Abstract
Key message
QTL analyses of two bi-parental mapping populations with AC Barrie as a parent revealed numerous FHB-resistance QTL unique to each population and uncovered novel variation near Fhb1.
Abstract
Fusarium head blight (FHB) is a destructive disease of wheat worldwide, leading to severe yield and quality losses. The genetic basis of native FHB resistance was examined in two populations: a recombinant inbred line population from the cross Cutler/AC Barrie and a doubled haploid (DH) population from the cross AC Barrie/Reeder. Numerous QTL were detected among the two mapping populations with many being cross-specific. Photoperiod insensitivity at Ppd-D1 and dwarfing at Rht-B1 and Rht-D1 was associated with increased FHB susceptibility. Anthesis date QTL at or near the Vrn-A1 and Vrn-B1 loci co-located with major FHB-resistance QTL in the AC Barrie/Reeder population. The loci were epistatic for both traits, such that DH lines with both late alleles were considerably later to anthesis and had reduced FHB symptoms (i.e., responsible for the epistatic interaction). Interestingly, AC Barrie contributed FHB resistance near the Fhb1 locus in the Cutler population and susceptibility in the Reeder population. Analyses of the Fhb1 candidate genes PFT and TaHRC confirmed that AC Barrie, Cutler, and Reeder do not carry the Sumai-3 Fhb1 gene. Resistance QTL were also detected at the expected locations of Fhb2 and Fhb5. The native FHB-resistance QTL detected near Fhb1, Fhb2, and Fhb5 do not appear to be as effective as Fhb1, Fhb2, and Fhb5 from Sumai-3. The presence of awns segregated at the B1 awn inhibitor locus in both populations, but was only associated with FHB resistance in the Cutler/AC Barrie population suggesting linkage caused the association rather than pleiotropy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Fusarium head blight (FHB), primarily caused by the fungus Fusarium graminearum Schwabe (teleomorph: Gibberella zeae (Schwein.) Petch), is one of the most damaging diseases of wheat worldwide, leading to severe yield and quality losses (Gilbert and Tekauz 2000). The pathogen produces harmful mycotoxins such as deoxynivalenol (DON) during infection, and consumption of DON-contaminated grain is harmful to human and animal health (Ferrigo et al. 2016). In Canada, serious outbreaks of FHB were first reported in eastern Canada in 1980 and in Manitoba in 1993 (Gilbert and Haber 2013). Despite the significant efforts to develop more tolerant cultivars and agronomic techniques to manage the disease, FHB has become more widespread in western Canada and elsewhere over the last several years (Gilbert and Tekauz 2000; Gilbert and Haber 2013). In 2016, farmers confronted potentially the worst FHB damage in most wheat growing areas of the western Canada, particularly in the province of Saskatchewan (Canadian Grain Commission 2019).
The development of FHB resistant wheat cultivars is recognized as the most practical and effective solution for minimizing FHB damage in wheat (Bai and Shaner 2004; Gilbert and Haber 2013). Resistance to FHB in wheat is quantitatively inherited, and symptom expression is often modulated by environmental factors (Miedaner et al. 2001; Buerstmayr et al. 2012). The genetic basis of FHB resistance in wheat has been studied extensively, and more than 100 QTL associated with FHB resistance have been reported on the 21 wheat chromosomes (Buerstmayr et al. 2009; Liu et al. 2009; Löffler et al. 2009; Giancaspro et al. 2016). Although several mechanisms of FHB resistance have been proposed, the commonly studied types are resistance to initial infection (type I) and resistance to spread of infection within the spike (type II) (Schroeder and Christensen 1963; Bai and Shaner 2004; Buerstmayr et al. 2009). The other types include resistance to accumulation of mycotoxins such as deoxynivalenol (DON) (type III), resistance to kernel infection (type IV), and yield reduction (type V) (Mesterházy 1995). Fhb1 (QFhs.ndsu-3BS) on chromosome arm 3BS, derived from Sumai-3 and its derivative Ning 7840, was described as the strongest and best-validated FHB-resistance QTL and is primarily associated with type II resistance (Buerstmayr et al. 2002; Liu et al. 2006, 2008). Despite significant efforts to identify the candidate gene for Fhb1 and define its biological mechanism through genomic and transcriptome-based analyses, functional validation of candidate genes remains challenging due to genome size and the polyploid nature of wheat (Schweiger et al. 2013, 2016; Xiao et al. 2013). Recently, Fhb1 region was sequenced and a pore-forming toxin-like (PFT) gene was reported as the gene responsible for FHB resistance at the Fhb1 locus (Rawat et al. 2016; He et al. 2018; Su et al. 2018). Subsequently, another gene encoding a putative histidine-rich calcium-binding protein (TaHRC) was proposed as the gene for Fhb1 (Schweiger et al. 2016; Su et al. 2018, 2019; Li et al. 2019).
Despite extensive work on the inheritance of FHB resistance and disease-related morphological traits (Bai and Shaner 2004; Miedaner et al. 2006; Buerstmayr et al. 2009), little is understood about the genetic basis of native FHB resistance in Canadian spring wheat (i.e., FHB resistance not introduced from Sumai-3 and other Asian spring wheats). Breeding for FHB resistance in wheat is often hampered by a poor understanding of the genetics of resistance, particularly native FHB resistance. Fully exploiting the FHB resistance in Sumai-3 and related Asian germplasm requires a thorough understanding of the genetic basis of FHB resistance of not only the donor (e.g., Sumai-3) but also the recipient (i.e., local elite) germplasm. This study aims to enhance our understanding of the FHB resistance in the latter group of germplasm. AC Barrie is a hard red spring wheat cultivar in the Canada Western Red Spring marketing class that possesses an intermediate level of FHB resistance and an important parent for many Canadian cultivars. The Canadian cultivar Neepawa was the main contributor of the genetics to AC Barrie (see the pedigrees in Materials and Methods), but Mexican and American germplasm is also present in its pedigree.
The objective of the present study was to examine the genetic basis of FHB resistance in the Cutler/AC Barrie (C/B) recombinant inbred line (RIL) and AC Barrie/Reeder (B/R) doubled haploid (DH) populations.
Materials and methods
Plant populations
Two mapping populations were developed to study FHB resistance from the spring wheat line AC Barrie. The first was a RIL population of 212 lines from the cross Cutler/AC Barrie (C/B). AC Barrie (pedigree: Neepawa/Columbus//BW90) (McCaig et al. 1996) is a hard red spring wheat that possesses an intermediate level of FHB resistance and is photoperiod sensitive. Columbus has the pedigree Neepawa*6/RL4137, and BW90 has the complex pedigree BW15/BW38//RL4359/RL4353 (where BW15 = Manitou/Tobari-66, BW38 = Sonora-64/Tezanos-Pintos-Precoz//Neepawa, RL4359 = Sonora-64/Tezanos-Pintos-Precoz//Neepawa, and RL4353 = WORLD-SEEDS-1809/Neepawa//Era/Park). Cutler (pedigree: Ciano'S'/4/Sonora-64/Yaqui-50E5//Gaboto/3/Inia'S') (Briggs et al. 1992) is a Canadian spring wheat derived from CIMMYT germplasm that is susceptible to FHB and photoperiod insensitive. The second mapping population was a DH population of 188 lines developed from the cross AC Barrie/Reeder (B/R), where Reeder (PI-613586) is a moderately susceptible American spring wheat developed by North Dakota State University (pedigree: IAS20*4/H567.71//Stoa/3/ND674) (Oelke and Kolmer 2004).
Phenotyping and statistical analysis
The C/B RIL population and parents were phenotyped in eight environments (i.e., field nurseries) over 2013 and 2014 (Carman MB 2013 and 2014, Winnipeg MB 2013, Morden MB 2014, Charlottetown PEI 2013 and 2014, and Ottawa ON 2013 and 2014). The check lines were Shaw, Neepawa, AC Morse, AC Vista, CDC Teal, Roblin, AC Cora, 5602 HR, 93FHB37, and ND2710. Entries were grown in an alpha lattice design with 12 incomplete blocks and three replicates per environment. The B/R DH population and parents were phenotyped in eight environments over 2015 and 2016 (Carman MB 2015 and 2016, Morden MB 2015 and 2016, Charlottetown PEI 2015 and 2016, and Ottawa ON 2015 and 2016). The check lines were Shaw, Neepawa, CDC Teal, Roblin, 93FHB37, and ND2710. Field experiments were arranged in an alpha lattice design with 12 incomplete blocks and three replicates per environment. The experimental plot was a single one metre row.
Plant height, anthesis date, FHB incidence, and FHB severity data were collected. Plant height was measured from the soil surface to the top of main tiller spikes (excluding awns if present). Anthesis date was recorded when 50% of the main tillers in the row had begun anthesis. Plant height data were not collected in Charlottetown PEI nurseries in 2013, 2014, and 2015. Anthesis date was not collected in any of the Charlottetown PEI nurseries.
The FHB nurseries in all locations were inoculated with F. graminearum isolates as described in McCartney et al. (2016). The Carman and Winnipeg FHB nurseries were spray inoculated twice (on the recorded anthesis date and 2–3 days later) with backpack sprayers. In PEI, plots were spray inoculated three times per week over the course of flowering in the trial with standard pesticide sprayer. Inoculum consisted of a mixture of F. graminearum isolates prepared to 50,000 macroconidia per ml in these nurseries. Carman nurseries were intermittently mist irrigated overnight. PEI nurseries were intermittently mist irrigated day and night. The Winnipeg nursery was irrigated with sprinkler heads to maintain wet soil to promote a humid micro-environment. The Morden and Ottawa FHB nurseries were inoculated with F. graminearum-infected corn and barley kernels on the soil surface. The infested grain was spread in plots about six weeks after seeding. Plots were irrigated with sprinklers to maintain wet soil to promote a humid micro-environment.
FHB incidence was the percentage of spikes with visual FHB symptoms. FHB severity was the percentage of the spike with visual FHB symptoms, when only considering diseased spikes. FHB incidence and severity data were converted into FHB visual rating index (VRI), VRI = (FHB incidence × FHB severity)/100. Deoxynivalenol (DON) content was determined (Sinha et al. 1995) on the C/B RIL population from grain samples harvested utilizing low air speeds to prevent loss of Fusarium-damaged kernels.
The software multi-environment trial analysis with R for Windows (META-R) (Alvarado et al. 2017) was used for analysis of variance (ANOVA) and to calculate best linear unbiased predictors (BLUPs) for plant height, anthesis date, FHB incidence, FHB severity, and DON content. Genotypes, environment, replicates, and blocks were considered random. Heritability was also calculated using META-R.
Genotyping
Genomic DNA was extracted from lyophilized leaf tissue with the DNeasy 96 Plant Kit (Qiagen, Toronto, Canada) and quantified using PicoGreen stain (Molecular Probes, Inc., Eugene, Oregon, USA). Single-nucleotide polymorphism (SNP) markers were genotyped on the RIL and DH populations and parents using the Illumina Infinium 90 K wheat SNP beadchip (Illumina, San Diego, CA) (Cavanagh et al. 2013; Wang et al. 2014). The raw data were analyzed with genotyping module of GenomeStudio V2011.1 software (Illumina, San Diego, USA) using default clustering parameters.
Construction of genetic maps
A total of 195 RILs and 176 DHs were used to calculate genetic linkage maps of the C/B RIL and B/R DH populations, and mapping QTL for FHB resistance and disease-related morphological traits. Before constructing genetic maps, SNPs were filtered by excluding those with poor quality data. All SNPs that were monomorphic between the two parents and those with > 10% missing data were excluded from linkage analysis. Each marker was tested for deviation from the expected 1:1 ratio using Chi-squared test. Markers showing significant (p < 0.01) segregation distortions were discarded. Markers were placed into preliminary linkage bins using the BIN module in QTL IciMapping version 4.0.6.0 (Li et al. 2007). A single marker with the least missing data was selected from each linkage bin and was used for linkage analysis with MapDisto version 1.7.7 (Lorieux 2012). A minimum logarithm of the odds (LOD) score of 4.0 and maximum recombination fraction of 0.2 were used to identify linkage groups. Loci were ordered using a combination of the “AutoMap”, “Order sequence”, and “Compare all order” functions. The best order of markers was generated using both “Auto Check Inversions” and “Auto Ripple” commands. The “Branch and Bound II” and “Seriation II” ordering methods were used in combination with the sum of adjacent recombination fractions (SARF) and count of crossover events (COUNT) as fitting criteria. For each linkage group, the shortest linkage map was selected from the linkage map solutions generated using these different mapping algorithms and criteria. Recombination fractions were converted into map distances using the Kosambi mapping function (Kosambi 1943). Linkage groups were assigned to chromosomes based on existing high-density SNP maps of wheat (Maccaferri et al. 2014; Wang et al. 2014).
DNA markers and candidate genes were located on IWGSC Chinese Spring RefSeq v1.0 (Appels et al. 2018) by basic local alignment search tool (BLAST). The candidate genes included the dwarfing genes Rht-B1 and Rht-D1 (Peng et al. 1999), the photoperiod sensitivity gene Ppd-D1 (Beales et al. 2007), the vernalization genes Vrn-A1 and Vrn-B1 (Yan et al. 2003), and the Fhb1 candidate genes PFT (Rawat et al. 2016) and TaHRC (Li et al. 2019; Su et al. 2019).
QTL analysis
QTL analysis was conducted with interval mapping (IM) and inclusive composite interval mapping (ICIM) using QTL IciMapping version 4.1.0.0 (Li et al. 2007, 2008). For all traits, QTL analyses were carried out using BLUPs (i.e., adjusted means) for test entries in each individual environment and pooled over all environments. Analysis for additive-effect QTL was conducted with 0.1 cM steps, and the 5% LOD significance threshold was calculated with 10,000 permutations. The LOD significance threshold was 3.11 for both IM and ICIM for the C/B RIL population and 3.09 for the B/R DH population for additive-effect QTL analysis based upon permutation analyses. Additive-effect QTL were declared when the LOD score exceeded the LOD significance threshold in two or more environments, or one or more environments plus the pooled dataset, based upon IM or ICIM. For these QTL, QTL statistics were also reported for additional environments in which the LOD score exceeded 2.5.
Analysis for epistatic QTL was conducted with 2.0 cM steps and a default LOD significance threshold of 3.5. The proportion of phenotypic variance explained by single QTL was determined by the square of the partial correlation coefficient (r2). For anthesis date and plant height, epistatic QTL were reported when the LOD exceeded 3.5 in four or more environments, or three environments plus the pooled dataset, based upon IM or ICIM. For FHB resistance, epistatic QTL were reported when the LOD exceeded 3.5 in seven or more combinations of environment (individual environments or pooled dataset) by traits (VRI, FHB incidence, FHB severity, and DON content).
Characterization of Fhb1 candidate genes in parental lines
A panel of twenty wheat cultivars varying in FHB resistance, including AC Barrie, Cutler, and Reeder, were included in this analysis to characterize two recently reported Fhb1 candidate genes, pore-forming toxin like (PFT) gene and histidine-rich calcium-binding protein (TaHRC) gene (He et al. 2018; Su et al. 2018, 2019; Li et al. 2019). Some of the cultivars in the panel are known to have Fhb1 or high levels of FHB resistance, and others were randomly selected cultivars with unknown FHB resistance (Supplementary Table S13). For the characterization of Fhb1 candidate gene PFT, three pairs of primers covering the entire PFT open reading frame (ORF) and part of the promoter region (P1F/P1R, P2F/P2R, and P3F/P3R) were used (He et al. 2018). Genomic DNA was extracted as previously described in the genotyping section. PCR reactions were carried out in a final volume of 50 μl containing 100 ng of total genomic DNA and 0.2 μM each primer using HotStarTaq Master Mix Kit (Qiagen, Toronto, Canada). PCR reactions were performed using the following conditions: an initial denaturation of 15 min at 95 °C, followed by 35 cycles at 94 °C for 30 s, 54 °C for 30 s, 72 °C for 1.30 min, and a final extension of 10 min at 72 °C. Three independent PCR reactions were performed for each genotype. PCR products from each reaction/genotype were separated on a 1% agarose gel to verify amplicon specificity.
Characterization of the TaHRC gene in the same wheat panel was performed using the gene specific primer pair TaHRC-GSM-F/TaHRC-GSM-R designed to capture the deletion polymorphism between the two TaHRC alleles (TaHRC-R and TaHRC-S) (Su et al. 2018). PCR reactions were performed using the following conditions: an initial denaturation of 15 min at 95 °C, followed by 35 cycles at 94 °C for 30 s, 54 °C for 30 s, 72 °C for 2 min, and a final extension of 10 min at 72 °C. Four independent PCR reactions were performed for each genotype, and aliquots of pooled PCR products were separated by agarose gel (1%) electrophoresis to verify amplicon specificity. PCR amplicons were purified with QIAquick PCR Purification Kit according to the manufacturer’s instructions (Qiagen, Toronto, Canada) and were sequenced with Sanger sequencing (National Research Council, Canada). DNA sequence trace files from the ABI 3730xl Genetic Analyzer were processed using PHRED (Ewing et al. 1998) and assembled with CAP3 (Huang and Madan 1999) as implemented in the internal data pipeline called SOOMOS v0.6 (T. Banks, personal communication). Sequence alignments were performed with ClustalW (Thompson et al. 1994).
Results
Phenotypic analyses
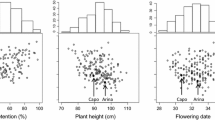
The frequency distributions of pooled datasets for anthesis date, plant height, FHB incidence, FHB severity, VRI, and DON content in the C/B RIL and B/R DH populations are illustrated in Fig. 1a, b, respectively. FHB resistance traits and related morphological traits were approximately normally distributed in the C/B RIL population. Cutler flowered 3 days earlier and was 14 cm shorter than AC Barrie. The average plant height among the 195 RILs was 86 cm, and average anthesis date was Julian date 198. Heritability estimates were 0.92 for anthesis date, 0.86 for plant height, 0.86 for incidence, 0.81 for severity, 0.84 for VRI, and 0.82 for DON in C/B RIL population (Table 1). There was some transgressive segregation for plant height and FHB resistance in the B/R cross, but most DHs were within the means of the parents. Plant height was essentially bi-modal in the B/R population. Reeder flowered 2 days earlier and was 14 cm shorter than AC Barrie. The average plant height among the 176 DHs was 80 cm, and average anthesis date was Julian date 203 (Fig. 1a, b, Supplementary Table S1 and S7). Heritability estimates were 0.94 for anthesis date, 0.97 for plant height, 0.87 for incidence, 0.84 for severity, and 0.89 for VRI in B/R DH population (Table 2).
a Frequency distribution of best linear unbiased predictors (BLUPs) of pooled data of anthesis date, plant height, FHB incidence, FHB severity, FHB visual rating index, and DON content in the Cutler/AC Barrie RIL population from eight environments over 2013 and 2014. Means of the parents are indicated. b Frequency distribution of best linear unbiased predictors (BLUPs) of pooled data of anthesis date, plant height, FHB incidence, FHB severity, and FHB visual rating index in the AC Barrie/Reeder DH population from eight environments over 2015 and 2016. Means of the parents are indicated
The resistant checks 93FHB37 and ND2710 had the lowest FHB incidence, severity, VRI, and DON content in the FHB field nurseries in both RIL and DH populations (Supplementary Tables S1 and S7). AC Morse, AC Vista, CDC Teal, and Roblin of C/B RIL experiments, and Shaw, CDC Teal, and Roblin of B/R DH experiments were the most susceptible checks to FHB based upon FHB incidence, severity, VRI, and DON content. Correlation analysis revealed a significant positive correlation between plant height and anthesis date, and a negative correlation between plant height and FHB symptoms (incidence, severity, VRI, and DON) in both the C/B and B/R populations (Tables 3, 4). Anthesis date and FHB symptoms (incidence, severity, and VRI) were negatively correlated in the B/R DH population (Table 4). In the C/B RIL population, anthesis date was negatively correlated with incidence and VRI, but not with severity or DON content (Table 3). Analysis of variance showed significant (p < 0.001) differences among genotypes for all traits (Tables 1, 2, Supplementary Tables S2 and S8).
Linkage maps
Two high-density whole genome linkage maps were developed using 90 K wheat Infinium SNPs tested on the C/B RIL and B/R DH populations. A total of 10,178 SNPs were mapped in the C/B population. The C/B linkage map consisted of 26 linkage groups and spanned 2438 cM (Supplementary Table S5). The number of linkage bins varied from 18 on chromosome 5D to 229 on chromosome 5B, with an average of 70 linkage bins per linkage group. The B/R linkage map consisted of 10,878 SNPs. The total map length across the 28 linkage groups was 2845 cM (Supplementary Table S11). The B1 awn gene was mapped onto chromosome arm 5AL of both populations (Supplementary Table S5 and S11). In some instances, a chromosome consisted of two linkage groups (e.g., linkage groups 2A.1 and 2A.2, 5A.1 and 5A.2, 5D.1 and 5D.2, 6D.1 and 6D.2, and 7B.1 and 7B.2) in the C/B RIL population.
Identification of QTL for morphological traits
In order to differentiate the QTL detected in each population, the abbreviation ‘mcb’ was included in the names of QTL detected in C/B population, whereas ‘mbr’ identified QTL in B/R population.
Anthesis date
Four additive-effect QTL for anthesis date were identified in the C/B RIL population (Table 5). The QTL were located on chromosomes 2D, 4A, and 7D and were named QAnth.mcb-2D, QAnth.mcb-4A.1, QAnth.mcb-4A.2, and QAnth.mcb-7D. AC Barrie alleles delayed anthesis at QAnth.mcb-2D and Q1Anth.mcb-4A.1, and QAnth.mcb-7D, and Cutler allele delayed anthesis at QAnth.mcb-4A.2. These results are consistent with Cutler being earlier flowering than AC Barrie. QAnth.mcb-2D maps to the expected location of photoperiod sensitivity gene Ppd-D1. Three digenic epistatic interactions were identified between loci on chromosomes 1A and 3B, 1B and 3A, and 4A and 7A (Supplementary Table S6). The additive-effect QTL QAnth.mcb-4A.1 was coincident with an epistatic QTL for anthesis date involving chromosome 7A.
Three additive-effect QTL for anthesis date were identified in the B/R DH population (Table 6). The QTL were located on chromosomes 5A, 5B, and 6B and were named QAnth.mbr-5A, QAnth.mbr-5B, and QAnth.mbr-6B. The AC Barrie allele delayed anthesis for QAnth.mbr-5B, while the Reeder allele delayed anthesis for QAnth.mbr-5A and QAnth.mbr-6B. QAnth.mbr-5A and QAnth.mbr-5B map to the expected locations of Vrn-A1 and Vrn-B1, respectively. Analysis for epistatic QTL identified five additional QTL for anthesis date (Supplementary Table S12). The additive-effect QTL QAnth.mbr-5A and QAnth.mbr-5B were consistently epistatic to each other, having large LOD scores for both QIME (i.e., interval mapping) and QICE (i.e., inclusive composite interval mapping) epistasis modules. Box plots show this interaction (Fig. 2a). Other epistatic interactions were identified among loci on chromosomes 1A and 7A, 1B and 1D, 2A and 7A, and 3B and 7A.
Plant height
Three additive-effect QTL for plant height were identified in the C/B RIL population (Table 5). These QTL were located on chromosomes 2D, 4D, and 5A and were named QHt.mcb-2D, QHt.mcb-4D and QHt.mcb-5A. The AC Barrie allele increased plant height for each of these QTL, which is consistent with AC Barrie being 14 cm taller than Cutler in field tests. Of the three QTL, QHt.mcb-4D was consistently detected over environments and the explained phenotypic variation ranged from 7.7% to 42.0% and is consistent with the location of Rht-D1. QHt.mcb-2D overlapped with QAnth.mcb-2D (i.e., Ppd-D1). Three epistatic QTL interactions were detected between loci on chromosomes 1A and 1D, 1A and 4B, and 3A and 4A (Supplementary Table S6). These interactions were consistently identified by IM, but only the epistatic interaction between 3A and 4A was identified by the epistasis module of ICIM.
Four additive-effect QTL for plant height were identified in the B/R DH population (Table 6). These QTL were located on chromosome 2B, 4A, 4B, and 7A and named QHt.mbr-2B, QHt.mbr-4A, QHt.mbr-4B, and QHt.mbr-7A. The AC Barrie allele at loci QHt.mbr-2B and QHt.mbr-4B increased plant height, while the Reeder allele at QHt.mbr-4A and QHt.mbr-7A increased plant height. The location of QHt.mbr-4B is consistent with the location of Rht-B1. QHt.mbr-4B had a much larger effect than the other QTL, which explains why AC Barrie was 14 cm taller than Reeder in field tests. Seven epistatic QTL interactions were detected among loci on chromosomes 1A and 2A, 2B and 5B, 2B and 7A, 2D and 6B, 3A and 7A, 4B and 5A, and 4B and 6D (Supplementary Table S12). Six of these interactions were consistently identified by IM, but only the epistatic interaction between 2B and 7A was identified by the ICIM epistasis module.
Identification of QTL for FHB resistance
C/B RIL population
Twelve additive-effect QTL for FHB resistance (FHB incidence, FHB severity, FHB visual rating index, and DON content) were identified using IM and ICIM analysis with the additive effect module of QTL IciMapping (Table 5). AC Barrie alleles contributed FHB resistance at nine of these QTL, which is consistent with AC Barrie being more FHB resistant than Cutler. Many of the FHB-resistance QTL were consistently detected by both IM and ICIM. However, FHB-resistance QTL QFhb.mcb-2A and QFhb.mcb-4A were less consistently detected across environments. AC Barrie contributed resistance alleles at QFhb.mcb-3B (chromosome 3B at 7.6 cM) and QFhb.mcb-6B (chromosome 6B at 33.9 cM), which are the expected locations of Fhb1 and Fhb2, respectively. Similarly, QFhb.mcb-5A.1 mapped to the expected location of Fhb5.
FHB-resistance QTL on chromosomes 2D, 4A, 4D, and 5A were coincident for two or more traits. QFhb.mcb-2D.1 mapped near the distal end of chromosome arm 2DS and coincided with QTL for anthesis date and plant height. Likewise, QFhb.mcb-4A was associated with the anthesis date QTL QAnth.mcb-4A.1. At the plant height QTL QHt.mcb-4D (i.e., Rht-D1), the reduced height allele was consistently associated with increased FHB damage. Finally, QFhb.mcb-5A.2 co-located with the B1 awn inhibitor gene.
Additional QTL for FHB resistance were identified by digenic epistasis QTL analysis. Five digenic epistatic interactions were identified between loci on chromosomes 1A and 2A, 1D and 3B, 3A and 7D, 4A and 4B, and 4A and 7A. Epistatic interaction between 4A and 4B was also reported as additive-effect QTL for FHB resistance on chromosome 4A (QFhb.mcb-4A) (Supplementary Table S6).
B/R DH population
IM and ICIM analyses identified nine additive-effect QTL controlling FHB resistance in the B/R DH population (Table 6). AC Barrie contributed resistance alleles at six of these QTL, which is consistent with AC Barrie being generally more resistant to FHB than Reeder. However, Reeder alleles contributed two major FHB-resistance QTL on chromosome 3B (QFhb.mbr-3B at 5.1 cM) at the expected location of Fhb1, and on chromosome 5A (QFhb.mbr-5A) at ~ 137 cM (Table 6). Despite the strong effect of QFhb.mbr-5A, this QTL was not located at the expected location of Fhb5 and was not associated with the presence or absence of awns. The B1 awn inhibitor mapped to 208.6 cM in the B/R population. The third QTL from Reeder was detected only by ICIM on chromosome 7A (QFhb.mbr-7A), and its effect was lower and less consistent than the effects found for the QTL regions on chromosomes 3B and 5A. The most prominent resistance QTL from AC Barrie was found on chromosome 5B (QFhb.mbr-5B) explaining up to 36% of the phenotypic variation. This 5B QTL was consistently detected in all the experiments. The resistance QTL QFhb.mbr-4B coincided with the plant height QTL QHt.mbr-4B mapped at 54 cM (i.e., Rht-B1). Additional QTL for FHB resistance were identified by digenic epistasis QTL analysis. Five digenic epistatic interactions were identified between loci on chromosomes 1A and 5B, 1D and 7B, 2A.2 and 7A, 2B and 5A, and 5A and 5B. Epistatic interaction between 5A and 5B was also detected as additive-effect QTL for FHB resistance (QFhb.mbr-5A and QFhb.mbr-5B) (Supplementary Table S12), at the same location as the epistatic interaction between the anthesis date QTL QAnth.mbr-5A and QAnth.mbr-5B. DH lines with the late anthesis allele at QAnth.mbr-5A and QAnth.mbr-5B interaction were considerably later to anthesis than the three other genotypic classes (Fig. 2a) and had reduced VRI (Fig. 2b), FHB incidence, and FHB severity (data not shown).
Comparison of mapping populations
Three FHB-resistance QTL on chromosomes 2D, 3B, and 5B were in common between the C/B RIL and B/R DH populations. QTL on chromosomes 2D (QFhb.mcb-2D.2 and QFhb.mbr-2D) and 5B (QFhb.mcb-5B and QFhb.mbr-5B) had resistant alleles derived from AC Barrie in both populations. However, contradictory results were obtained for the QTL on chromosome 3B at the expected location of Fhb1. AC Barrie contributed FHB resistance at the Fhb1 locus in the C/B RIL population and susceptibility in the B/R DH population. This result is suggestive of an allelic series in which the Cutler allele was the most susceptible, the AC Barrie allele was intermediate, and the Reeder allele was most resistant.
Cutler contributed FHB resistance at QFhb.mcb-5A.2, which mapped to the location of the B1 awn inhibitor gene in the C/B RIL population, while there was no FHB-resistance QTL detected at the B1 locus in the B/R DH population despite segregation of the presence/absence of awns at the B1 locus in the B/R DH population.
Characterization of Fhb1 candidate genes in parental lines
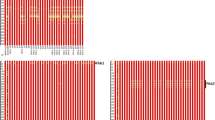
PFT and TaHRC genes were previously reported as candidate genes that confer FHB resistance at the Fhb1 locus (Rawat et al. 2016; Su et al. 2018, 2019; Li et al. 2019). Characterization of PFT and TaHRC was carried out in a panel of 20 wheat genotypes varying in FHB resistance including the parents AC Barrie, Cutler, and Reeder. Among the tested wheat genotypes, the complete PFT gene was amplified in five genotypes (DH181, FL62R1, Nyubai, Sumai-3, Wangshuibai), but not in the remaining 15 genotypes (including the three parents AC Barrie, Cutler, and Reeder) (Fig. 3a). The PCR product of TaHRC-GSM-F/TaHRC-GSM-R from Sumai-3 is shorter than that from Chinese Spring due to the large deletion in Sumai-3 (Su et al. 2018, 2019). The same amplification pattern was observed between genotypes carrying Sumai-3 alleles and non-Sumai-3 alleles in the germplasm tested (Fig. 3b). Sequence alignments of TaHRC among 20 wheat genotypes clearly showed DNA polymorphism between resistant and susceptible genotypes and agreed with the PCR amplification pattern in Fig. 3b (Supplementary Figure S1). The Fhb1-resistant haplotypes (Hap_Ning) (Su et al. 2019) showed identical sequences and carried a large deletion (25 bp) including the start codon (ATG), a 22 bp sequence downstream of the original ATG in the predicted ORF region of TaHRC (TaHRC-R), whereas the other three haplotypes (Hap_CS, Hap_Funo, and Hap_Clark) (Su et al. 2019) carried complete ORF (TaHRC-S) despite the other sequence variations such as SNPs and indels among the haplotypes (Supplementary Figure S1, Supplementary Table S13, 14). Amino acid substitution from arginine (R) to cysteine (C) at residue 169 in the deduced amino acid sequence placed Cutler and Reeder into the Hap_Funo haplotype, and AC Barrie into the Hap_Clark haplotype as shown in the Supplementary Table S13.
a Characterization of the pore-forming toxin like (PFT) gene in 24 selected wheat cultivars including three parents AC Barrie, Cutler, and Reeder. Three pairs of primers were used for identification of the PFT; a, b, and c indicate that PCR products amplified with primer pairs P1F/P1R, P2F/P2R, and P3F/P3R, UTR—untranslated region (He et al. 2018). b Characterization of the histidine-rich calcium-binding protein (TaHRC) gene in 24 selected wheat cultivars including three parents AC Barrie, Cutler, and Reeder using the gene specific primer pair TaHRC-GSM-F/TaHRC-GSM-R designed to capture the deletion polymorphism between the two TaHRC alleles (Su et al. 2018)
Discussion
The genetic basis of native FHB resistance is not well understood in Canada and other parts of the world. This lack of information may limit progress in breeding FHB-resistant cultivars. Knowledge of the FHB-resistance QTL present in local germplasm allows strategic introgression of QTL from exotic wheat lines. Without this knowledge, breeders may introgress FHB resistance alleles already present in the local germplasm or even the recurrent parent, which would provide limited or no opportunity for enhancing FHB resistance in the targeted germplasm. To explore this issue, the genetic basis of FHB resistance in Canadian spring wheat cultivar AC Barrie was characterized in two mapping populations derived from crosses to the Canadian cultivar Cutler and the American cultivar Reeder.
Genetic characterization of FHB resistance is challenging due to the complexity of the disease phenotyping and the underlying genetic basis of resistance. Resistance to FHB in wheat is controlled by polygenes that usually have moderate to small effects and are often influenced by environmental factors (Miedaner et al. 2001; Buerstmayr et al. 2012). QTL analysis in the present study identified 12 QTL controlling FHB resistance in the C/B RIL population on chromosomes 1B, 2A, 2B, 2D, 3B, 4A, 4D, 5A, 5B, and 6B, and nine QTL controlling FHB resistance in the B/R DH population on chromosomes 1B, 2B, 2D, 3B, 4B, 5A, 5B, 7A, and 7D (Tables 5, 6). AC Barrie contributed resistant alleles at most of these loci in both populations, which is consistent with AC Barrie being more resistant to FHB in the field nurseries. Only three of these QTL have common map locations despite AC Barrie being a parent of each population: QFhb.mcb-2D.2 and QFhb.mbr-2D, QFhb.mcb-3B and QFhb.mbr-3B, and QFhb.mcb-5B and QFhb.mbr-5B. The QTL QFhb.mcb-2B and QFhb.mbr-2B may also have an underlying gene in common since the location of QFhb.mbr-2B was somewhat inconsistent, ranging from 39 to 70 cM on the B/R chromosome 2B linkage map. The QTL peaks at position 67 to 70 cM in the B/R population may coincide with QFhb.mcb-2B in the C/B population.
FHB-resistance QTL co-located with major genes affecting plant height in the C/B and B/R populations. There was a strong association between the dwarfing alleles at Rht-B1 and Rht-D1 and increased FHB susceptibility in the B/R (QFhb.mbr-4B) and C/B (QFhb.mcb-4D) populations, respectively. These associations have been reported in past studies for Rht-1 loci and Rht8 (Draeger et al. 2007; Handa et al. 2008; Holzapfel et al. 2008; Srinivasachary et al. 2008, 2009). QFhb.mcb-4D was the largest effect QTL in the C/B population, while QFhb.mbr-4B was an intermediate QTL in the B/R population. This relative of ranking of Rht-D1b and Rht-D1b associated FHB susceptibility is consistent with previous studies. Oddly, the additive effects suggest that QHt.mcb-4D had less of an impact on plant height in the C/B population than QHt.mbr-4B in the B/R population, despite the strong impact of QFhb.mcb-4D on FHB resistance. Cumulatively, these loci and the Ppd-D1 locus (outlined below) explained the negative correlation between plant height and the FHB traits (INC, SEV, VRI, DON) in both populations.
QTL affecting anthesis date also co-located with QTL for FHB resistance resulting in the negative correlation between anthesis date and the FHB traits (INC, SEV, VRI, DON) in both populations. Cutler carries the photoperiod insensitivity allele Ppd-D1a, which was associated with increased FHB susceptibility in the C/B RIL population (QFhb.mcb-2D.1) and reduced plant height allele (QHt.mcb-2D). The Ppd-D1 locus had a similar effect in the Kenyon/86ISMN 2137 RIL population (McCartney et al. 2016).
In the B/R DH population, two major anthesis date QTL were identified (QAnth.mbr-5A and QAnth.mbr-5B), which co-located with two major QTL for FHB resistance (QFhb.mbr-5A and QFhb.mbr-5B). In both cases, earlier anthesis was associated with increased FHB susceptibility. QAnth.mbr-5A mapped to ~ 136 cM on chromosome 5A, but LOD peaks were occasionally as far away at 146.7 cM based on ICIM. LOD peaks for QAnth.mbr-5A ranged from 139.0 to 143.4 cM based on IM. The Vrn-A1 locus (587.416 Mbp in Chinese Spring RefSeq v1.0) is located in the interval from 136.8 to 152.7 cM (approximately 570.939 to 608.604 Mbp in Chinese Spring RefSeq v1.0) based upon BLAST locations of the Vrn-A1 gene and the 90 K Infinium SNPs in IWGSC Chinese Spring reference genome sequence (Appels et al. 2018). AC Barrie and Reeder have the same haplotype based upon 90 K Infinium SNPs in this interval (data not shown), which explains the 16 cM interval (136.8 to 152.7 cM) in the chromosome 5A map with no polymorphic markers and suggests that AC Barrie and Reeder carry the same spring habit allele at the Vrn-A1 locus. The haplotype data suggests that variation at the Vrn-A1 locus is not responsible for the variation in anthesis date and FHB resistance at this region of chromosome 5A; however, the map location of QAnth.mbr-5A does not exclude the Vrn-A1 locus as a candidate. Cutler and AC Barrie are reported to carry Vrn-A1a, vrn-B1, and vrn-D1 (i.e., spring habit at Vrn-A1 and winter alleles at Vrn-B1 and Vrn-D1 loci) (Iqbal et al. 2006; Chen et al. 2016; Perez-Lara et al. 2016), while Reeder carries Vrn-A1, Vrn-B1, and vrn-D1 (i.e., spring habit at Vrn-A1 and Vrn-B1 loci, and winter habit at Vrn-D1) (Sherman et al. 2004; Blake et al. 2009, 2011). QAnth.mbr-5B maps to ~ 115 cM on chromosome 5B. The 90 K Infinium SNPs at 114.5 to 115.0 cM on chromosome 5B flank the Vrn-B1 locus based on the BLAST locations of the Vrn-B1 gene and the 90 K Infinium SNPs. AC Barrie and Reeder are polymorphic for many SNPs in this region of chromosome 5B. These results, combined with the previous reports of the spring habit allele at Vrn-B1 in Reeder, suggest that variation at the Vrn-B1 locus is responsible for the variation in anthesis date and FHB resistance at this region of chromosome 5B. The four genotypic classes defined by QAnth.mbr-5A or QAnth.mbr-5B consisted of spring habit plants, although the DH lines with both late alleles were 4.9 days later to anthesis than the population mean and the VRI of these DH lines was 38% lower than the population mean (Fig. 2). There was no statistically significant segregation distortion at either QAnth.mbr-5A or QAnth.mbr-5B, but there were fewer DH lines with both late alleles than the other genotypic classes. Epistasis was reported between these loci for both anthesis date and FHB resistance (Table S12, Fig. 2). The 5B QTL region had a slightly larger effect on both anthesis date and FHB susceptibility than the 5A QTL region. Wheat researchers may wish to explore the merit of the wheat lines carrying both late alleles given the reduction in FHB symptoms observed in this study. This reduction in FHB damage may be due to less favorable environmental conditions for F. graminearum infection later in the growing season.
In the C/B RIL population, the FHB-resistance QTL QFhb.mcb-5B mapped to the location of the Vrn-B1 locus, or slightly proximal, with AC Barrie carrying the resistant allele. However, in this population, no QTL for anthesis date was identified at the Vrn-B1 locus despite many markers mapping to that region of the chromosome 5B linkage map. In the C/B population, Vrn-A1 is located in the gap that separates linkage groups 5A.1 and 5A.2, which is consistent with Cutler and AC Barrie carrying the same Vrn-A1 allele. QFhb.mcb-5B was a relatively weak FHB-resistance QTL in the C/B RIL population in comparison with QFhb.mbr-5B in the B/R DH population. In the Kenyon/86ISMN 2137 RIL population, an anthesis date QTL were detected at the Vrn-B1 locus and an FHB-resistance QTL was detected about 10–30 cM distal of the anthesis date QTL (McCartney et al. 2016). Kenyon is closely related to AC Barrie and both share the same 90 K Infinium SNP haplotype at Vrn-B1. Additional research is needed to determine whether the FHB resistance in this genomic region is the result of the Vrn-B1 locus and/or linked gene(s).
The Fhb1 region of chromosome 3B showed very interesting QTL analysis results. FHB-resistance QTL were located at the expected location of Fhb1 (Cuthbert et al. 2006; Liu et al. 2008; Rawat et al. 2016; Bai et al. 2018; He et al. 2018) in both populations. These QTL were not consistently detected in all FHB nurseries as would be expected for Fhb1 from Sumai-3. The detection of FHB resistance near the expected location of Fhb1 was surprising given that AC Barrie and Reeder do not have clear ancestry to Asian spring wheats such as Sumai-3. AC Barrie had the resistant allele in the C/B RIL population, but had the susceptible allele in the B/R DH population (i.e., the Reeder allele was more resistant than the AC Barrie allele). This contrasted with AC Barrie carrying the resistant alleles for the two other QTL segregating in both populations (i.e., QFhb.mcb-2D.2 = QFhb.mbr-2D, and QFhb.mcb-5B = QFhb.mbr-5B). Additional experiments were undertaken to explore these findings at the Fhb1 locus. Three primer pairs for the Fhb1 candidate gene PFT failed to amplify a PCR product in AC Barrie, Cutler, and Reeder, while strongly amplifying products in positive controls (Sumai-3, Wangshuibai, Nyubai, DH181, and FL62R1). This suggested that PFT had no role in the FHB resistance segregating in these populations. Subsequently, the Fhb1 candidate gene TaHRC gene was sequenced in the parents of the mapping populations and control genotypes. This revealed that AC Barrie had the Hap_Clark haplotype, while Cutler and Reeder had the Hap_Funo haplotype using the haplotype nomenclature in Su et al. (2019). Reeder and Cutler had the most and least resistant alleles at the Fhb1 locus in this study, respectively, but had the same TaHRC haplotype. Also, the Hap_Clark and Hap_Funo TaHRC haplotypes are not typical of Fhb1 carriers. Therefore, variation in the PFT and TaHRC genes did not explain the variation observed at the Fhb1 locus in the C/B RIL and B/R DH populations, as expected based on pedigree. These results suggest that at least one additional gene is involved in FHB resistance near the Fhb1 locus.
AC Barrie contributed FHB resistance on chromosome 6B in the C/B RIL population, while Cutler contributed FHB resistance on chromosome 5A. QFhb.mcb-6B coincides with the expected location of the major FHB-resistance gene Fhb2 (Cuthbert et al. 2007). Fhb2 also originates from Sumai-3 and related Asian germplasm, so the identification of an FHB-resistance QTL in this genetic region from AC Barrie was unexpected. However, FHB resistance has been detected in the Fhb2 region in non-Asian germplasm, such as the Swiss cultivar Arina (Draeger et al. 2007; Semagn et al. 2007), the French cultivar Apache (Holzapfel et al. 2008), the American cultivar Patton (Bonin and Kolb 2009), and T. carthlicum Blackbird (Somers et al. 2006). The relative effectiveness of each QTL allele remains to be determined. Similarly, QFhb.mcb-5A.1 maps to the expected location of Fhb5 in Wangshuibai, CM-82036, Frontana, and other wheats (Steiner et al. 2004, 2019; Xue et al. 2011; Buerstmayr et al. 2018). However, QFhb.mcb-5A.1 was significant in two of eight FHB nurseries in this study, suggesting this QTL is not as strong as Fhb5 from Sumai-3. The detection of native FHB-resistance QTL at the locations of Fhb2 and Fhb5 in AC Barrie and Cutler underlines the importance of understanding the genetic basis of FHB resistance in local breeding germplasm.
A QTL for FHB resistance (QFhb.mcb-5A.2) was detected at the B1 awn inhibitor locus in the C/B RIL population. RIL lines with the awned allele had improved FHB resistance in this population. This contrasts with observations made in wheat breeding germplasm in which awned genotypes were more susceptible to FHB (Mesterházy 1995). Note that the B/R DH population also segregated for awns but no QTL for FHB resistance was detected at the B1 locus. The results of Mesterházy (1995) may have been complicated by population structure that was not accounted for (i.e., potential false positive) or due to a linked gene. If the B1 locus has no effect on FHB resistance, then QFhb.mcb-5A.2 in the C/B population must have been due to a linked gene. This possibility would explain why an FHB-resistance QTL was not detected at the B1 locus in B/R DH population despite segregation for awns at the locus.
FHB-resistance QTL on chromosome 2B have also been detected in spring wheat but have received little attention due to their minor effect and occasional occurrence in spring wheat (Zhang et al. 2018). Zhang et al. (2018) identified a locus from chromosome 2BL originating from the Canadian spring wheat cultivar Stettler providing type II resistance. The 2B QTL from Stettler was flanked by SNP markers that overlap with SNP markers spanning the 2B QTL from AC Barrie (C/B RIL population) identified in the present study. AC Barrie also contributed a major QTL for FHB resistance in the B/R DH population, but the location of the QTL peak varied in different field trials and different measures of FHB symptoms. It is not clear if the FHB-resistance QTL on chromosome 2B are the same in the two AC Barrie mapping populations. A 2BL QTL was also detected from the Brazilian spring wheat cultivar Frontana (Steiner et al. 2004), which is in the distant pedigree of AC Barrie.
A number of other minor FHB-resistance QTL were identified in these populations. AC Barrie contributed FHB-resistance QTL on chromosome 1B in the C/B (QFhb.mcb-1B) and B/R (QFhb.mbr-1B) populations. These QTL mapped to different locations, and each was detected in multiple field tests. The FHB-resistance QTL QFhb.mcb-2D.2 and QFhb.mbr-2D derived from AC Barrie in both RIL and DH populations mapped to the similar location as FHB-resistance QTL derived from Wuhan-1 (Somers et al. 2003; Hu et al. 2019). AC Barrie contributed the resistance alleles at QFhb.mcb-4A and QFhb.mbr-7D, Cutler at QFhb.mcb-2A, and Reeder at QFhb.mbr-7A.
In summary, only part of the FHB resistance can be attributed to putatively identical resistance QTL among individual populations. While the two populations shared three or four QTL in common, several other genomic regions controlling FHB resistance were identified that were unique to each population. FHB resistance is very complex and regulated by many different genomic regions with major- or minor-effect QTL, even in wheat cultivars with intermediate levels of resistance. These results indicate that a complete understanding of the FHB resistance in a wheat genotype cannot be determined with a single bi-parental mapping population. A full understanding of the FHB resistance of a highly resistant wheat may require a nested association mapping (NAM) population (Yu et al. 2008), in which the FHB resistant wheat is the common parent crossed to genetically diverse wheats.
Availability of data and material
Data supporting the current study can be obtained by contacting the corresponding author (curt.mccartney@canada.ca).
Abbreviations
- ANOVA:
-
Analysis of variance
- BLAST:
-
Basic local alignment search tool
- BLUPs:
-
Best linear unbiased predictors
- C/B:
-
Cutler/AC Barrie
- DH:
-
Doubled haploid
- ICIM:
-
Inclusive composite interval mapping
- IM:
-
Interval mapping
- LOD:
-
Logarithm of odds
- META-R:
-
Multi-environment trial analysis with R
- PVE:
-
Phenotypic variation explained
- QTL:
-
Quantitative trait loci
- B/R:
-
AC Barrie/Reeder
- RIL:
-
Recombinant inbred line
- SNP:
-
Single-nucleotide polymorphism
References
Alvarado G, López M, Vargas M, Pacheco Á, Rodríguez F, Burgueño J, Crossa J (2017) META-R (Multi Environment Trail Analysis with R for Windows) Version 6.01. CIMMYT Research Data & Software Repository Network. https://hdl.handle.net/11529/10201
Appels R, Eversole K, Feuillet C, Keller B, Rogers J, Stein N, Pozniak CJ, Stein N, Choulet F, Distelfeld A et al (2018) Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science. https://doi.org/10.1126/science.aar7191
Bai G, Shaner G (2004) Management and resistance in wheat and barley to Fusarium head blight. Annu Rev Phytopathol 42:135–161. https://doi.org/10.1146/annurev.phyto.42.040803.140340
Bai G, Su Z, Cai J (2018) Wheat resistance to Fusarium head blight. Can J Plant Pathol 40:336–346. https://doi.org/10.1080/07060661.2018.1476411
Beales J, Turner A, Griffiths S, Snape JW, Laurie DA (2007) A Pseudo-Response Regulator is misexpressed in the photoperiod insensitive Ppd-D1a mutant of wheat (Triticum aestivum L.). Theor Appl Genet 115:721–733. https://doi.org/10.1007/s00122-007-0603-4
Blake NK, Lanning SP, Berg JE, Bruckner PL, Sherman JD, Talbert LE (2011) Registration of spring- and winter-habit wheat lines derived from elite cultivars of the alternate growth habit. J Plant Registr 5:418–421. https://doi.org/10.3198/jpr2011.01.0003crg
Blake NK, Lanning SP, Martin JM, Doyle M, Sherman JD, Naruoka Y, Talbert LE (2009) Effect of variation for major growth habit genes on maturity and yield in five spring wheat populations. Crop Sci 49:1211–1220. https://doi.org/10.2135/cropsci2008.08.0505
Bonin CM, Kolb FL (2009) Resistance to fusarium head blight and kernel damage in a winter wheat recombinant inbred line population. Crop Sci 49:1304–1312. https://doi.org/10.2135/cropsci2008.08.0459
Briggs KG, Kibite S, Kutschera K (1992) Cutler red spring wheat. Can J Plant Sci 72:229–233
Buerstmayr H, Ban T, Anderson JA (2009) QTL mapping and marker-assisted selection for Fusarium head blight resistance in wheat: a review. Plant Breed 128:1–26. https://doi.org/10.1111/j.1439-0523.2008.01550.x
Buerstmayr H, Lemmens M, Hartl L, Doldi L, Steiner B, Stierschneider M, Ruckenbauer P (2002) Molecular mapping of QTLs for Fusarium head blight resistance in spring wheat. I. Resistance to fungal spread (type II resistance). Theor Appl Genet 104:84–91. https://doi.org/10.1007/s001220200009
Buerstmayr M, Huber K, Heckmann J, Steiner B, Nelson JC, Buerstmayr H (2012) Mapping of QTL for Fusarium head blight resistance and morphological and developmental traits in three backcross populations derived from Triticum dicoccum x Triticum durum. Theor Appl Genet 125:1751–1765. https://doi.org/10.1007/s00122-012-1951-2
Buerstmayr M, Steiner B, Wagner C, Schwarz P, Brugger K, Barabaschi D, Volante A, Valè G, Cattivelli L, Buerstmayr H (2018) High-resolution mapping of the pericentromeric region on wheat chromosome arm 5AS harbouring the Fusarium head blight resistance QTL Qfhs.ifa-5A. Plant Biotechnol J 16:1046–1056. https://doi.org/10.1111/pbi.12850
Canadian Grain Commission (2019) Frequency and severity of Fusarium damaged kernels (FDK) in Harvest Sample Program red spring wheat samples. https://www.grainscanada.gc.ca/en/grain-research/export-quality/cereals/wheat/western/annual-fusarium-damage/canada-western-red-spring/. Accessed 13 Dec 2019
Cavanagh CR, Chao S, Wang S, Huang BE, Stephen S, Kiani S, Forrest K, Saintenac C, Brown-Guedira GL, Akhunova A, See D, Bai G, Pumphrey M, Tomar L, Wong D, Kong S, Reynolds M, da Silva ML, Bockelman H, Talbert L, Anderson JA, Dreisigacker S, Baenziger S, Carter A, Korzun V, Morrell PL, Dubcovsky J, Morell MK, Sorrells ME, Hayden MJ, Akhunov E (2013) Genome-wide comparative diversity uncovers multiple targets of selection for improvement in hexaploid wheat landraces and cultivars. P Natl Acad Sci USA 110:8057–8062. https://doi.org/10.1073/pnas.1217133110
Chen H, Moakhar NP, Iqbal M, Pozniak C, Hucl P, Spaner D (2016) Genetic variation for flowering time and height reducing genes and important traits in western Canadian spring wheat. Euphytica 208:377–390. https://doi.org/10.1007/s10681-015-1615-9
Cuthbert PA, Somers DJ, Brule-Babel A (2007) Mapping of Fhb2 on chromosome 6BS: a gene controlling Fusarium head blight field resistance in bread wheat (Triticum aestivum L.). Theor Appl Genet 114:429–437. https://doi.org/10.1007/s00122-006-0439-3
Cuthbert PA, Somers DJ, Thomas J, Cloutier S, Brulé-Babel A (2006) Fine mapping Fhb1, a major gene controlling fusarium head blight resistance in bread wheat (Triticum aestivum L.). Theor Appl Genet 112:1465–1472. https://doi.org/10.1007/s00122-006-0249-7
Draeger R, Gosman N, Steed A, Chandler E, Thomsett M, Srinivasachary SJ, Buerstmayr H, Lemmens M, Schmolke M, Mesterhazy A, Nicholson P (2007) Identification of QTLs for resistance to Fusarium head blight, DON accumulation and associated traits in the winter wheat variety Arina. Theor Appl Genet 115:617–625. https://doi.org/10.1007/s00122-007-0592-3
Ewing B, Hillier L, Wendl MC, Green P (1998) Base-calling of automated sequencer traces using phred I Accuracy assessment. Genome Res 8:175–185
Ferrigo D, Raiola A, Causin R (2016) Fusarium toxins in cereals: Occurrence, legislation, factors promoting the appearance and their management. Molecules. https://doi.org/10.3390/molecules21050627
Giancaspro A, Giove SL, Zito D, Blanco A, Gadaleta A (2016) Mapping QTLs for Fusarium head blight resistance in an interspecific wheat population. Front Plant Sci 7:1381. https://doi.org/10.3389/fpls.2016.01381
Gilbert J, Haber S (2013) Overview of some recent research developments in Fusarium head blight of wheat. Can J Plant Pathol 35:149–174. https://doi.org/10.1080/07060661.2013.772921
Gilbert J, Tekauz A (2000) Review: Recent developments in research on Fusarium head blight of wheat in Canada. Can J Plant Pathol 22:1–8. https://doi.org/10.1080/07060660009501155
Handa H, Namiki N, Xu D, Ban T (2008) Dissecting of the FHB resistance QTL on the short arm of wheat chromosome 2D using a comparative genomic approach: from QTL to candidate gene. Mol Breed 22:71–84. https://doi.org/10.1007/s11032-008-9157-7
He Y, Zhang X, Zhang Y, Ahmad D, Wu L, Jiang P, Ma H (2018) Molecular characterization and expression of PFT, an FHB resistance gene at the Fhb1 QTL in wheat. Phytopathology 108:730–736. https://doi.org/10.1094/PHYTO-11-17-0383-R
Holzapfel J, Voss HH, Miedaner T, Korzun V, Häberle J, Schweizer G, Mohler V, Zimmermann G, Hartl L (2008) Inheritance of resistance to Fusarium head blight in three European winter wheat populations. Theor Appl Genet 117:1119–1128. https://doi.org/10.1007/s00122-008-0850-z
Hu X, Rocheleau H, McCartney C, Biselli C, Bagnaresi P, Balcerzak M, Fedak G, Yan Z, Valè G, Khanizadeh S, Ouellet T (2019) Identification and mapping of expressed genes associated with the 2DL QTL for fusarium head blight resistance in the wheat line Wuhan 1. BMC Genet 20:47. https://doi.org/10.1186/s12863-019-0748-6
Huang X, Madan A (1999) CAP3: a DNA sequence assembly program. Genome Res 9:868–877
Iqbal M, Navabi A, Salmon DF, Yang RC, Spaner D (2006) A genetic examination of early flowering and maturity in Canadian spring wheat. Can J Plant Sci 86:995–1004. https://doi.org/10.4141/P06-002
Kosambi DD (1943) The estimation of map distances from recombination values. Ann Eugenic 12:172–175. https://doi.org/10.1111/j.1469-1809.1943.tb02321.x
Li G, Zhou J, Jia H, Gao Z, Fan M, Luo Y, Zhao P, Xue S, Li N, Yuan Y, Ma S, Kong Z, Jia L, An X, Jiang G, Liu W, Cao W, Zhang R, Fan J, Xu X, Liu Y, Kong Q, Zheng S, Wang Y, Qin B, Cao S, Ding Y, Shi J, Yan H, Wang X, Ran C, Ma Z (2019) Mutation of a histidine-rich calcium-binding-protein gene in wheat confers resistance to Fusarium head blight. Nat Genet. https://doi.org/10.1038/s41588-019-0426-7
Li H, Ribaut JM, Li Z, Wang J (2008) Inclusive composite interval mapping (ICIM) for digenic epistasis of quantitative traits in biparental populations. Theor Appl Genet 116:243–260. https://doi.org/10.1007/s00122-007-0663-5
Li H, Ye G, Wang J (2007) A modified algorithm for the improvement of composite interval mapping. Genetics 175:361–374. https://doi.org/10.1534/genetics.106.066811
Liu S, Hall MD, Griffey CA, McKendry AL (2009) Meta-Analysis of QTL associated with Fusarium head blight resistance in wheat. Crop Sci 49:1955–1968. https://doi.org/10.2135/cropsci2009.03.0115
Liu S, Pumphrey M, Gill B, Trick H, Zhang J, Dolezel J, Chalhoub B, Anderson J (2008) Toward positional cloning of Fhb1, a major QTL for Fusarium head blight resistance in wheat. Cereal Res Commun 36:195–201. https://doi.org/10.1556/CRC.36.2008.Suppl.B.15
Liu S, Zhang X, Pumphrey MO, Stack RW, Gill BS, Anderson JA (2006) Complex microcolinearity among wheat, rice, and barley revealed by fine mapping of the genomic region harboring a major QTL for resistance to Fusarium head blight in wheat. Funct Integr Genomic 6:83–89. https://doi.org/10.1007/s10142-005-0007-y
Löffler M, Schön CC, Miedaner T (2009) Revealing the genetic architecture of FHB resistance in hexaploid wheat (Triticum aestivum L.) by QTL meta-analysis. Mol Breed 23:473–488. https://doi.org/10.1007/s11032-008-9250-y
Lorieux M (2012) MapDisto: fast and efficient computation of genetic linkage maps. Mol Breeding 30:1231–1235. https://doi.org/10.1007/s11032-012-9706-y
Maccaferri M, Cane MA, Sanguineti MC, Salvi S, Colalongo MC, Massi A, Clarke F, Knox R, Pozniak CJ, Clarke JM, Fahima T, Dubcovsky J, Xu S, Ammar K, Karsai I, Vida G, Tuberosa R (2014) A consensus framework map of durum wheat (Triticum durum Desf.) suitable for linkage disequilibrium analysis and genome-wide association mapping. BMC Genomics 15:873. https://doi.org/10.1186/1471-2164-15-873
McCaig T, DePauw R, Clarke J, McLeod J, Fernandez M, Knox R (1996) AC Barrie hard red spring wheat. Can J Plant Sci 76:337–339
McCartney CA, Brûlé-Babel AL, Fedak G, Martin RA, McCallum BD, Gilbert J, Hiebert CW, Pozniak CJ (2016) Fusarium head blight resistance QTL in the spring wheat cross Kenyon/86ISMN 2137. Front Microbiol 7:1542. https://doi.org/10.3389/fmicb.2016.01542
Mesterházy A (1995) Types and components of resistance to Fusarium head blight of wheat. Plant Breed 114:377–386. https://doi.org/10.1111/j.1439-0523.1995.tb00816.x
Miedaner T, Reinbrecht C, Lauber U, Schollenberger M, Geiger HH (2001) Effects of genotype and genotype-environment interaction on deoxynivalenol accumulation and resistance to Fusarium head blight in rye, triticale, and wheat. Plant Breeding 120:97–105. https://doi.org/10.1046/j.1439-0523.2001.00580.x
Miedaner T, Wilde F, Steiner B, Buerstmayr H, Korzun V, Ebmeyer E (2006) Stacking quantitative trait loci (QTL) for Fusarium head blight resistance from non-adapted sources in an European elite spring wheat background and assessing their effects on deoxynivalenol (DON) content and disease severity. Theor Appl Genet 112:562–569. https://doi.org/10.1007/s00122-005-0163-4
Oelke LM, Kolmer JA (2004) Characterization of leaf rust resistance in hard red spring wheat cultivars. Plant Dis 88:1127–1133. https://doi.org/10.1094/PDIS.2004.88.10.1127
Peng J, Richards DE, Hartley NM, Murphy GP, Devos KM, Flintham JE, Beales J, Fish LJ, Worland AJ, Pelica F, Sudhakar D, Christou P, Snape JW, Gale MD, Harberd NP (1999) 'Green revolution' genes encode mutant gibberellin response modulators. Nature 400:256–261. https://doi.org/10.1038/22307
Perez-Lara E, Semagn K, Chen H, Iqbal M, N'Diaye A, Kamran A, Navabi A, Pozniak C, Spaner D (2016) QTLs associated with agronomic traits in the Cutler × AC Barrie spring wheat mapping population using single nucleotide polymorphic markers. PLoS ONE. https://doi.org/10.1371/journal.pone.0160623
Rawat N, Pumphrey MO, Liu S, Zhang X, Tiwari VK, Ando K, Trick HN, Bockus WW, Akhunov E, Anderson JA, Gill BS (2016) Wheat Fhb1 encodes a chimeric lectin with agglutinin domains and a pore-forming toxin-like domain conferring resistance to Fusarium head blight. Nat Genet 48:1576–1580. https://doi.org/10.1038/ng.3706
Schroeder HW, Christensen JJ (1963) Factors affecting resistance of wheat to scab caused by Gibberella zeae. Phytopathology 53:831–838
Schweiger W, Steiner B, Ametz C, Siegwart G, Wiesenberger G, Berthiller F, Lemmens M, Jia H, Adam G, Muehlbauer GJ, Kreil DP, Buerstmayr H (2013) Transcriptomic characterization of two major Fusarium resistance quantitative trait loci (QTLs), Fhb1 and Qfhs.ifa-5A, identifies novel candidate genes. Mol Plant Pathol 14:772–785. https://doi.org/10.1111/mpp.12048
Schweiger W, Steiner B, Vautrin S, Nussbaumer T, Siegwart G, Zamini M, Jungreithmeier F, Gratl V, Lemmens M, Mayer KF, Berges H, Adam G, Buerstmayr H (2016) Suppressed recombination and unique candidate genes in the divergent haplotype encoding Fhb1, a major Fusarium head blight resistance locus in wheat. Theor Appl Genet 129:1607–1623. https://doi.org/10.1007/s00122-016-2727-x
Semagn K, Skinnes H, Bjørnstad Å, Marøy AG, Tarkegne Y (2007) Quantitative trait loci controlling fusarium head blight resistance and low deoxynivalenol content in hexaploid wheat population from ‘arina’ and NK93604. Crop Sci 47:294–303. https://doi.org/10.2135/cropsci2006.02.0095
Sherman JD, Yan L, Talbert L, Dubcovsky J (2004) A PCR marker for growth habit in common wheat based on allelic variation at the VRN-A1 gene. Crop Sci 44:1832–1838. https://doi.org/10.2135/cropsci2004.1832
Sinha RC, Savard ME, Lau R (1995) Production of monoclonal antibodies for the specific detection of deoxynivalenol and 15-acetyldeoxynivalenol by ELISA. J Agr Food Chem 43:1740–1744. https://doi.org/10.1021/jf00054a061
Somers DJ, Fedak G, Clarke J, Cao W (2006) Mapping of FHB resistance QTLs in tetraploid wheat. Genome 49:1586–1593. https://doi.org/10.1139/g06-127
Somers DJ, Fedak G, Savard M (2003) Molecular mapping of novel genes controlling Fusarium head blight resistance and deoxynivalenol accumulation in spring wheat. Genome 46:555–564. https://doi.org/10.1139/g03-033
Srinivasachary GN, Steed A, Hollins TW, Bayles R, Jennings P, Nicholson P (2009) Semi-dwarfing Rht-B1 and Rht-D1 loci of wheat differ significantly in their influence on resistance to Fusarium head blight. Theor Appl Genet 118:695–702. https://doi.org/10.1007/s00122-008-0930-0
Srinivasachary GN, Steed A, Simmonds J, Leverington-Waite M, Wang Y, Snape J, Nicholson P (2008) Susceptibility to Fusarium head blight is associated with the Rht-D1b semi-dwarfing allele in wheat. Theor Appl Genet 116:1145–1153. https://doi.org/10.1007/s00122-008-0742-2
Steiner B, Buerstmayr M, Wagner C, Danler A, Eshonkulov B, Ehn M, Buerstmayr H (2019) Fine-mapping of the Fusarium head blight resistance QTL Qfhs.ifa-5A identifies two resistance QTL associated with anther extrusion. Theor Appl Genet 132:2039–2053. https://doi.org/10.1007/s00122-019-03336-x
Steiner B, Lemmens M, Griesser M, Scholz U, Schondelmaier J, Buerstmayr H (2004) Molecular mapping of resistance to Fusarium head blight in the spring wheat cultivar Frontana. Theor Appl Genet 109:215–224. https://doi.org/10.1007/s00122-004-1620-1
Su Z, Bernardo A, Tian B, Chen H, Wang S, Ma H, Cai S, Liu D, Zhang D, Li T, Trick H, St Amand P, Yu J, Zhang Z, Bai G (2019) A deletion mutation in TaHRC confers Fhb1 resistance to Fusarium head blight in wheat. Nat Genet. https://doi.org/10.1038/s41588-019-0425-8
Su Z, Jin S, Zhang D, Bai G (2018) Development and validation of diagnostic markers for Fhb1 region, a major QTL for Fusarium head blight resistance in wheat. Theor Appl Genet. https://doi.org/10.1007/s00122-018-3159-6
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Wang S, Wong D, Forrest K, Allen A, Chao S, Huang BE, Maccaferri M, Salvi S, Milner SG, Cattivelli L, Mastrangelo AM, Whan A, Stephen S, Barker G, Wieseke R, Plieske J, Lillemo M, Mather D, Appels R, Dolferus R, Brown-Guedira G, Korol A, Akhunova AR, Feuillet C, Salse J, Morgante M, Pozniak C, Luo MC, Dvorak J, Morell M, Dubcovsky J, Ganal M, Tuberosa R, Lawley C, Mikoulitch I, Cavanagh C, Edwards KJ, Hayden M, Akhunov E, International Wheat Genome Sequencing Consortium (2014) Characterization of polyploid wheat genomic diversity using a high-density 90,000 single nucleotide polymorphism array. Plant Biotechnol J 12:787–796. https://doi.org/10.1111/pbi.12183
Xiao J, Jin X, Jia X, Wang H, Cao A, Zhao W, Pei H, Xue Z, He L, Chen Q, Wang X (2013) Transcriptome-based discovery of pathways and genes related to resistance against Fusarium head blight in wheat landrace Wangshuibai. BMC Genomics 14:197. https://doi.org/10.1186/1471-2164-14-197
Xue S, Xu F, Tang M, Zhou Y, Li G, An X, Lin F, Xu H, Jia H, Zhang L, Kong Z, Ma Z (2011) Precise mapping Fhb5, a major QTL conditioning resistance to Fusarium infection in bread wheat (Triticum aestivum L.). Theor Appl Genet 123:1055–1063. https://doi.org/10.1007/s00122-011-1647-z
Yan L, Loukoianov A, Tranquilli G, Helguera M, Fahima T, Dubcovsky J (2003) Positional cloning of the wheat vernalization gene VRN1. Proc Natl Acad Sci U S A 100:6263–6268. https://doi.org/10.1073/pnas.0937399100
Yu J, Holland JB, McMullen MD, Buckler ES (2008) Genetic design and statistical power of nested association mapping in maize. Genetics 178:539–551. https://doi.org/10.1534/genetics.107.074245
Zhang W, Francis T, Gao P, Boyle K, Jiang F, Eudes F, Cuthbert R, Sharpe A, Fobert PR (2018) Genetic characterization of type II Fusarium head blight resistance derived from transgressive segregation in a cross between Eastern and Western Canadian spring wheat. Mol Breeding. https://doi.org/10.1007/s11032-017-0761-2
Acknowledgements
The authors thank technical staff from the participating laboratories for their contributions to this research. The authors wish to acknowledge Dr. Mark Jordan for providing laboratory facilities to carry out FHB candidate gene sequencing work. This project was funded by AAFC Growing Forward II, Western Grains Research Foundation, and as part of CTAG and CTAG2, Genome Prairie projects funded by Genome Canada, Manitoba Agriculture, Saskatchewan Ministry of Agriculture, and Western Grain Research Foundation.
Author information
Authors and Affiliations
Contributions
DT developed the linkage maps, conducted QTL analysis, characterized candidate genes, interpreted the data, and co-wrote the manuscript. AB, BB, GF, AF, JG, MH, DM, RM, BM, DS, MI, CP, and AN designed and conducted the field experiments and generated Fusarium head blight data. CM planned and organized the study, participated in the analysis and interpretation of data, and co-wrote the manuscript. All authors contributed to and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Human and animal rights
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Communicated by Hermann Buerstmayr.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary file 2: Figure S1
CLUSTAL alignment of (a) DNA sequences and (b) deduced amino acid sequences of the predicted complete ORF of TaHRC gene amplified from a panel of wheat cultivars including three parents AC Barrie, Cutler, and Reeder using gene specific primer pair TaHRC-GSM-F/TaHRC-GSM-R (Su et al. 2018). Identical residues indicated by asterisks (*) and gaps are identified by dashes. Conserved amino acid substitutions are denoted with colon (:) and semi-conserved substitutions are indicated by a dot (.). Numbers on the right indicate the position number. The red rectangular frames show the locations of the start (ATG (+1)) and stop (TAA) codons, respectively. Fhb1-resistant haplotypes similar to Hap_Ning (Su et al. 2019) carried a large deletion including the start codon (ATG (+1)) and a 22 bp sequence downstream of the original ATG (shown in figure S1a), and part of upstream sequence (not shown in the Figure S1a) (PDF 134 kb)
Rights and permissions
About this article
Cite this article
Thambugala, D., Brûlé-Babel, A.L., Blackwell, B.A. et al. Genetic analyses of native Fusarium head blight resistance in two spring wheat populations identifies QTL near the B1, Ppd-D1, Rht-1, Vrn-1, Fhb1, Fhb2, and Fhb5 loci. Theor Appl Genet 133, 2775–2796 (2020). https://doi.org/10.1007/s00122-020-03631-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-020-03631-y