Abstract
This article describes a paper-based low cost single cell HaloChip assay that can be used to assess drug- and radiation-induced DNA damage at point-of-care. Printing ink on paper effectively blocks fluorescence of paper materials, provides high affinity to charged polyelectrolytes, and prevents penetration of water in paper. After exposure to drug or ionizing radiation, cells are patterned on paper to create discrete and ordered single cell arrays, embedded inside an agarose gel, lysed with alkaline solution to allow damaged DNA fragments to diffuse out of nucleus cores, and form diffusing halos in the gel matrix. After staining DNA with a fluorescent dye, characteristic halos formed around cells, and the level of DNA damage can be quantified by determining sizes of halos and nucleus with an image processing program based on MATLAB. With its low fabrication cost and easy operation, this HaloChip on paper platform will be attractive to rapidly and accurately determine DNA damage for point-of-care evaluation of drug efficacy and radiation condition.

Single cell HaloChip on paper
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
DNA damage is the fundamental basis for many cancer therapies including chemotherapy and radiation therapy [1, 2]. In the context of point-of-care diagnosis, quantifying drug- and radiation-induced DNA damage in extracted cancer cells allows doctor to identify the best available treatment for each patient before prescription which will significantly minimize adversary effect and enhance efficacy of treatment [3]. However, most existing DNA damage assays are not suitable for point-of-care in terms of accuracy, sensitivity, availability, rapidness, and cost [4]. Micronucleus assay can detect tiny micronuclei formation due to chromosome breakage and loss, but it cannot be used on non-dividing cells, and also requires time-consuming scoring of small micronuclei [5]. Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay detects DNA fragmentations by labeling terminal ends of nucleic acids. It is expensive and takes long time to fix and stain cells [6]. Single cell assays have been developed to evaluate DNA damage, but these methods are limited to centralized research laboratories and costly equipment. Flow cytometry detects only DNA double strand breaks by measuring phosphorylation of histone [7]. Comet assay (single cell gel electrophoresis) can detect both single and double strand breaks by assessing geometries of comet tails, but it is limited by low throughput and random distribution of cells and comets [8]. The random cell distribution could lead to cell overlapping and requires users to frequently adjust focus of microscopes to find cells at different height and location. Comet chip has been developed to solve random distribution issue by trapping cells in microfabricated wells, but each comet has to be measured individually due to complicated comet shape [9]. Halo assay is a single cell assay. Instead of using electrical power to pull damaged DNAs out of nucleus core, damaged DNAs will self-diffuse out of nucleus and form halos inside a gel matrix [10]. The halo radius is proportional to the level of DNA damage. But cells in traditional halo assay are randomly dispersed inside a gel matrix that leads to random distribution and overlapping of cells and halos [11]. Meanwhile, these DNA damage assays need trained operators and sophisticated equipment which are not available in resource-limiting environment or at point-of-care.
Paper or membrane materials have been used for biochemical analyses such as dipstick assay, lateral flow assay, and microfluidic analytical devices [12, 13]. Two major types of paper materials that are useful at point-of-care are cellulose fiber materials such as filter paper and chromatography paper that are used as substrates for dipstick and microfluidic device [14–16], and nitrocellulose membrane for lateral flow assay [17]. The porosity, surface chemistry, as well as optical properties of paper are critical for biochemical analyses [18]. Briefly, surface chemistry can affect molecule or particle immobilization, non-specific adsorption, and color expression. Porosity and surface chemistry affect wetting behavior of paper materials. Paper’s optical properties can affect accuracy of colorimetric and fluorescent-based readouts, as many commercially available paper materials contain brightening agents which produce high fluorescence background [19]. Despite the facts that paper materials are affordable, user-friendly, robust, and scalable for manufacturing, current paper-based diagnostic techniques are only limited to polymerase chain reaction or enzyme-linked immune sorbent assay [20, 21]. Although paper has been used as substrate for cell culture, it has not been used in cell-based in vitro toxicity assays [22, 23]. The lack of such experimental demonstration is likely because of several reasons: (1) paper has high fluorescence background, which prevent the use of fluorescent-based DNA damage assay; (2) porous structure of paper materials has low affinity to cells and prevents cell attachment and observation; and (3) there is no reliable and accurate way to detect DNA damage of cells attached on paper.
This paper describes a simple yet powerful way to create single cell array on paper and the use of single cell array to detect DNA damage with HaloChip assay [24]. The method starts with patterning drug or radiation-treated cells on paper, capping cells in an agarose gel, alkaline lysis of cells to liberate damaged DNA, and fluorescence observation of diffusing halos. An image processing software based on MATLAB has been developed to quantify DNA damage in HaloChip. The software can resolve halo and nucleus core automatically based on the fluorescence intensity, extract halo and nucleus radii, and determine the level of DNA damage. Instead of forming cell arrays on silicon or glass substrates, single cell arrays are generated on normal printing paper. Instead of using labile chemicals for surface modification, printing ink is used to modify paper surface to provide excellent water-repelling ability. Cell attracting regions are generated with dip-coating of polyelectrolyte multilayers. Cell attachment is easily achieved through attraction between positively charged islands made by soft lithography and negatively charged cell membranes. Paper-based HaloChip assay is easy to use and does not require power source, expensive equipment (other than microscope), and reagents. This method can be used for quick diagnostic testing of drug efficacy at point of care or point of collection.
Materials and methods
The following chemicals were from Aldrich (St. Louis, MO): polydiallyldimethyl ammonium chloride (PDAC) (100,000–200,000 Da), sodium chloride, ammonium hydroxide, sulfuric acid, sodium hydroxide, hydrogen peroxide, and ethanol. Polydimethylsiloxane (PDMS Sylgard 184) was from Dow-Corning (Midland, MI). PDMS stamps were prepared by casting PDMS prepolymer and curing agent against solid masters with opposite features generated using photolithography. RPMI-1640 medium, penicillin/streptomycin, trypsin/EDTA solution, and fetal bovine serum were obtained from Thermo Scientific (Logan, UT). Low melting point agarose, live/dead viability assay kit, and SYBR green I were from Invitrogen (Carlsbad, CA). Trypan blue and phosphate-buffered saline (PBS) were obtained from VWR (West Chester, PA). Two anticancer drugs had been used. Doxorubicin hydrochloride (579.99 g/mol) was from TOCRIS Bioscience and irinotecan hydrochloride (CPT-11, 623.14 g/mol) was from Sigma-Aldrich. Fibroblast cells, human glioblastoma cells (A172), and breast cancer cells (MCF7) were from American Type Culture Collection (Manassas, VA), and cultured in standard conditions (5 % CO2 in air at 37 °C) in RPMI-1640 medium supplemented with 10 % (v/v) fetal bovine serum and 1 % (v/v) penicillin/streptomycin. After monolayer reached 70–80 % confluence, cells were trypsinized with 0.25 % trypsin-0.53 mM EDTA solution at 37 °C for 3 min, followed by adding fresh medium at room temperature to neutralize trypsin. After centrifugation and re-suspension in fresh medium, cell viability was tested by staining with Trypan blue, and cell number was counted with hemocytometer (Horsham, PA).
Multiple ink layers were printed on paper prior to microcontact printing. Polyelectrolyte solutions (0.5 % wt, 0.15 M NaCl) were made with 18.2 MΩ · cm−1 millipore water without adjusting pH. Unmodified PDMS stamp was immersed in PDAC solution for 15 min at room temperature, rinsed by DI water, and dried in a gentle nitrogen stream. After deposition of a layer of PDAC on the surface of PDMS stamp, the stamp was brought into contact with the ink-covered paper. A slight pressure was applied on the stamp manually for 15 s to ensure conformal contact between the stamp and ink-covered paper, and then the stamp was peeled off from the paper. Subsequently, cells were seeded on the paper at density of 1 × 106 cells/ml. After incubating cells on the paper for 30 min, unattached cells were rinsed away with PBS, followed by adding 1.5 ml 1 % low melting point (LGT) agarose on paper. The substrate was kept at room temperature for 10 min to allow gel solidification. A Mini-X X-ray tube from Amptek (Bedford, MA) with a silver anode operating at 40 kV and 100 μA was used to generate X-ray [25]. The tube was fitted with a brass collimator (2 mm diameter pinhole) to focus X-rays onto the paper. After X-ray exposure, cells on the paper were immersed in 0.3 M NaOH for 30 min at room temperature and stained with diluted ×10,000 SYBR green I solution for 15 min. After removing unbound dye, fluorescent images of cells were taken using an Olympus IX81 microscope. The images were analyzed by home-made image analysis software to quantify DNA damage. Each data point was averaged from at least 50 individual cells.
In order to exclude the possibility that the surface attachment of cells on the paper might change cell cycle, cells patterned on the ink-covered paper were detached, collected by centrifugation at 1200 rpm for 4 min at room temperature, and suspended in 1 ml ice cold PBS buffer. Cell suspension was added drop-wisely to 9 ml of 70 % ethanol and stored at 4 °C to for 2 h. Cells were collected from ethanol by centrifugation at 1200 rpm for 10 min at 4 °C. The collected cells were stained with 500 μl PI (20 μg/ml) containing 0.1 % Triton X-100 for 15 min at 37 °C and assessed with a BD Accuri C6 flow cytometer (BD Biosciences). The data were processed with OriginPro 8.5 and presented as the mean with a standard deviation. The statistical significance of results was determined by means of an analysis of variance using SPSS software (SPSS 19.0, IBM, Armonk, NY). Comparisons between control group and treatment group were based on t test. A result is considered statistically significant difference when P ≤ 0.05. The results were the mean of three independent experiments.
Results and discussions
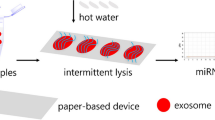
Figure 1a shows the procedure of forming single cells array on the surface of ink-covered paper. Bare paper is covered with ink by printing ink using a commercial printer (Cannon MF4890dw) for few times, where the ink-covered region is negatively charged. Positively charged PDAC layer is formed on a PDMS stamp that has microposts with diameter of 10 μm. The PDMS stamp is then brought into contact with an ink-covered paper for 15 s. After removing PDMS stamp, PDAC layer is transferred onto the ink-covered paper due to electrostatic attraction between ink and the positively charged PDAC. In order to catch cells, the polymer layer that is in direct contact with PDMS stamp is selected to be positively charged. Figure 1b shows the process of halo assay on ink-covered paper. After exposure to DNA damaging drugs or X-ray radiation, cells are patterned onto ink-covered paper to form ordered single cell arrays, embedded inside an agarose gel, and treated with an alkaline solution, where damaged DNAs diffuse into the gel. After fluorescent staining of DNAs, characteristic halos around cell are observed with a fluorescence microscope.
Normal paper is hydrophilic with a water contact angle of 40° as shown in optical image (Fig. 2a), where water spreading is seen in few minutes (Fig. 2b), while paper covered with five layers of ink is hydrophobic with a water contact angle of 90° due to wax and resin added in ink (Fig. 2c). The ink-covered paper can repel cell culture media and buffers as well. Due to the high salt content, the water contact angle is increased to 110°. The hydrophobic ink prevents water spreading on and penetration into paper. Water drop remains on the ink-covered paper even after 30 min (Fig. 2d). In order to exclude the possibility that ink on paper might change cell cycle, after cells are seeded on the ink-cover paper for 0.5 h, cells are detached from ink-covered paper, fixed with 70 % ethanol for 2 h at 4 °C, stained with 20 μg/ml of propidium iodide, and checked with a flow cytometer. Cells from petri dish are used as a control. DNA content within cell is taken as a marker of cellular maturity. Figure 2e, f shows that DNA content in each cell cycle is similar for cells patterned on the ink-covered paper and those on the petri dish.
Paper with different layers of ink has different roughness. Figure 3a–d shows the scanning electron microscope (SEM) images of bare paper, paper covered with one, three, and five layers of ink. As layers of ink increase, paper surface becomes smoother, and fibers in paper cannot be seen clearly. The quality of single cell array is primarily determined by surface roughness (Fig. 3e–h). As the paper surface becomes smoother, more cells are attached, and the array is more order. The attachment probability is derived as the ratio between the number of adsorbed cells and the number of micro-patches. The probability of single cell array increases as the number of ink layers increases from 25 ± 3.1 %, 70 ± 3.5 %, to 87 ± 6.4 % for one, three, and five layers of ink, respectively. Five layers of ink can provide a high quality substrate for single cell array formation, making this method ideal for paper-based halo assay. After incubation for 30 min, unattached cells are rinsed away using PBS. In order to confirm that arrayed cells are still alive, cells are tested with Live/Dead viability/cytotoxicity assay, in which dead cells are labeled as red, and living cells are labeled as green. Fluorescent images (Fig. 3e–h) show that all arrayed cells are alive (green), and round after patterning on ink-covered paper for 1 h.
A MATLAB-based imaging processing software is developed for HaloChip assay. In order to isolate individual cells, images are threshold using Otsu’s method into four levels [26]: the first level is considered background; the second and third levels are combined to label halo; the fourth level is used to label cell nucleus. All non-background levels are combined to form a binary image of cell. The eccentricity of each binary cell object is measured on a scale of 0 to 1, where a score of 0 corresponds to a perfectly circular object and a score of 1 corresponds to a line. Cell objects with eccentricity >0.5 are removed from the image based on the assumption that they contain overlapping cells (due to deposition of un-patterned cells). Figure 4a–e shows the process of automatic analysis of single cell halo assay using the software. The original image of halo assay (Fig. 4a) is transferred to grayscale image (Fig. 4b). The contrast of image is needed to quantify the amount of stained DNA. The contrast can be clearly observed in grayscale image than in color image. The image is threshold into four levels (Fig. 4c), where red corresponds with cell nuclei, yellow and cyan with halos, and blue with background. Cell objects are outlined and shown next to an eccentricity measurement. Cell objects passing the criteria with a measure of 0.500 or less are marked with a black circle at their centers (Fig. 4d). DNA damages are measured based on the halos and nuclei labeled for each cell. Nuclear diffusion factor (NDF) is a measure of the relative surface area of the entire cell (halo and nucleus) to the nucleus alone. Computationally, NDF is found by measuring the area of the halo, S h, and the area of the nucleus, S n, and relating them with the following formula:
A relative NDF (rNDF) is derived by subtracting from the NDF of a control experiment (no DNA damage occur). DNA damage measurement is shown for each cell (Fig. 4e) and an enlarged cell is shown in Fig. 4f. The result can be derived from automate imaging analysis without user intervention, special equipment, or complex software owing to clear boundary, symmetric shapes of halos and nuclei, and non-overlapping nature of cells/halos.
The cell array is exposed to X-ray radiation and embedded in 1 % low-melting-point agarose. After gel solidification (10 min at room temperature), cells are stained with SYBR Green I which inserts into DNA, and the fluorescent intensity is proportional to amount of DNA (because DNA is stained randomly with the dye). Figure 5a–e shows the fluorescent images of cells exposed to different dose of X-ray (0, 0.25, 0.75, 1.25, and 2.5 Gy). The doses and the energy (40 keV) of X-ray used in this experiment are lower than those used in radiation therapy (∼20 Gy and ∼1 MeV), but the responses of cells to low dose and energy can provide DNA damage information for high dose radiations. The control cells show no DNA diffusion from nucleus, where DNA is entirely localized within nucleus and appears as a bright circle (Fig. 5a). As X-ray dose increases, the amount of damaged DNA increases, and nucleus area becomes dimmer and halo becomes bigger (Fig. 5b–e). Figure 5f shows the rNDF values of MCF7 cells exposed to different dose of X-ray, where NDF increases from 0.74 to 2.94 when dose increases from 0.25 to 2.5 Gy.
Cancer is a genetic disease. Many treatments such as chemotherapy and radiation therapy will have to end up with damages to the genetic materials of cells (DNA). A high throughput HaloChip is used to measure DNA damage induced by different treatments. Figure 6a shows an image of multiple samples loaded onto different region of ink-covered surface, where each black region is a patterned array of PDAC islands. Each sample is loaded on an ink-covered square to form single cell array. After embedding cell arrays inside agarose gel, cells are treated with alkaline solution, stained with fluorescence dye, and washed to remove unbound dye. Figure 6b, c shows the fluorescent images of the sample edges, where ink-covered area block cells well, and there is no cross contamination between two adjacent samples. This method has been used to quantify DNA damages simultaneously in two cancer cells (MCF7 and A172) and normal fibroblast cells. Figure 6d shows the NDF of cells after exposure to X-ray radiation, where when X-ray dose increases from 0.25 to 2.5 Gy, rNDF value increases from 0.83 ± 0.12 to 2.61 ± 0.21 for MCF7 cells, 0.75 ± 0.14 to 2.95 ± 0.31 for A172 cells, and 1.17 ± 0.13 to 3.27 ± 0.24 for fibroblast cells. The errors come mostly from cell-to-cell variation Higher X-ray dose causes more DNA damage. Figure 6e shows the NDF values of cells after exposure to doxorubicin hydrochloride at different concentration. As the drug concentration increases from 0 to 50 μM, NDF of MCF7 cells increases from 0 to 3.95, that of A172 cells increases from 0 to 3.91, and that of fibroblast cells increases from 0 to 4.63. The drug concentrations used in clinical settings (50 μM) are close to the ones studied here. Figure 6f shows rNDF values of cells after exposure to CPT-11 at different concentration. When CPT-11 concentration increases from 0 to 50 μM, NDF of MCF7 cells increases from 0 to 4.05, that of A172 cells increases from 0 to 4.19, and that of fibroblast cells increases from 0 to 4.34. These results indicate that higher concentrations of drugs can cause more DNA damage.
Conclusions
Single cell array can be formed on ink-covered paper using microcontact printing. The printing ink effectively blocks fluorescent signal of paper materials, provides high affinity to charged polyelectrolytes, and prevents penetration of water into paper. The HaloChip assay in its high throughput format has been performed successfully on paper after loading multiple types of cells and testing different treatment conditions. DNA damages induced by X-ray radiation and anticancer drugs can be rapidly assessed with an imaging processing software based on MATLAB without user intervention. With its low fabrication cost and easy operation, the HaloChip on paper platform will be more attractive to determine DNA damage for personalized treatment at point-of-care.
References
Fedier A, Fink D. Mutations in DNA mismatch repair genes: implications for DNA damage signaling and drug sensitivity (review). Int J Oncol. 2004;24(4):1039–47.
Frankenberg-Schwager M. Review of repair kinetics for DNA damage induced in eukaryotic cells in vitro by ionizing radiation. Radiother Oncol J Eur Soc Ther Radiol Oncol. 1989;14(4):307–20.
Zhang P, Qiao Y, Wang C, Ma L, Su M. Enhanced radiation therapy with internalized polyelectrolyte modified nanoparticles. Nanoscale. 2014;6(17):10095–9.
Wan J, Johnson M, Schilz J, Djordjevic MV, Rice JR, Shields PG. Evaluation of in vitro assays for assessing the toxicity of cigarette smoke and smokeless tobacco. Cancer Epidemiol Biomarkers Prev. 2009;18(12):3263–304.
Fenech M. The advantages and disadvantages of the cytokinesis-block micronucleus method. Mutat Res. 1997;392(1–2):11–8.
Watanabe M, Hitomi M, van der Wee K, Rothenberg F, Fisher SA, Zucker R, et al. The pros and cons of apoptosis assays for use in the study of cells, tissues, and organs. Microsc Microanal. 2002;8(5):375–91.
Nusse M, Marx K. Flow cytometric analysis of micronuclei in cell cultures and human lymphocytes: advantages and disadvantages. Mutat Res. 1997;392(1–2):109–15.
Slamenova D, Gabelova A, Ruzekova L, Chalupa I, Horvathova E, Farkasova T, et al. Detection of MNNG-induced DNA lesions in mammalian cells; validation of comet assay against DNA unwinding technique, alkaline elution of DNA and chromosomal aberrations. Mutat Res. 1997;383(3):243–52.
Wood DK, Weingeistb DM, Bhatia SN, Engelwardb BP. Single cell trapping and DNA damage analysis using microwell arrays. Proc Natl Acad Sci U S A. 2010;107:10008–13.
Gichner T, Mukherjee A, Wagner ED, Plewa MJ. Evaluation of the nuclear DNA diffusion assay to detect apoptosis and necrosis. Mutat Res. 2005;586(1):38–46.
Sestili P, Martinelli C, Stocchi V. The fast halo assay: an improved method to quantify genomic DNA strand breakage at the single-cell level. Mutat Res. 2006;607(2):205–14.
Yu WW, White IM. Inkjet-printed paper-based SERS dipsticks and swabs for trace chemical detection. Analyst. 2013;138(4):1020–5.
Martinez AW, Phillips ST, Wiley BJ, Gupta M, Whitesides GM. FLASH: a rapid method for prototyping paper-based microfluidic devices. Lab Chip. 2008;8(12):2146–50.
Zhou M, Yang M, Zhou F. Paper based colorimetric biosensing platform utilizing cross-linked siloxane as probe. Biosens Bioelectron. 2014;55:39–43.
Jokerst JC, Adkins JA, Bisha B, Mentele MM, Goodridge LD, Henry CS. Development of a paper-based analytical device for colorimetric detection of select foodborne pathogens. Anal Chem. 2012;84(6):2900–7.
Yang X, Forouzan O, Brown TP, Shevkoplyas SS. Integrated separation of blood plasma from whole blood for microfluidic paper-based analytical devices. Lab Chip. 2012;12(2):274–80.
Miranda BS, Linares EM, Thalhammer S, Kubota LT. Development of a disposable and highly sensitive paper-based immunosensor for early diagnosis of Asian soybean rust. Biosens Bioelectron. 2013;45:123–8.
Pelton R. Bioactive paper provides a low-cost platform for diagnostics. Trends Anal Chem. 2009;28(8):925–42.
Hu J, Wang S, Wang L, Li F, Pingguan-Murphy B, Lu TJ, et al. Advances in paper-based point-of-care diagnostics. Biosens Bioelectron. 2014;54:585–97.
Jefferies R, Ryan UM, Irwin PJ. PCR-RFLP for the detection and differentiation of the canine piroplasm species and its use with filter paper-based technologies. Vet Parasitol. 2007;144(1–2):20–7.
Cheng CM, Martinez AW, Gong J, Mace CR, Phillips ST, Carrilho E, et al. Paper-based ELISA. Angew Chem. 2010;49(28):4771–4.
Juvonen H, Maattanen A, Lauren P, Ihalainen P, Urtti A, Yliperttula M, et al. Biocompatibility of printed paper-based arrays for 2-D cell cultures. Acta Biomater. 2013;9(5):6704–10.
Derda R, Tang SKY, Laromaine A, Mosadegh B, Hong E, Mwangi M, et al. Multizone paper platform for 3D cell cultures. PLoS One. 2011;6(5):1–14.
Qiao Y, Wang C, Su M, Ma L. Single cell DNA damage/repair assay using HaloChip. Anal Chem. 2012;84(2):1112–6.
Hossain M, Luo Y, Sun Z, Wang C, Zhang M, Fu H, et al. X-ray enabled detection and eradication of circulating tumor cells with nanoparticles. Biosens Bioelectron. 2012;38:348–54.
Otsu N. A threshold selection method from gray-level histograms. IEEE Trans Syst Man Cybern B Cybern. 1979;9:62–6.
Acknowledgments
This work has been supported by a New Investigator Award from Bankhead-Copley Cancer Research Program and a seed grant from Kennedy Space Center to Liyuan Ma. This work is partially supported by a Director’s New Innovator Award from National Institute of Health (NIH) to Ming Su (1DP2EB016572).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no potential conflict of interest.
Rights and permissions
About this article
Cite this article
Ma, L., Qiao, Y., Jones, R. et al. Single cell HaloChip assay on paper for point-of-care diagnosis. Anal Bioanal Chem 408, 7753–7759 (2016). https://doi.org/10.1007/s00216-016-9872-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-016-9872-6