Abstract
Fusarium head blight (FHB) is a major disease of wheat in many wheat-growing regions affecting wheat yield and quality. Glenn is arguably the most FHB-resistant spring wheat cultivar in the USA. To decipher the genetics of FHB resistance in Glenn, a population of 112 recombinant inbred lines was developed from the cross Glenn × MN00261-4 and evaluated for various FHB related traits at multiple locations in two states (North Dakota and Minnesota) over 3 years. Quantitative trait loci (QTL) analysis detected 15 QTL for FHB resistance related traits and heading date (HD). One QTL for FHB incidence (INC; type I resistance), six QTL for FHB severity (SEV; type II resistance), one QTL for Fusarium damaged kernels (FDK; type IV resistance), four QTL for disease index (DI), and three QTL for HD were detected. Major QTL defined as those consistent across multiple environments and explaining > 10% of the phenotypic variation were detected on chromosomes 5BL, 6BS, and 7AS. They were associated with multiple FHB variables and HD. The stable 6BS QTL with a large effect for FHB SEV resistance corresponded to the Fhb2 gene in Sumai 3. In addition, potentially novel QTL were identified on 1AS, 5BL and 7DS. Our study showed that FHB resistance in the GM population was contributed by loci other than Fhb1. The major QTL detected for FHB resistance have potential for use in marker-assisted breeding for FHB resistance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Fusarium head blight (FHB) is a devastating disease of wheat (Triticum aestivum L.). Caused by Fusarium graminearum it is a major disease of wheat in warm and humid regions of the US upper Midwest, particularly the states of Minnesota and North Dakota. It caused hundreds of millions of dollars in losses due to epidemics in the 1990s (Nganje et al. 2004). In 2000 alone, the loss from this disease was estimated at US$160 million (Nganje et al. 2004). Asexually derived conidiospores or sexually produced ascospores are spread by wind and rain (Gilbert and Fernando 2004). One of the major characteristics of the fungus is that it produces a mycotoxin, deoxynivalenol (DON) that facilitates its spread by subcuticular growth within the plant host (Gunnaiah et al. 2012). The presence of DON in grains is harmful to humans and animals. Heavily infected grains reduce quality or may be unsuitable for human and animal consumption (Shin et al. 2012). Breeding for FHB resistance is considered a sustainable and economically and environmentally beneficial strategy to reduce FHB damage in wheat and barley (Lu et al. 2013). However, breeding for FHB resistance is not easy due to quantitative inheritance of resistance and significant environmental effects.
FHB resistance in wheat was divided into five discrete types, (1) type I: resistance to initial infection measured by incidence (INC); (2) type II: which relates to pathogen spread within a single spike and measured by severity (SEV); (3) type III: related to toxin (DON) accumulation; (4) type IV: a measure of Fusarium damaged kernels (FDK); and (5) type V: yield loss (Mesterhazy 1995). Although all types have been investigated, type II resistance (SEV) is the most widely studied because it is considered the most effective measure of disease response and is less affected by environment (Bai and Shaner 1996). Consistency in measures of type II resistance is based on minimizing infection escape which is usually achieved by using controlled environments and precise inoculation methods (Buerstmayr et al. 2013). FHB resistance in wheat was also found to be associated with morpho-physiological traits including plant height and heading date (HD) (Buerstmayr et al. 2009).
Many QTL for different types of FHB resistance were identified in wheat cultivars using bi-parental populations or/and association mapping panels (Buerstmayr et al. 2009). Major QTL for resistance were detected on chromosomes 2D (Lin et al. 2006), 3A (Yu et al. 2008), 3BS (Fhb1; Anderson et al. 2001), 4B (Fhb4; Randhawa et al. 2013), 5A (Fhb5; Xue et al. 2011), and 6B (Fhb2; Cuthbert et al. 2007). Many of these QTL were associated with both types I and II resistance, and were most commonly derived from the resistant cultivar Sumai 3 (PI 481542). Various Sumai derived cultivars such as DH181 (Yang et al. 2005), CJ9306 (Jiang et al. 2007), Ning7840 (Zhou et al. 2002), CM82036 (Buerstmayr et al. 2002), and Line685 (Lu et al. 2011) carry FHB resistance QTL. Overall, genetic data thus far indicate that Sumai 3 is the most valuable and widely used source of FHB resistance in wheat, especially for type II resistance.
Molecular markers tightly linked to FHB resistance can expedite development of FHB resistant cultivars through marker-assisted breeding (MAB). However, the effectiveness of MAB depends on: (1) the degree of linkage between the marker and QTL; (2) polymorphism of the marker in different genetic backgrounds; and (3) the stability, economic value, and efficiency of the assessment methods used (Randhawa et al. 2013). The Mendelised Fhb1 allele in Sumai 3, located on chromosome 3BS, has the strongest effect, and is thus widely used in breeding for FHB resistance (Buerstmayr et al. 2009). Liu et al. (2008) fine mapped the Fhb1 region and found that the sequence-tagged site (STS) marker UMN10 was located 1 cM away. Furthermore, many scientists worldwide worked on Fhb1 and finally cloned it (Rawat et al. 2016; Liu et al. 2008; Su et al. 2019). It is believed that the Fhb1 allele can reduce the type II response by up to 50% relative to related lines with the alternative allele. However, depending on genetic backgrounds a 20–25% reduction was commonly reported (Pumphrey et al. 2007). Other evidence indicated that the effectiveness of FHB resistance is enhanced when Fhb1 is combined with other major effect QTL (Zhuang et al. 2013).
FHB is a major threat to wheat production in the upper Midwest region of the USA (states Minnesota and North Dakota). Therefore, FHB resistance is a major objective of wheat breeding programs in the region. Glenn (PI 639273), released in 2005 for the upper Midwest USA, is a valuable source of FHB resistance developed and released by the Hard-Red Spring Wheat (HRSW) breeding program at North Dakota State University in Fargo, North Dakota. In addition to FHB resistance Glenn was released for high end-use quality and superior agronomic performance (Mergoum et al. 2006). Glenn’s pedigree presumes the source of FHB resistance was Sumai 3. However, detailed analysis showed that Glenn did not possess the Fhb1 resistant allele based on lack of the flanking marker UMN10 identified in Sumai 3. Therefore, a RIL population was developed from the cross Glenn × MN00261-4 (hereafter referred as the GM population) and evaluated for different FHB resistance types and heading date (HD) under field and greenhouse environments over 3 years (2010–2012). The objectives of the study were to (1) investigate the genetic factors controlling FHB resistance in Glenn, (2) determine if FHB resistance loci from Sumai 3, particularly Fhb1, contributed to FHB resistance in Glenn, and (3) investigate the association between FHB resistance and HD based on phenotypic and genotypic analyses.
Materials and methods
Mapping population development
Glenn was selected among the progeny of a cross between ND2831 (PI 665,931; Mergoum et al. 2012) and Steele-ND (PI 634,981; Mergoum et al. 2005). ND2831 is an NDSU experimental line with Sumai 3 background (SUMAI-3/STOA; SUMAI-3/WHEATON//GRANDIN/3/ND-688; ND-2709/ND-688). Glenn had a high level of FHB resistance in addition to its superior agronomic and yield-related traits (Mergoum et al. 2006). Given that, molecular marker analysis showed that Glenn did not possess flanking marker allele umn10 from Sumai 3 and therefore presumably the Fhb1 resistance allele it was crossed with the FHB susceptible line, MN00261-4, and a RIL population of 112 lines was developed. MN00261-4 (MN95286/MN94155//VERDE) is an experimental line developed by the University of Minnesota. Seven HRSW check cultivars were also included in field and greenhouse evaluations. The checks included FHB resistant line ND2710 (PI 633976; Frohberg et al. 2004), two moderately resistant cultivars Faller (PI 648350; Mergoum et al. 2008) and Alsen (PI 615543; Frohberg et al. 2006), two moderately susceptible cultivars Steele-ND and Barlow (PI 658018; Mergoum et al. 2011), susceptible line Vida (PI 642366; Lanning et al. 2006), and highly susceptible line 2398.
Field and greenhouse evaluation for FHB response
Parents, checks and RILs were evaluated under field experiments for 3 years (2010–2012) at Minneapolis in Minnesota (MN), and three locations in North Dakota (ND), viz. Carrington, Prosper, and Langdon. “Grain spawn” inocula (Rudd et al. 2001) was used for field inoculations. In this method, infected grain produces ascospores as primary inoculum. The infested corn grain was spread in the experimental fields at heading stage (Feekes growth stage 10) as it generates ascospores over a longer period of time compared to the discrete events involved with spray and point inoculations (Dill-Macky 2003). Overhead mist irrigation to support disease development was provided during the grain spawn inoculation period. FHB reactions were recorded 21 days post flowering. Field experiments were arranged in randomized complete block design (RCBD) with four replications in ND and two replications in MN. Approximately 50–60 seeds in 0.3 m hill plots were planted per entry per replication and 10–15 heads were randomly selected as an experimental unit for data collection.
Parents, checks, and RILs were also tested under greenhouse (GH) conditions during 2011 and 2012 using an RCBD design. Five seeds were planted in 20 cm diameter pots in four replications. The plant materials were grown in Sunshine Mix #1 soil (Sun Gro Horticulture, Agawam, MA, USA) and about 20 g Osmocote® slow release fertilizer (Scott’s Company LLC, Marysville, OH, USA) was added to each pot after planting. Artificial inoculations by injection of single mid-spike florets (Zadoks et al. (1974) growth stage 55) with conidiospore suspensions as described in Bekele (1995). Approximately 5 µl of spore suspension (100 k spores/ml) was injected into each floret of 5–10 spikes per pot) using a needleless syringe. Briefly, the middle floret of a single spikelet of each spike was manually injected. Inoculated spikes were individually covered with plastic bags for 72 h to maintain humidity. FHB severity was assessed about three weeks post inoculation.
Each genotype was evaluated for reaction to FHB in all replications. Heading dates (HD) were recorded in both GH and field experiments. FHB INC was assessed in field trials by dividing the number of infected spikes by the total spikes per plot. FHB INC data were obtained from five field environments (year by location combinations) including MN 2010 and 2012 (M10 and M12), Carrington 2011 (C11) and Prosper 2011 and 2012 (P11 and P12). FHB SEV data were recorded as the proportion of infected spikelets per spike averaged over 10 random spikes per plot and 5–10 spikes per pot in the field and GH experiments, respectively. The seven environments where SEV data was recorded included M10, M12, Prosper 2010 (P10), P11, P12, C11, and GH 2011 (G11). Data on FDK was recorded as the proportion of infected kernels in a 200-kernel random sample from each genotype and replicate. The five environments for FDK included M10, MN 2011 (M11), M12, G11, and GH 2012 (G12). Disease index (DI) was recorded by multiplying the INC and SEV percentages for each genotype per replicate. HD from seven environments (M10, M12, P10, P11, P12, C11, and Langdon 2011 (L11) was recorded as the number of days from planting to Zadoks growth stage 55, where 50% of spikes in each plot were fully emerged. The phenotypic data utilized in the QTL analysis were plot means within environments for all FHB-related traits and HD.
Statistical analysis of phenotypic data
Phenotypic data recorded for all FHB-related traits and HD were subjected to analysis of variance (ANOVA) using the Mixed Procedure in Statistical Analysis System 9.3® (SAS Institute, Cary, NC, 2004). Genotypes (G) were considered a fixed effect whereas environments (E) and replicates (rep) were considered random effects. ANOVA was performed for each environment (field and GH) and also the combined data across all environments to estimate G × E interaction. Fmax ratios (Tabachnick and Fidell 2001) were calculated to compare variances between environments. If the Fmax ratio between the major and minor experimental errors in an individual environment was less than 10 the error terms were combined and considered valid at p ≤ 0.05 and individual environments were combined. Correlation coefficients (r) calculated individually for traits at each location, and for means of combined environments for each trait using PROC CORR in SAS 9.3® were considered significant at p ≤ 0.05. Differences in genotypic means were determined using F-protected least significant differences (LSD, p ≤ 0.05) generated from the Proc Mixed output file of the SAS analysis. Broad sense heritability (H2) was calculated based on family means (Holland et al. 2003) using the output file of the ANOVA random model of SAS Proc Mixed analysis excluding the parental and check means. In calculating H2 the means of combined environments per trait with non-significant F-max homogeneity were used (Otto et al. 2002), where H2\(=\frac{{\upsigma }^{2}G}{{\upsigma }^{2}G+\frac{{\upsigma }^{2}GE}{e}+\frac{{\upsigma }^{2}E}{re}}\) and \({\upsigma }^{2}G\) = genotypic variance, \({\upsigma }^{2}GE\) = variance due to genotype × environment interaction, \({\upsigma }^{2}E\) = experimental error variance, e = total number of environments, and r = total number of replicates within each environment.
Construction of genetic only map and QTL analysis
The DNA was extracted from young leaf samples taken from both parental lines, RILs, and checks using a Qiagen DNeasy Plant mini kit (Cat# 69106). Thirty µl of DNA (80 ng/µl) from each entry was sent to Triticarte Pvt. Ltd (Canberra, Australia; https://www.triticate.com.au) for Diversity Array Technology (DArT) genotyping (Akbari et al. 2006). Scores of all polymorphic DArT markers were converted to genotypic codes according to the parental scores. Conformity with expected 1:1 segregation ratio was determined by χ2 tests. Genetic maps were constructed using both Mapmaker 3.0 (Lander et al. 1987) and CarthaGene V.1.2.3R (De Givry et al. 2005) with a minimum LOD score of 3.0 and maximum recombination frequency of 40%. MapMaker was used to construct the groups based on anchor markers from each consensus mapped chromosome. CarthaGene was used to construct final map orders using build10, greedy search, genetic algorithm, annealing, flips, and polish functions. Genetic distances were determined using the Kosambi mapping function (Kosambi 1944). Final maps were compared with the DArT consensus maps (Huang et al. 2012) using Autograph (Derrien et al. 2007; https://autograph.genouest.org/) for accuracy of the marker maps.
QTL analysis was conducted using composite interval mapping (CIM) (Zeng 1994) in QGene v.4.3.10 (Joehanes and Nelson 2008). The parameters used to detect significant QTL included a scanning interval of 1 cM between markers with a window size of 10 cM. Five markers were set to control background by forward and reverse regression. The LOD threshold to identify significant QTL was calculated using 1000 permutations. Only the QTL detected above threshold LOD score (2.5) were included. Additive QTL effects, positive and negative signs indicate respectively, the contributions of Glenn and MN00261-4 as parents, toward higher trait values. The proportion of phenotypic variation (PV) explained by each QTL was determined by R2, the squared partial correlation coefficient. QTL contributing ≥ 10% of PV were considered major QTL (Xu et al. 2000). QTL detected in at least two environments were considered stable/consistent. Confidence intervals (CI) were determined using positions of markers at ± 2 LOD from the QTL peak. QTL with overlapping CIs or located within a 10 cM region were considered as one QTL. Graphical representation of linkage groups and QTL was composed using MapChart 2.2 (Voorrips 2002).
Results
Phenotypic analysis of the parental lines and GM population
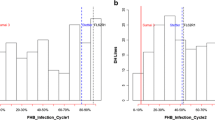
Significant differences in reaction to FHB-related traits and HD between RILs and their parental genotypes were observed (Table S1). Glenn showed lower values than MN00261-4 for FHB INC, SEV, DI and FDK but the differences were not significant. This indicated that both parents possess some level of FHB resistance. The RILs varied significantly for INC (range 66.3–100%) and their mean (94.9%) was lower than both the parents and checks. For SEV and DI, the RILs varied significantly from 22.3 and 14.4 to 88.1 and 86.5, respectively. Their means (41.5% and 38.1%, respectively) were lower than those for MN00261-4 and susceptible check 2398. These means were all higher than those for Glenn and resistant checks Alsen and Parshall. FDK and HD varied significantly in the RIL population with means slightly higher than both parents and all checks.
The GM population displayed wide variation for all FHB related traits and HD (Fig. 1). There were significant G × E interactions for most traits except for FDK (Table S2). Although significant F-values for E and G × E variances attributed variation to environments genotypes caused the largest proportion of variation (Table S2). There were high H2 values (0.69–0.93) for SEV, FDK, DI, and HD (Table S2) and moderate H2 values (0.48–0.53) for INC and FDK. The coefficients of correlation for individual traits across different pairs of environments were mostly positive, except for SEV (P10, P12, and G11) which were negative or non-significant between any of these three environments and other environments (data not shown). There was a wide range of correlation coefficients (r = 0.02–0.94) for traits in the combined environments analysis (Table 1). FDK under greenhouse conditions (G11 and G12) was negatively correlated with the INC, SEV, DI and HD traits, but positively correlated in MN field environments. SEV, INC, and DI were negatively correlated with HD suggesting that earliness was associated with lower values possibly caused by disease escape.
Development of linkage map
Of 2,289 polymorphic DArT markers, 659 were polymorphic between the parents and therefore were used to generate the linkage maps. The informative markers were represented by 458 unique loci that were mapped to 37 linkage groups on 19 different chromosomes (Table S3). Five chromosomes (2B, 4B, 6B, 7A and 7D) had three linkage groups, eight chromosomes (1B, 2A, 2D, 4A, 5B, 6A, 6D and 7B) had two linkage groups, and six chromosomes (1A, 1D, 3A, 3B, 3D and 5A) had one. No marker was mapped to chromosomes 4D or 5D. The total map distance was 2,229 cM with an average distance of 3.46 cM between adjacent markers. Total map lengths for genomes A, B, and D were 879.4, 1,106, and 243.6 cM, respectively (Table S3).
Detection of QTL for FHB resistance
QTL analysis detected a single major QTL on chromosome 1AS for FHB INC resistance and the QTL was detected in three environments (Table 2, Fig. 2). This QTL, designated Qinc.ndsu- 1AS, explained 11.8% of the phenotypic variation (PV) in the GM population. The additive value of the QTL was − 4.1, indicating that this QTL was contributed by the resistant parent Glenn. Six QTL were detected for FHB SEV resistance with PV > 10% and were located on chromosomes 1BL, 5BL, 6BS, 7AS, and 7DS (Table 2; Fig. 2). The PV explained by these QTL ranged from 10.2 to 23.8% (Qfhb.ndsu-5BL1b1), and four of the six QTL were contributed by the resistant parent Glenn (Table 2). Similarly, four QTL with PV > 10% were detected for FHB index (DI) on chromosomes 5BL, 6BS, and 7AS (Table 2, Fig. 2). The PV explained by these QTL ranged from 10.0 to 19.1%. Of the four QTL detected for DI, three were contributed by Glenn and one was contributed by MN00261-4. For FDK resistance, a single major QTL, Qfdk.ndsu-5BL2, was detected in three environments (Table 2, Fig. 2). The QTL was contributed by MN00261-4 and explained 18.6% of the variation in the GM population.
Detection of QTL for HD
Four QTL controlling HD were detected on chromosomes 5BL, 1DS, 6AS, and 6DS (Table 2, Fig. 2). The most stable QTL, Qfhb.ndsu-5BL1b, was detected in all tested environments and explained 23.8% of the PV in our population. Similarly, QTL Qhd.ndsu-6AS was detected in six environments whereas Qhd.ndsu-6DS and Qhd.ndsu-1DS were detected in five and three environments, respectively. The PV explained by these QTL ranged from 8.4 to 12.2% and the range of additive effect was between 0.8 and 1.3. The main Qfhb.ndsu-5BL1b was contributed by Glenn whereas the remaining three QTL were contributed by MN00261-4.
Association between FHB resistance and HD
Correlation analysis showed that FHB resistance was negatively correlated with HD (Table 1). The correlation between HD and FHB-related traits ranged from − 0.34 to − 0.66 (Table 1). Furthermore, QTL analysis detected a common locus for both, FHB resistance and HD (Table 2). Indeed, the major QTL, Qfhb.ndsu-5BL1b, detected on long arm of chromosome 5BL was common for SEV, DI and HD with overlapping confidence intervals (CI) (Table 2, Fig. 2). Alleles of this QTL were located in a common genomic region indicating that a single QTL was involved. Furthermore, all alleles of enhanced FHB resistance at this region were contributed by MN00261-4.
Discussion
Among the different types of FHB resistance studied, type II resistance is the most effective because the effects of environment are comparatively low (Bai and Shaner 1996). Type II is the most frequently deployed form of FHB resistance and most released resistant cultivars in the spring wheat region of the USA possess type II resistance conferred by Fhb1 (Mergoum unpublished). The consistency of Type II results was based on minimizing the likelihood of disease escape with experiments in controlled conditions and direct spore placement for infection (Buerstmayr et al. 2013). Detailed genetic knowledge of the different types of FHB resistance is useful for breeders and geneticists in developing FHB resistant cultivars. This study was designed to dissect the genetics and trait associations of FHB resistance, especially type II, but also I, DI, and HD.
Type II resistance effects were, in general, positively correlated with all other resistance types (I, IV and DI) except under greenhouse conditions, where types II and IV seemed to have a negative but not significant correlation. However, this correlation was positive and significant for all MN field environments (Table 1). Similar findings were reported previously reports where single-floret injection (as applied in GH) experiments conferred a lower correlation between type II resistance and some agronomic traits rather than the correlation conferred by the grain spawn inoculation method used in the field) (Buerstmayr et al. 2013). The difference in correlation between GH and field between types II and IV resistance could be due to the inoculation method. Furthermore, we found that the correlation between the field and GH data was relatively low indicating low effectiveness of GH evaluation data when applied to the field situation. Based on phenotypic observations, a few resistant RILs showed highly consistent resistant type I and/or II reactions, while conferring inconsistent type IV reactions. This likely reflects genetic differences controlling FHB resistance mechanisms and/or environmental impacts. Other reports similarly describe negative correlations between different FHB resistance types under field conditions and HD (Emrich et al. 2008). In this study, Glenn matured 1–2 days earlier than MN00261-4, yet only one RIL (GM-19) under ND conditions and three RIL (GM-9, 42 and 86) in MN environments headed earlier than Glenn. Controlled inoculation and assessment methods used for the GH population mitigated the interaction of HD and disease development.
Three QTL, QFhb.ndsu-1AS, QFhb.ndsu-1BL, and QHd.ndsu-1DS, were associated with FHB resistance and HD, each accounting more than 10% of the phenotypic variation in the GM population (Table 2). Draeger et al. (2007) also reported QTL in the vicinity of QFhb.ndsu-1BL for resistance to FHB SEV, FDK, and yield loss in winter cv. Arina. However, the yield drag reported for Arina could be due to unfavorable alleles associated with resistance type V from the genetic background of winter wheat. In contrast, Glenn is a spring cultivar with comparatively high yield and quality (Mergoum et al. 2006), suggesting that Glenn has a superior allelic combination for FHB resistance and yield and quality traits. Haberle et al. (2009) also reported a major effect QTL, Qfhs.lX-1BL in four European winter wheat lines. Based on an integrated genetic map (Maccaferri et al. 2015), QFhb.ndsu-1BL and Qfhs.lX-1BL linked markers, wPt8168 and Xwmc728, respectively, were located 95.23 cM apart, indicating genetic independence. Qinc.ndsu-1AS enhanced the FHB resistance (INC) and was genetically independent of HD, and was therefore, presumed to be a true FHB resistance locus. A FHB QTL in this region was reported by Li et al. (2011) in Chinese landrace Haiyanzhong; however, it had a wide genome coverage (18–38 cM). Several other QTL in the region were also reported (Liu et al. 2009; Löffler et al. 2009; Venske et al. 2019). The repeated reports of QTL in this region across different populations and cultivars suggests that the region is important for FHB resistance and worthy of fine mapping for favorable FHB alleles.
Chromosome 5BL harbored six QTL for resistance to FHB SEV, FDK, DI and HD (Table 2; Fig. 2). Similar marker alleles in the vicinity of these QTL were reported for resistance to FHB SEV in both tetraploid and hexaploid genotypes (Buerstmayr et al. 2009; Ruan et al. 2012). A QTL on chromosome 5B was also reported for HD (Vrn-B1) and plant height in a a RIL population derived from the soft red winter wheat cross NC-Neuse × AGS 2000 (Petersen et al. 2016). Interestingly, Qfhb.ndsu-5BL1b had the most significant effect on SEV, DI, HD and explained up to 23.8% of phenotypic variation in the GM population. This region has not been reported previously and could represent a novel QTL for FHB resistance. Another QTL, Qfdk.ndsu-5BL2, on chromosome 5BL had major effects for FDK resistance but was detected only in MN environments. This QTL region was also not previously reported and therefore could be a novel QTL for FHB resistance.
Qfhb.ndsu-6BS had significant effects on FHB SEV and DI resistance suggesting the presence of Fhb2 from Sumai 3. Previous studies also detected a QTL for resistance to FHB SEV from durum (Somers et al. 2006) and hexaploid wheat cultivars (Buerstmayr et al. 2012; Cuthbert et al. 2007; Venske et al. 2019) in this region. Qfhb.ndsu-6BS was contributed by Glenn, most likely from ND2831, the source of any Sumai 3 contribution in Glenn. This QTL was independent of HD and is therefore likely to be a true FHB resistance locus that could coincide with Fhb2.
Qsev.ndsu-7DS had a major effect on FHB resistance (SEV) in the GM population (Table 2). This QTL could be the same as one reported in the same region in a DH population from Arina/Riband (Draeger et al. 2007; Venske et al. 2019). Three QTL (Qhd.ndsu-1DS, Qhd.ndsu-6AS, and Qhd.ndsu-6DS) were detected only for HD in our GM population (Table 2). The QTL on chromosome 1DS was detected only in MN environments, however the other two QTL were detected in both the MN and ND environments (Table 2). The QTL on chromosomes 1DS and 6DS were not previously reported, and therefore might be novel loci for HD.
It is well documented that the most stable QTL with major FHB resistance in wheat is Fhb1. This QTL was reported to confer types I, II, III and IV resistance by many researchers (Buerstmayr et al. 2009) and was thus used extensively in wheat breeding programs worldwide to develop FHB resistant germplasm (Cuthbert et al. 2006). Recent map-based cloning showed that Fhb1 produces a pore-forming toxin (PFT) (Rawat et al. 2016). Based on its pedigree and high level of FHB resistance Glenn was presumed to carry Fhb1. However, no QTL was detected on chromosome 3B in the GM population. This was consistent with an earlier molecular analysis showing that the diagnostic SSR marker umn10 was not present in Glenn (data not shown). Moreover, the pedigree and FHB response of MN00261-4 did not support the presence of Fhb1. Thus, the available evidence showed that Glenn did not possess Fhb1. Our results support the claim by Liu et al. (2009) that a high level of FHB resistance, equivalent to that conferred by Fhb1, can be achieved by pyramiding FHB QTL with relatively minor effects. In addition to its desirable agronomic characteristics Glenn is a valuable alternative to Fhb1 in breeding for resistance to FHB.
Conclusions
Development of FHB resistant wheat cultivars is challenging mainly because of the quantitative nature of the resistance. This study was a detailed dissection of the genetic control of FHB resistance in cv. Glenn, a widely grown hard red spring type. Twelve stable QTL for FHB resistance were identified and should be amenable for marker assisted breeding. Some of the QTL were reported previously. However, a new, major QTL (Qfhb.ndsu-5BL1b) conferred FHB SEV and DI resistance. A second QTL (Qfdk.ndsu-5BL2) was identified for FDK resistance. Additionally, other QTL on chromosomes 6AS, 6DS, 7AS and 7DS were effective against more than two disease types. Most importantly, this study showed that Fhb1 was not part of the highly effective FHB resistance displayed by Glenn and demonstrated that there are sources of resistance equally effective to that conferred by Fhb1. Both parents, Glenn and MN00261-4, contributed resistance alleles. Lines possessing favorable alleles from both parents were selected and used in breeding.
References
Akbari M, Wenzl P, Caig V, Carling J, Xia L, Yang SY, Uszynski G, Mohler V, Lehmensiek A, Kuchel H, Hayden MJ, Howes N, Sharp P, Vaughan P, Rathmell B, Huttner E, Kilian A (2006) Diversity arrays technology (DArT) for high-throughput profiling of the hexaploid wheat genome. Theor Appl Genet 113:1409–1420
Anderson JA, Stack RW, Liu S, Waldron BL, Fjeld AD, Coyne C, Moreno-Sevilla P, Fetch JM, Song QJ, Cregan PB, Frohberg RC (2001) DNA markers for Fusarium head blight resistance QTLs in two wheat populations. Theor Appl Genet 102:1164–1168
Bai GH, Shaner G (1996) Variation in Fusarium graminearum and cultivar resistance to wheat scab. Plant Dis 80:975–979
Bekele GT (1995) Head SCAB screening methods used in CIMMYT. In: Wheats for more tropical environments. CIMMYT, Mexico D.F., Mexico, pp 169–173
Buerstmayr H, Lemmens M, Hartl L, Doldi L, Steiner B, Stierschneider M, Ruckenbauer P (2002) Molecular mapping of QTLs for Fusarium head blight resistance in spring wheat. I. Resistance to fungal spread (type II resistance). Theor Appl Genet 104:84–91
Buerstmayr H, Ban T, Anderson JA (2009) QTL mapping and marker-assisted selection for Fusarium head blight resistance in wheat: a review. Plant Breed 128:1–26
Buerstmayr M, Huber K, Heckmann J, Steiner B, Nelson JC, Buerstmayr H (2012) Mapping of QTL for Fusarium head blight resistance and morphological and developmental traits in three backcross populations derived from Triticum dicoccum × Triticum durum. Theor Appl Genet 125:1751–1765
Buerstmayr M, Alimari A, Steiner B, Buerstmayr H (2013) Genetic mapping of QTL for resistance to Fusarium head blight spread (type 2 resistance) in a Triticum dicoccoides × Triticum durum backcross-derived population. Theor Appl Genet 126:2825–2834
Cuthbert PA, Somers DJ, Thomas J, Cloutier S, Brule-Babel A (2006) Fine mapping Fhb1, a major gene controlling Fusarium head blight resistance in bread wheat (Triticum aestivum L.). Theor Appl Genet 112:1465–1472
Cuthbert P, Somers D, Brulé-Babel A (2007) Mapping of Fhb2 on chromosome 6BS: a gene controlling Fusarium head blight field resistance in bread wheat (Triticum aestivum L.). Theor Appl Genet 114:429–437
Derrien T, Andre C, Galibert F, Hitte C (2007) AutoGRAPH: an interactive web server for automating and visualizing comparative genome maps. Bioinformatics 23:498–499
Dill-Macky R (2003) Inoculation methods and evaluation of Fusarium head blight resistance in wheat. In: Leonard KJ, Bushnell WR (eds) Fusarium head blight in wheat and barley. APS Press, St Paul, pp 184–210
Draeger R, Gosman N, Steed A, Chandler E, Thomsett M, Srinivasachary SJ, Buerstmayr H, Lemmens M, Schmolke M, Mesterhazy A, Nicholson P (2007) Identification of QTLs for resistance to Fusarium head blight, DON accumulation and associated traits in the winter wheat variety Arina. Theor Appl Genet 115:617–625
Emrich K, Wilde F, Miedaner T, Piepho HP (2008) REML approach for adjusting the Fusarium head blight rating to a phenological date in inoculated selection experiments of wheat. Theor Appl Genet 117:65–73
Frohberg RC, Stack RW, Mergoum M (2004) Registration of spring wheat germplasm ND2710 resistant to Fusarium head blight. Crop Sci 44:1498–1499
Frohberg RC, Stack RW, Olson T, Miller JD, Mergoum M (2006) Registration of ‘Alsen’ wheat. Crop Sci 46:2311–2312
Gilbert J, Fernando W (2004) Epidemiology and biological control of Gibberella zeae/Fusarium graminearum. Can J Plant Pathol 26:464–472
de Givry S, Bouchez M, Chabrier P, Milan D, Schiex T (2005) CarthaGene: multipopulation integrated genetic and radiation hybrid mapping. Bioinformatics 21:1703–1704
Gunnaiah R, Kushalappa AC, Duggavathi R, Fox S, Somers DJ (2012) Integrated metabolo-proteomic approach to decipher the mechanisms by which wheat QTL (Fhb1) contributes to resistance against Fusarium graminearum. PLoS ONE 7:e40695
Haberle J, Holzapfel J, Schweizer G, Hartl L (2009) A major QTL for resistance against Fusarium head blight in European winter wheat. Theor Appl Genet 199:325–332
Holland JB, Nyquist WE, Cervantes-Martínez CT (2003) Estimating and interpreting heritability for plant breeding: an update. Plant Breed Rev 22:9–111
Huang BE, George AW, Forrest KL, Kilian A, Hayden MJ, Morell MK, Cavanagh CR (2012) A multiparent advanced generation inter-cross population for genetic analysis in wheat. Plant Biotechnol J 10:826–839
Jiang GL, Dong Y, Shi J, Ward RW (2007) QTL analysis of resistance to Fusarium head blight in the novel wheat germplasm CJ 9306. II. Resistance to deoxynivalenol accumulation and grain yield loss. Theor Appl Genet 115:1043–1052
Joehanes R, Nelson JC (2008) QGene 4.0, an extensible Java QTL-analysis platform. Bioinformatics 24:2788–2789
Kosambi DD (1944) The estimation of map distances from recombinant values. Ann Eugen 12:172–175
Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181
Lanning SP, Carlson GR, Nash D, Wichman DM, Kephart KD, Stougaard RN, Kushnak GD, Eckhoff JL, Grey WE, Dyer A, Talbert LE (2006) Registration of ‘Vida’ wheat. Crop Sci 46:2315–2316
Li T, Bai G, Wu S, Gu S (2011) Quantitative trait loci for resistance to Fusarium head blight in a Chinese wheat landrace Haiyanzhong. Theor Appl Genet 122:1497–1502
Lin F, Xue SL, Zhang ZZ, Zhang CQ, Kong ZX, Yao GQ, Tian DG, Zhu HL, Li CJ, Cao Y, Wei JB, Luo QY, Ma ZQ (2006) Mapping QTL associated with resistance to Fusarium head blight in the Nanda2419 × Wangshuibai population. II: type I resistance. Theor Appl Genet 112:528–535
Liu S, Pumphrey MO, Gill BS, Trick HN, Zhang JX, Dolezel J, Chalhoub B, Anderson JA (2008) Toward positional cloning of Fhb1, a major QTL for Fusarium head blight resistance in wheat. Cereal Res Commun 36:195–201
Liu SY, Hall MD, Griffey CA, McKendry AL (2009) Meta-analysis of QTL associated with fusarium head blight resistance in wheat. Crop Sci 49:1955–1968
Löffler M, Schon CC, Miedaner T (2009) Revealing the genetic architecture of FHB resistance in hexaploid wheat (Triticum aestivum L.) by QTL meta-analysis. Mol Breed 23:473–488
Lu QX, Szabo-Hever A, Bjørnstad A, Lillemo M, Semagn K, Mesterhazy A (2011) Two major resistances QTL were required to counteract the increased susceptibility to Fusarium head blight at the Rht-D1b gene in wheat. Crop Sci 51:2430–2438
Lu QX, Lillemo M, Skinnes H, He XY, Shi JR, Ji F, Dong YH, Bjornstad A (2013) Anther extrusion and plant height are associated with Type I resistance to Fusarium head blight in bread wheat line ‘Shanghai-3/Catbird’. Theor Appl Genet 126:317–334
Maccaferri M, Zhang J, Bulli P, Abate Z, Chao S, Cantu D, Bossolini E, Chen X, Pumphrey M, Dubcovsky J (2015) A genome-wide association study of resistance to stripe rust (Puccinia striiformis f. sp. tritici) in a worldwide collection of hexaploid spring wheat (Triticum aestivum L.). G3 (Bethesda) 5:449–465
Mergoum M, Frohberg RC, Miller JD, Stack RW (2005) Registration of ‘Steele-ND’ wheat. Crop Sci 45:1163–1164
Mergoum M, Frohberg RC, Stack RW, Olson T, Friesen TL, Rasmussen JB (2006) Registration of ‘Glenn’ wheat. Crop Sci 46:473–474
Mergoum M, Frohberg RC, Stack RW, Rasmussen JW, Friesen TL (2008) Registration of ‘Faller’ spring wheat. J Plant Reg 2:224–229
Mergoum M, Simsek S, Frohberg RC, Rasmussen JB, Friesen TL, Adhikari T (2011) ‘Barlow’: a high-quality and high-yielding hard red spring wheat cultivar adapted to the North Central Plains of the USA. J Plant Reg 5:62–67
Mergoum M, Frohberg RC, Stack RW, Singh PK, Adhikari TB, Rasmussen JB, Alamri MS, Friesen TL (2012) ND 803 spring wheat germplasm combining resistance to scab and leaf diseases with good agronomic and quality traits. J Plant Reg 7:113–118
Mesterhazy A (1995) Types and components of resistance to Fusarium head blight. Plant Breed 114:377–386
Nganje WE, Bangsund DA, Leistritz FL, Wilson WW, Tiapo NM (2004) Regional economic impacts of Fusarium head blight in wheat and barley. Rev Agric Econ 26:332–347
Otto CD, Kianian SF, Elias EM, Stack RW, Joppa LR (2002) Genetic dissection of a major Fusarium head blight QTL in tetraploid wheat. Plant Mol Biol 48:625–632
Petersen S, Lyerly JH, Maloney PV, Brown-Guedira G, Cowger C, Costa JM, Dong Y, Murphy JP (2016) Mapping of Fusarium head blight resistance quantitative trait loci in winter wheat cultivar NC-Neuse. Crop Sci 56:1473–1483
Pumphrey MO, Bernardo R, Anderson JA (2007) Validating the Fhb1 QTL for Fusarium head blight resistance in near isogenic wheat lines developed from breeding populations. Crop Sci 47:200–206
Randhawa HS, Asif M, Pozniak C, Clarke JM, Graf RJ, Fox SL, Humphreys DJ, Knox RE, DePauw RM, Singh AK, Cuthbert RD, Hucl P, Spaner D (2013) Application of molecular markers to wheat breeding in Canada. Plant Breed 132:458–471
Rawat N, Pumphrey MO, Liu S, Zhang X, Tiwari VK, Ando K, Trick HN, Bockus WW, Akhunov E, Anderson JA, Gill BS (2016) Wheat Fhb1 encodes a chimeric lectin with agglutinin domains and a pore-forming toxin-like domain conferring resistance to Fusarium head blight. Nat Genet 12:1576–1580
Ruan Y, Comeau A, Langevin F, Hucl P, Clarke JM, Brule-Babel A, Pozniak CJ (2012) Identification of novel QTL for resistance to Fusarium head blight in a tetraploid wheat population. Genome 55:853–864
Rudd JC, Horsley RD, McKendry AL, Elias EM (2001) Host plant resistance genes for Fusarium head blight: sources, mechanisms and utility in conventional breeding systems. Crop Sci 41:620–627
SAS Institute Inc (2004) SAS online doc, version 9.1.2 SAS Inst., Cary, NC
Shin S, Torres Acosta JA, Heinen SJ, McCormick Susan P, Lemmens M, Kovalsky Paris MP, Berthiller F, Adam G, Muehlbauer GJ (2012) Transgenic Arabidopsis thaliana expressing a barley UDPglucosyltransferase exhibit resistance to the mycotoxin deoxynivalenol. J Exp Bot 63:4731–4740
Somers DJ, Fedak G, Clarke J, Wenguang C (2006) Mapping of FHB resistance QTLs in tetraploid wheat. Genome 49:1586–1593
Su Z, Bernardo A, Tian B, Chen H, Wang S, Ma H, Cai S, Liu D, Zhang D, Li T, Trick H, Amand PS, Yu J, Zhang Z, Bai G (2019) A deletion mutation in TaHRC confers Fhb1 resistance to Fusarium head blight in wheat. Nat Genet 51:1099–1105
Tabachnick BG, Fidell LS (2001) Using multivariate statistics. Allyn and Bacon, Inc., Boston, p 85
Venske E, Dos Santos RS, Farias DDR, Rother V, da Maia LC, Pegoraro C, Costa de Oliveira A (2019) Meta-analysis of the QTLome of Fusarium head blight resistance in bread wheat: refining the current puzzle. Front Plant Sci 10:727
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78
Xu W, Subudhi PK, Crasta OR, Rosenow DT, Mullet J, Nguyen HT (2000) Molecular mapping of QTLs conferring stay-green in grain sorghum (Sorghum bicolor L. Moench). Genome 43:461–469
Xue S, Xu F, Tang M, Zhou Y, Li G, An X, Lin F, Xu H, Jia H, Zhang L (2011) Precise mapping Fhb5, a major QTL conditioning resistance to Fusarium infection in bread wheat (Triticum aestivum L.). Theor Appl Genet 123:1055–1063
Yang Z, Gilbert J, Fedak G, Somers D (2005) Genetic characterization of QTL associated with resistance to Fusarium head blight in a doubled-haploid spring wheat population. Genome 48:187–196
Yu JB, Bai GH, Zhou WC, Dong YH, Kolb FL (2008) Quantitative trait loci for Fusarium head blight resistance in a recombinant inbred population of Wangshuibai/Wheaton. Phytopathology 98:87–94
Zadoks JC, Chang TT, Konzak CF (1974) A decimal code for the growth stages of cereals. Weeds Res 14:415–421
Zeng ZB (1994) Precision mapping of quantitative trait loci. Genetics 136:1457–1468
Zhou W, Kolb F, Bai G, Shaner G, Domier L (2002) Genetic analysis of scab resistance QTL in wheat with microsatellite and AFLP markers. Genome 45:719–727
Zhuang Y, Gala A, Yen Y (2013) Identification of functional genic components of major Fusarium head blight resistance quantitative trait loci in wheat cultivar Sumai 3. Mol Plant Microbe Interact 26:442–450
Acknowledgements
The material used in this study was based upon work supported by the U.S. Department of Agriculture, under Agreement No. 59-0206-9-066. This is a cooperative project with the U.S. Wheat & Barley Scab Initiative. Any opinions, findings, conclusions, or recommendations expressed in this publication are those of the author(s) and do not necessarily reflect the view of the U.S. Department of Agriculture. The authors thank J. Underdahl, M. Abdallah, A. Walz, B. Schatz, J. Hegstad, and A. Peckrul for technical assistance.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethical approval
The research was conducted with the ethical and professional standards of North Dakota State University.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
ElDoliefy, A.E.A., Kumar, A., Anderson, J.A. et al. Genetic dissection of Fusarium head blight resistance in spring wheat cv. ‘Glenn’. Euphytica 216, 71 (2020). https://doi.org/10.1007/s10681-020-02610-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-020-02610-0