoric acid (85%, w/w) to prepare Coomassie G-250 solution (0.01%, w/v). 20 mg of the BBPI, HBBPI or HBBPI-G samples were dispersed in 4 mL of phosphate buffer (100 mM, pH 7.0), respectively, to make the sample concentration of 5 mg/mL. All these samples were stirred for 2 h and centrifuged at 12,000g for 20 min at 20 °C. The supernatant was collected and diluted (1:100 v/v) by deionized water. 5 mL Coomassie G-250 solution was added to 1 mL above sample solution. Then, the protein content in supernatant was determined by measuring the absorbance at 595 nm. Bovine serum albumin (BSA) standard solutions at varying concentrations (0.01–0.1 mg/mL) were prepared, and then a standard curve was built by their absorbance measurements at 595 nm. The standard equation was as follow: Y = 7.9286X + 0.0649, R2 = 0.996, where Y is the absorbance at 595 nm and X is the sample concentrations (mg/mL). The total protein content in sample was determined using the Kjeldahl method (N × 5.8). Protein solubility was calculated as follows:
Emulsifying property
The emulsifying activity index (EAI) and emulsifying stability index (ESI) were determined according to the method described by Pearce and Kinsella [22] with some modifications. The protein samples were dissolved in phosphate buffer (10 mM, pH 7.0) (final protein concentration, 2 mg/mL). The 24 mL of the protein solution and 8 mL of soybean oil were homogenized at 10,000 g for 1 min using the homogenizer (AE300L-H Shanghai Angni Instruments Co., Shanghai, China). After homogenisation for 0 min and 10 min, 50 µL of the emulsion was immediately taken from the bottom of the beaker, and diluted at 1:100 with 0.1% (w/v) SDS solution. The absorbance was immediately measured at 500 nm. The EAI and ESI were calculated as follows:
where T is 2.303, A0 and A10 are the absorbance of the emulsion at 0 and 10 min, respectively. N is dilution factor (100), ϕ is the oil volume fraction (0.25), L is path length of cuvette (1 cm), and C is the weight of protein per unit volume (g/mL).
Evaluation of antioxidant activity
Iron chelating capacity
Iron chelating capacity of the BBPI and its modified samples were determined according to the method described by Dinis et al. [23] with some modifications. 1 mL of the protein sample (5 mg/mL), 1.85 mL of distilled water and 0.05 mL of ferrous chloride (2 mM) were placed in test tube for 30 s. The mixture was then reacted with 0.1 mL of ferrozine (5 mM) at 25 °C for 10 min. The distilled water was treated under the same condition as a control. The absorbance value of the protein samples (A) and the distilled water (A0) were measured at 562 nm. Iron chelating capacity was calculated as follows:
where A0 is the absorbance of the distilled water and A is the absorbance of the protein sample at 562 nm.
Reducing power
Reduction capacity was determined according to the method described by Oyaizu [24] with some modifications. 1 mL of protein sample (10 mg/mL) was mixed with 1.0 mL of sodium phosphate buffer (100 mM, pH 6.6) and 1.0 mL of potassium ferricyanide (1%). After incubation at 50 °C in a temperature-controlled water bath for 20 min, 1.0 mL of trichloroacetic acid (10%) was added to the mixture after cooling to room temperature. The mixtures were centrifuged at 2500g for 10 min at 25 °C, and 2.0 mL of supernatant was added to 2.0 mL of distilled water and 400 µL of ferric chloride (0.1%). After mixing, the samples were left to stand for 10 min. The absorbance of the reaction mixture was measured at 700 nm. An increase in absorbance was used as the enhancement of reducing power.
Hydroxyl radical scavenging rate
Hydroxyl radical scavenging rate was determined according to the method described by Amarowicz et al. [25] with some modifications. For the sample, 2 mg/mL of protein sample (1 mL) was added to 3 mM ferrous sulfate (1 mL) and 3 mM hydrogen peroxide (1 mL), put in dark for 10 min, and then added to 3 mM salicylic acid ethanol solution (1 mL) and put in dark for 30 min. The blank was prepared in the same manner, except that distilled water was used instead of the sample. For the control, the distilled water was treated under the same condition instead of the salicylic acid ethanol solution. The absorbance was finally determined at 510 nm. The hydroxyl radical scavenging rate was calculated according to the following formula:
where Asample is the absorbance of sample, Acontrol is the absorbance of control and Ablank is the absorbance of the blank at 510 nm.
Statistical analysis
All treatments were performed in triplicate. The mean value of triplicate with the standard deviation (error bars) were used in the tables and figures represented. Statistical analysis was performed using SPSS (20.0) software. Significant differences (P < 0.05) between means were determined using Duncan’s multiple range test.
Results and discussion
Glycosylation of the hydrolysates of black bean protein isolate
The DG and browning values are frequently used to evaluation the degree of Maillard reactions [26]. Glycosylation of HBBPI (6.5% DH) was selected, in this condition, the hydrolysate had good emulsifying property and antioxidant activity (data not shown) was carried out with glucose at different incubation time (1, 2, 3, 4, 5 and 6 h) at 80 °C. The variations in DG and browning values of the HBBPI-G conjugates obtained under different treatment conditions are shown in Fig. 1. The pattern showed that the DG and browning values of different HBBPI-G conjugate obviously increased with the increase of incubation time (P < 0.05). Maillard reaction occurs by covalent attachment between amino groups of proteins or peptides and reducing end carbonyl groups of sugars, consequently, as the amount of free amino groups decreased, the degree of glycation was increased [27]. During the Maillard reaction, some intermediate products might undergo polymerization to form the brown polymeric compounds referred to as “melanoidins” when the reaction time extended, which usually symbolized an advanced stage of the Maillard reaction [28]. Hence, there was a dramatic increase in browning values of HBBPI-G conjugates from 1 to 6 h incubation compared with that of HBBPI in Fig. 1, which was in agreement with the result of Pirestani et al. [11].
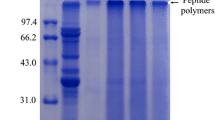
Confirmation of combined modification of BBPI by SDS-PAGE
SDS-PAGE is one of the reliable methods to verify the covalent coupling between HBBPI and glucose during the Maillard reaction. Figure 2 illustrates the SDS-PAGE patterns of BBPI, HBBPI, and HBBPI-G conjugates obtained under different treatment conditions. The bands with much lower molecular weights shown in lane 2 manifested that the molecular weights of BBPI decreased after hydrolysis. The decreased electrophoretic mobility of HBBPI bands after glycosylation resulted in a shift to higher molecular weight area (Fig. 2, lanes 3–8), indicating the formation of the HBBPI-G conjugates. The similar electrophoretic patterns were observed in other study with soy protein–sugar conjugates [12]. Meanwhile, the molecular weight gradually increased with the increase of Maillard reaction time, which also indicated the growth of the DG between the protein and glucose.
Secondary structure analysis by FTIR spectroscopy
FTIR spectroscopy was used to determine the changes in secondary structure of native BBPI, HBBPI and HBBPI-G conjugates. The detailed data of α-helix, β-sheet, β-turns and random coil levels are shown in Table 1. Compared with the untreated BBPI, the proportions of α-helix, β-sheet, β-turn, and random coil were changed after the enzymatic hydrolysis and glycosylation reaction, caused the increase of unordered structure content (β-turn + random coil), mainly at the expense of the ordered structure content (α-helix + β-sheet) compared with BBPI. It is fact that the HBBPI and sugar involves a condensation between the carbonyl group of a reducing carbohydrate with a ε-amino group, which is within the α-helix region or its neighbor of proteins. The decrease of α-helix is considered to be sugar bound to ε-amino group in the α-helix region [29]. In addition, α-helix and β-sheet of proteins are usually buried in the interior site of polypeptide chains in Maillard reaction [12]. As a result, it can be expected that the ordered structure content of HBBPI-G should become lower, when the DG increased. Meanwhile, the structural modifications allowing greater conformational flexibility of protein may improve the functionality [30]. Martínez et al. [31] has shown that the increase of unordered structure contributes to evident improvement of emulsifying properties.
Tertiary structure analysis by fluorescence spectroscopy
Fluorescence spectroscopy is primarily reflected to characterize changes in tryptophan surroundings of the proteins. It is an important index of the change in protein conformation and amino acid loss [32]. The fluorescence spectroscopic technique was used to characterize the tertiary structure changes of the BBPI during the enzymatic hydrolysis and glycation reaction. When excitated at wavelength 290 nm, BBPI exhibited a fluorescence emission maximum (λmax) at 364.5 nm, the λmax of HBBPI was 369.5 nm, and the λmax of HBBPI-G conjugates incubated for 1–6 h were 379, 377.5, 378, 379.5, 379, 379 nm, respectively (Fig. 3). The λmax of HBBPI-G generally exhibited bathochromic shifts compared with BBPI, which suggested that the microenvironment of the tryptophan group became more polar. However, the maximal fluorescence intensity seemed to be in contrast to the λmax (Fig. 3). A reasonable explanation for this “paradox” may be that the attachment of sugar moieties to the protein increases the polar nature of the microenvironment of Trp hydrophobic chromophores, but it renders the Trp hydrophobic chromophores more buried within the molecules. Therefore, it is usually recognized that a red-shift phenomenon is associated with a decrease in maximal fluorescence intensity during the Maillard reaction [33]. The fluorescence intensity of the HBBPI-G conjugates gradually decreased during the first 4-h glycosylation. This phenomenon was due to the shielding effect of the glucose bound, which revealed that the tryptophan hydrophobic chromophores of HBBPI-G were more buried within the molecules [34]. However, the fluorescence intensity of the HBBPI-G conjugates increased after incubation for 5–6 h. This phenomenon might be related to the protein thermal denaturation during the Maillard reaction. According to the study of Sheng et al. and Liu et al. [35, 36], it can be known that heat treatment could induce the exposure of tryptophan group, which will result in the increase of fluorescence intensity. Moreover, it was evident that the tryptophan progressive increase could be observed with extended heating time [36, 37]. Therefore, the stronger fluorescence intensity appeared at 5–6 h, which may be explained that more tryptophan hydrophobic chromophores were exposed to the surface of the protein after longer heat treatment.
Effect of combined modification on surface hydrophobicity
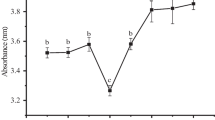
Protein surface hydrophobicity (H0) is an index of the number of hydrophobic groups on the protein surface in contact with the polar aqueous environment. The H0 profiles of BBPI, HBBPI, and different HBBPI-G conjugates are shown in Fig. 4. The pattern indicated that the surface hydrophobicity of the HBBPI-G conjugates was decreased, after the sugars attached with the black bean protein peptides, which was related to the degree of glycation [38]. Achouri et al. [39] found that surface hydrophobicity of soy proteins reduced observably as the degree of glycation increased. The decrease in surface hydrophobicity of the conjugates with the increase in reaction time could be attributed to the formation of aggregates as well as the blocking of lysine and/or arginine residues by glucose during the Maillard reaction [40]. Li et al. [13] also found that the binding of glucose to β-Lg resulted in a significant reduction of surface hydrophobicity. In addition, it should be noticed that the H0 value moderately increased after 5–6 h glycation, which was attributed to the structure change of the black bean protein as the formation of advanced glycation products as mentioned above.
Protein solubility
Solubility is the most utility indicator of protein denaturation and aggregation, therefore, it is an ideal index of protein functionality [34]. The solubility (pH 7.0) of BBPI, HBBPI, and different HBBPI-G conjugates at ambient temperature are shown in Fig. 5. The solubility of HBBPI samples increased compared to the BBPI (P < 0.05). Since the protein hydrolysates were more polar than polypeptides in non-hydrolyzed proteins, they could form stronger hydrogen bonds with water and thereby being more soluble in aqueous solutions [41]. The solubility of HBBPI after glycosylation was higher than that of the native protein, which was consistent with previous studies [13, 42]. The solubility of HBBPI-G conjugates was further increased. It might be due to the attachment of a hydrophilic saccharide to the surface of a hydrophobic protein, the hydrogen bonding capacity of sugar changes the surface hydrophobicity, leading to an increased solubility [43].
Emulsifying property
Emulsifying property contains the ability of an emulsifier to form and stabilize emulsions, and it is essentially determined by their conformational and physicochemical properties. The emulsifying activity index (EAI) and the emulsifying stability index (ESI) of native BBPI, HBBPI and different HBBPI-G conjugates are shown in Fig. 6. The increase in EAI and ESI of HBBPI-G conjugates were significant (P < 0.05), and the highest values were observed at 4 h. That may be because that the polysaccharide portion of the conjugate would exhibit a yield value high enough to resist the buoyant forces favoring upward movement and creaming [44]. But at 5–6 h, HBBPI-G might result in loss of binding spots of the protein for the O/W interface [13]. Therefore, higher extent of DG might decrease the EAI and ESI of HBBPI-G conjugates.
Evaluation of antioxidant activity
Iron chelating capacity
The effects of Maillard reaction on the iron chelating capacity of BBPI are shown in Fig. 7a. The HBBPI and HBBPI-G conjugates demonstrated obvious increase on iron chelating capacity compared with BBPI (P < 0.05). The results indicated that the iron chelating capacity of the black bean protein hydrolysate was increased by conjugating with the glucose. The increased ability of the HBBPI and HBBPI-G conjugates might be caused by the structural changes of BBPI, which opens and exposures the active amino acid residues and patches capable of reacting with oxidants [45]. Fan et al. [46] found that the antioxidative activity of soy protein hydrolysate was further enhanced when it was conjugated with curdlan. Some peptide reductants generated from the Maillard reaction may cause the improvement of the iron chelating capacity of HBBPI.
Reducing power
During the reducing power assay, the presence of reductants in the tested samples could induce the conversion of Fe3+/ferricyanide to the ferrous form (Fe2+) and was initially proposed to measure the total antioxidant capacity. The Fe2+ can thus be monitored by measuring the formation of Prussian blue at 700 nm [47]. The reducing power of BBPI, HBBPI and HBBPI-G conjugates are shown in Fig. 7b. The reducing power of HBBPI had no obvious improvement compared with that of BBPI. The result demonstrated a significant improvement (P < 0.05) of reducing power in the model systems by adding the HBBPI-G conjugates with the incubation time range from 1 to 2 h. However, the HBBPI-G conjugates of 3–6 h incubation showed obviously higher reducing power. This result was also in agreement with the previous study [12], which demonstrated that the reducing power partially depended on the melanoidins, which were usually produced in the advantage stage of the Maillard reaction [48].
Hydroxyl radical scavenging
The changes in the hydroxyl radical scavenging activity of BBPI, HBBPI and HBBPI-G conjugates are shown in Fig. 7c. The hydrolysis had no obvious improvement on the hydroxyl radical scavenging activity of BBPI. However, a significant (P < 0.05) increase in hydroxyl radical scavenging activity of HBBPI was observed with glucose glycosylation. Kim et al. [49] demonstrated that Maillard reaction products had radical scavenging activity. Fan et al. [46] suggested that the improved emulsifying property of soy protein hydrolysate conjugated with curdlan was a significant contributor for the improvement of its radical scavenging activity. They postulated that besides acting as a free radical scavenger, the radical scavenging activity of the soy protein isolate may be elevated by cover in oil surfaces with the soy protein hydrolysate-curdlan conjugate. Hence, the excellent emulsifying property may be helpful in improving the radical scavenging activity of HBBPI-G conjugates.
Conclusion
We demonstrate that the combination of limited hydrolysis and Maillard reaction could significantly improve the solubility, emulsifying properties and antioxidative activities of BBPI. Grafting HBBPI with glucose forms conjugates of higher molecular weight compared with HBBPI in electrophoresis profiles. HBBPI-G conjugates have a higher content of β-turn and random coil, but a lower content of α-helix and β-sheet structure than BBPI. On the other hand, HBBPI-G conjugates have lower fluorescence intensity and exhibited bathochromic shift compared with BBPI. The phenomenon can be explained that the Trp hydrophobic chromophores are assigned to be in the more polar environment, but they are also more buried within the molecules, due to the shielding effect of the glucose bound. In conclusion, the combination of limited hydrolysis and glycosylation is a promising way to improve functional properties of BBPI.
References
Inagaki S, Morimura S, Shigematsu T, KidaI K, Akutagama H (2005) Apoptosis induction by vinegar produced from boiled extract of black soybeans in human monoblastic leukemia U937 cells: difference in sensitivity to cell toxicity compared to normal lymphocytes. Food Sci Technol Res 11(3):311–317
Takahashi R, Ohmori R, Kiyose C, Momiyama Y, Ohsuzu F, Kondo K (2005) Antioxidant activities of black and yellow soybeans against low density lipoprotein oxidation. J Age Food Chem 53(11):4578–4582
Xu B, Chang SKC (2008) Antioxidant capacity of seed coat, dehulled bean, and whole black soybeans in relation to their distributions of total phenolics, phenolic acids, anthocyanins, and isoflavones. J Age Food Chem 56(18):8365–8373
Lee HI, Hung YH, Chou CC (2008) Solid-state fermentation with fungi to enhance the antioxidative activity, total phenolic and anthocyanin contents of black bean. Int J Food Microbiol 121(2):150–156
Evangelho JA, Vanier NL, Pinto VZ, Dias AR, Zavareze Eda R (2017) Black bean (Phaseolus vulgaris L.) protein hydrolysates: physicochemical and functional properties. Food Chem 214:460–467
Zhu D, Damodaran S, Lucey JA (2008) Formation of whey protein isolate (WPI)-dextran conjugates in aqueous solutions. J Age Food Chem 56:7113–7118
Jiang LZ, Wang J, Li Y, Wang ZJ, Liang J (2014) Effects of ultrasound on the structure and physical properties of black bean protein isolates. Food Res Int 62:595–601
David B, Teresita S, Jorge R, Maira S, Luis C (2014) Enzymatic hydrolysis of hard-to-cook bean (Phaseolus vulgaris L.) protein concentrates and its effects on biological and functional properties. Int J Food Sci Technol 49(1):2–8
Chen L, Chen J, Ren J, Zhao M (2011) Modifications of soy protein isolates using combined extrusion pre-treatment and controlled enzymatic hydrolysis for improved emulsifying properties. Food Hydrocoll 25(5):887–897
Kong BH, Xiong YLL (2006) Antioxidant activity of zein hydrolysates in a liposome system and the possible mode of action. J Agric Food Chem 54(16):6059–6068
Pirestani S, Nasirpour A, Keramat J, Desobry S (2017) Preparation of chemically modified canola protein isolate with gum Arabic by means of Maillard reaction under wet-heating conditions. Carbohydr Polym 155:201–207
Zhang YT, Tan C, Zhang XM, Xia SQ, Jia CS (2014) Effects of maltodextrin glycosylation following limited enzymatic hydrolysis on the functional and conformational properties of soybean protein isolate. Eur Food Res Technol 238(6):957–968
Li Y, Zhong F, Ji W, Yokoyama W, Shoemaker CF, Zhu S, Xia W (2013) Functional properties of Maillard reaction products of rice protein hydrolysates with mono-, oligo- and polysaccharides. Food Hydrocoll 30(1):53–60
Adler-nissen J (1986) Enzymic hydrolysis of food proteins. Elsevier Applied Science Publishers, London
Evangelho JA, Vanier NL, Pinto VZ, Berrios JJ, Dias AR (2017) Black bean (Phaseolus vulgaris L.) protein hydrolysates: physicochemical and functional properties. Food Chem 214:460–467
Church FC, Swaisgood HE, Porter DH, Catignani GL (1983) Spectrophotometric assay using o-phthaldialdehyde for determination of proteolysis in milk and isolated milk proteins. J Dairy Sci 66(6):1219–1227
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Wang YT, Wang ZJ, Handa CL, Xu J (2017) Effects of ultrasound pre-treatment on the structure of beta-conglycinin and glycinin and the antioxidant activity of their hydrolysates. Food Chem 218:165–172
Liu Y, Kitts DD (2011) Confirmation that the Maillard reaction is the principle contributor to the antioxidant capacity of coffee brews. Food Res Int 44(8):2418–2424
Haskard CA, Li-Chan ECY (1998) Hydrophobicity of bovine serum albumin and ovalbumin determined using uncharged (PRODAN) and anionic (ANS-) fluorescent probes. J Age Food Chem 46(7):2671–2677
Cattaneo F, Sayago JE, Alberto MR (2014) Anti-inflammatory and antioxidant activities, functional properties and mutagenicity studies of protein and protein hydrolysate obtained from Prosopis alba seed flour. Food Chem 161:391–399
Pearce KN, Kinsella JE (1978) Emulsifying properties of proteins: evaluation of a turbidimetric technique. J Agric Food Chem 26(3):716–723
Dinis TC, Madeira VM, Almeida LM (1994) Action of phenolic derivatives (acetaminophen, salicylate, and 5-aminosalicylate) as inhibitors of membrane lipid peroxidation and as peroxyl radical scavengers. Arch Biochem Biophys 315:161–169
Oyaizu M (1988) Antioxidative activities of browning products of glucosamine fractionated by organic solvent and thin-layer chromatography. J Jpn Soc Food Sci 35(11):771–775
Amarowicz R, Naczk M, Shahidi F (2000) Antioxidant activity of various fractions of non-tannin phenolics of canola hulls. J Agric Food Chem 48:2755–2759
Delphine L, Claude I, Berger C, Éric V, Laurent D, Fabienne G (2008) Kinetic study on the Maillard reaction. Consideration of sugar reactivity. Food Chem 111(4):1032–1042
Dickinson E, Euston SR (1991) Stability of food emulsions containing both protein and polysaccharide. In: Dickinson E (ed) Food polymers, gels and colloids. The Royal Society of Chemistry, London, pp 132–146
Morales FJ, Jimenez-Perez S (2001) Free radical scavenging capacity of Maillard reaction products as related to colour and fluorescence. Food Chem 72:119–125
Li CH, Xue Q, Wang X (2014) Comparative studies on the physicochemical properties of peanut protein isolate–polysaccharide conjugates prepared by ultrasonic treatment or classical heating. Food Res Int 57:1–7
Mu L, Zhao M, Yang B, Zhao H, Cui C (2010) Effect of ultrasonic treatment on the graft reaction between soy protein isolate and gum acacia and on the physicochemical properties of conjugates. J Agric Food Chem 58(7):4494–4499
Martínez KD, Sanchez CC, Ruíz-Henestrosa VP, Rodríguez Patino JM, Pilosof AM (2007) Effect of limited hydrolysis of soy protein on the interactions with polysaccharides at the air–water interface. Food Hydrocoll 21:813–822
Liu J, Ru Q, Ding Y (2012) Glycation a promising method for food protein modification: physicochemical properties and structure, a review. Food Res Int 49(1):170–183
Tang CH, Sun X (2011) Modulation of physicochemical and conformational properties of kidney bean vicilin (phaseolin) by glycation with glucose: implications for structure–function relationships of legume vicilind. J Agric Food Chem 59(18):10114–10123
Xue F, li C, Zhu X, Wang L, Pan S (2013) Comparative studies on the physicochemical properties of soy protein isolate–maltodextrin and soy protein isolate–gum acacia conjugate prepared through Maillard reaction. Food Res Int 51(2):490–495
Sheng L, Su P, Han K, Chen J (2017) Synthesis and structural characterization of lysozyme–pullulan conjugates obtained by the Maillard reaction. Food Hydrocoll 71:1–7
Liu Q, Geng R, Zhao J, Chen Q, Kong B (2015) Structural and gen textural properties of soy protein isolate when subjected to extreme acid pH-shifting and mild heating processes. J Agric Food Chem 63(19):4853–4861
Nikolaidis A, Andreadis M, Moschakis T (2017) Effect of heat, pH, ultrasonication and ethanol on the denaturation of whey protein isolate using a newly developed approach in the analysis of difference-UV spectra. Food Chem 232:425–433
Li W, Zhao H, He Z, Zeng M, Qin F (2016) Modification of soy protein hydrolysates by Maillard reaction: effects of carbohydrate chain length on structural and interfacial properties. Colloids Surf B Biointerfaces 138:70–77
Achouri A, Boye JI, Belanger D, Chiron T (2010) Functional and molecular properties of calcium precipitated soy glycinin and the effect of glycation with κ-carrageenan. Food Res Int 43(5):1494–1504
Gasymov OK, Glasgow BJ (2007) ANS fluorescence: potential to augment the identification of the external binding sites of proteins. Biochim Biophys Acta 1774(3):403–411
Liu Q, Kong BH, Xiong YL, Xia XF (2010) Antioxidant activity and functional properties of porcine plasma protein hydrolysate as influenced by the degree of hydrolysis. Food Chem 118(2):403–410
Li B, Bao Z, Xu W, Chi Y (2014) Influence of glycation extent on the physicochemical and gelling properties of soybean β-conglycinin. Eur Food Res Technol 240(2):399–411
Wang ZJ, Han FF, Sui B, Yang Y, Zhang H (2016) Effect of ultrasound treatment on the wet heating Maillard reaction between mung bean [Vigna radiate (L.)] protein isolates and glucose and on structural and physico-chemical properties of conjugates. J Sci Food Agric 96(5):1532–1540
Parker A, Gunning PA, Ng K, Robins MM (1995) How does xanthan stabilise salad dressing. Food Hydrocoll 9(4):333–342
Kong BH, Xiong YL (2006) Antioxidant activity of zein hydrolysates in a liposome system and the possible mode of action. J Agric Food Chem 54(16):6059–6068
Fan JF, Zhang YY, Tan S, Zhou M (2006) Improving functional properties of soy protein hydrolysate by conjugation with curdlan. J Food Sci 71(5):C285–C291
Gu F, Kim JM, Hayat K, Xia S, Feng B (2009) Characteristics and antioxidant activity of ultrafiltrated Maillard reaction products from a casein–glucose model system. Food Chem 117(1):48–54
Guerard F, Sumaya-Martinez MT (2003) Antioxidant effects of protein hydrolysates in the reaction with glucose. J Am Oil Chem Soc 80(5):467–470
Kim JS, Lee YS (2009) Antioxidant activity of Maillard reaction products derived from aqueous glucose/glycine, diglycine, and triglycine model systems as a function of heating time. Food Chem 116(1):227–232
Acknowledgements
This work was supported by the National Natural Science Foundation of China (no. 31301600), the Natural Science Foundation of Heilongjiang Province (no. LC2017010), Science and Technology Research Project of Education Department of Heilongjiang Province (no. 12541008) and Postdoctoral Project in Heilongjiang Province (no. LBH-Q16012), Academic Backbone Project of Northeast Agricultural University (no. 17XG02).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Compliance with ethics requirements
This article does not contain any studies with human or animal subjects.
Rights and permissions
About this article
Cite this article
Xu, J., Han, D., Chen, Z. et al. Effect of glucose glycosylation following limited enzymatic hydrolysis on functional and conformational properties of black bean protein isolate. Eur Food Res Technol 244, 1111–1120 (2018). https://doi.org/10.1007/s00217-018-3032-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00217-018-3032-5