Abstract
Key message
Rye genetic resources provide a valuable source of new alleles for the improvement of frost tolerance in rye breeding programs.
Abstract
Frost tolerance is a must-have trait for winter cereal production in northern and continental cropping areas. Genetic resources should harbor promising alleles for the improvement of frost tolerance of winter rye elite lines. For frost tolerance breeding, the identification of quantitative trait loci (QTL) and the choice of optimum genome-based selection methods are essential. We identified genomic regions involved in frost tolerance of winter rye by QTL mapping in a biparental population derived from a highly frost tolerant selection from the Canadian cultivar Puma and the European elite line Lo157. Lines per se and their testcrosses were phenotyped in a controlled freeze test and in multi-location field trials in Russia and Canada. Three QTL on chromosomes 4R, 5R, and 7R were consistently detected across environments. The QTL on 5R is congruent with the genomic region harboring the Frost resistance locus 2 (Fr–2) in Triticeae. The Puma allele at the Fr–R2 locus was found to significantly increase frost tolerance. A comparison of predictive ability obtained from the QTL-based model with different whole-genome prediction models revealed that besides a few large, also small QTL effects contribute to the genomic variance of frost tolerance in rye. Genomic prediction models assigning a high weight to the Fr–R2 locus allow increasing the selection intensity for frost tolerance by genome-based pre-selection of promising candidates.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Compared to related small grain cereals like wheat and barley, rye is more frost tolerant (Fowler and Limin 1987) and, therefore, constitutes an ideal model to investigate the genetic architecture of frost tolerance in cereals. Owing to its high degree of abiotic stress tolerance, rye is a valued crop in production areas where most small grain cereals are not profitable (Miedaner 2013). The high level of frost tolerance allows winter rye cultivation in northern and continental cropping areas of the temperate zones. As climate change proceeds, climatologists predict that cold winter extremes will occur more frequently in the northern hemisphere despite global warming (Petoukhov and Semenov 2010; Sorokina et al. 2016). In these high stress regions, winter rye production is only efficient if high yield is combined with a high level of frost tolerance.
The genetic basis underlying frost tolerance has been investigated in the Triticeae species barley, einkorn, and bread wheat. Two major determinants of frost tolerance have been identified on the Triticeae homoeologous group 5. Frost tolerance is mediated by the vernalization locus Vrn–1 and a quantitative trait locus (QTL) at this position explained up to 36.6% of the phenotypic variance for frost tolerance in a winter × spring cross in barley (Francia et al. 2004). The Frost resistance locus 2 (Fr–2) maps about 30 cM proximal to the Vrn–1 locus in wheat and harbors a cluster of transcription factor genes belonging to the C-repeat/dehydration-responsive element binding factor (Cbf) gene family (Båga et al. 2007; Pasquariello et al. 2014). In bread wheat and barley, a QTL at Fr–2 explained about 30–40% of the phenotypic variance for frost tolerance assessed in controlled freeze tests (Båga et al. 2007; Francia et al. 2004). The remaining variation is most likely determined by minor effect genes (Thomashow 1999; Zhao et al. 2013).
Gaining insight into the genetic architecture of frost tolerance in winter rye is vitally important for rye breeding programs. The frost tolerance level of high-yielding European elite winter rye varieties is not sufficient to supply the markets in Canadian and Russian cropping areas, which extend to climatically unfavorable geographic regions. Genetic resources adapted to these regions are expected to exhibit strong overwintering capabilities that could be used in breeding for frost tolerance in the Central European breeding programs. Identification of QTL for traits of interest enables marker-based selection for beneficial new alleles from the wider gene pool and aids in minimizing the linkage drag that often hampers the introgression of favorable alleles from genetic resources (Haussmann et al. 2004).
We carried out a QTL mapping study in a cross between a European winter rye inbred line and a frost tolerant selection from the Canadian cultivar (cv.) Puma (Shebeski et al. 1973), a genetic resource that is adapted to high frost stress environments. Lines per se and testcrosses of the mapping population were evaluated for frost tolerance. The correlation between line per se and testcross performance is an indication of the potential to accelerate development of frost tolerant hybrid rye varieties, for which demand is steadily growing in Europe (Geiger and Miedaner 2009). Due to the strong environmental effect on frost tolerance gene expression (Fowler and Limin 2004; Gray et al. 1997), reliable phenotyping in field trials requires extensive testing in multiple sites. Conditions for frost field tests can be unfavorable or technically demanding due to variable snow coverage, damage by snow mold or mild temperatures. Various types of freeze tests under controlled conditions have been developed to facilitate frost tolerance phenotyping and to increase the repeatability of results (Fowler et al. 1973; Pomeroy and Fowler 1973; Skinner and Mackey 2009). A close relationship between frost tolerance assessed in the field and in controlled freeze tests could reduce expensive field testing or replace it in case of low accuracy, thereby accelerating selection progress in breeding programs. Selection for frost tolerance can also be improved by informative molecular markers. The large effects of the QTL at the Vrn–1 and Fr–2 loci have been successfully exploited by marker-based selection for the improvement of frost tolerance in barley (Akar et al. 2009; Tóth et al. 2004).
In rye, genome analysis and detailed investigation of genomic regions involved in frost tolerance have been constrained by the large and highly repetitive genome (Doležel et al. 1998). The recent development of genome-wide molecular tools and the availability of sequence resources open new avenues for genomic research in rye (Bauer et al. 2017; Haseneyer et al. 2011). In this study, we identified genomic regions involved in frost tolerance in winter rye using phenotypic data assessed in field trials and in controlled freeze tests. The aims of our study were to (1) identify the number, location, and the effects of genomic regions involved in frost tolerance in a biparental winter rye population by QTL analysis, (2) determine the optimum selection strategy with regard to the phenotyping platform, the plant material, and the selection method given the genetic architecture of frost tolerance, and (3) assess the population structure of European breeding pools and genetic resources at the Fr–R2 and Vrn–R1 loci to evaluate their potential for frost tolerance breeding.
Methods
Plant material
Lo157 × Puma-SK mapping population
A biparental mapping population was developed from a cross between the European elite inbred line Lo157 and a Canadian genetic resource. In rye breeding, hybrids are generally produced by crossing a male-sterile mother from the so-called seed parent heterotic pool and a fertile father from the pollen parent pool (Geiger and Miedaner 2009). The self-fertile inbred line Lo157 belongs to the seed parent pool. The genetic resource was represented by one plant from a re-selected population of the frost tolerant Canadian open-pollinated cv. Puma (Shebeski et al. 1973). This re-selected fraction resulted from a recurrent selection program, exhibits enhanced frost tolerance and has been designated Puma–SK (Limin and Fowler, University of Saskatchewan, Canada; unpublished). For the mapping population, one F1 plant resulting from the Lo157 × Puma–SK cross was selfed to obtain 273 F2 individuals. These were advanced by single-seed descent up to the F4 generation. In higher selfing generations, some lines exhibited strong inbreeding depression leading to seed shortage. To provide sufficient seed for the F5 generation, three single plants per F4 line were randomly chosen and selfed to produce F5 lines. Testcross seed was obtained by crossing F3 single plants and F4 single plants, respectively, to a male-sterile single-cross tester of the seed parent pool (Lo115-P × Lo133-N).
Diversity panel
A diverse panel of 122 accessions composed of 38 and 46 elite inbred breeding lines from the seed and the pollen parent pool from a commercial rye breeding program and 38 genetic resources was used for molecular analyses. Lines from the seed parent pool were advanced by selfing to the F5 or F6 generation, while lines from the pollen parent pool were selfed only until the F3 generation and maintained as F3:4 bulks. The genetic resources were represented by single plants from Eastern European open-pollinated populations and from accessions from Germany, Canada, and USA (Table S1).
Quantitative genetic analysis of the Lo157 × Puma–SK mapping population
Phenotypic trait assessment
Frost tolerance of the Lo157 × Puma–SK population was assessed using two phenotyping platforms: a controlled freeze test and field trials. In the winter 2011/2012, F4 lines per se and testcrosses of their parental F3 single plants and in 2012/2013 and 2013/2014 F5 lines per se and testcrosses of their parental F4 single plants were phenotyped in controlled freeze tests and the field (Table S2).
In preparation for the freeze test, plants were vernalized for 7 weeks at 2–3 °C and 8 h light per day. At the three leave stage, plants were transferred to the freezer at 0 °C. Subsequently, the temperature was decreased to −9 °C in ~2 °C steps per day. At the fourth or fifth day, temperature was further decreased by 2 °C per hour to a minimum of −20 to −23 °C depending on the trial. This temperature was held for 1–2 h and then increased to −5 °C. In the following 2 days, the temperature was increased to 1 and 5 °C, respectively. Throughout the freezing cycle, plants were kept in the dark. Afterwards, plants were allowed to recover at 8–10 °C for 2 weeks until they were scored for the trait recovery after freezing (REC). A score of 1 corresponded to plants with fully necrotic leaves which did not recover from frost stress. A score of 9 corresponded to healthy and vital plants with fully green leaves which recovered completely. In 2012/2013, lines and testcrosses were evaluated at two minimum temperatures, −21 °C and −23 °C. In 2011/2012 and 2012/2013, the freeze test was carried out in two freezers. Since not all lines and testcrosses of the mapping population could be placed in the two freezers simultaneously, the freeze test in 2011/2012 and 2012/2013 was performed in several series. Series were connected by common entries (Lo157, experimental lines, and commercial checks). The freeze test in 2013/2014 was performed in a single climate chamber with plants arranged in a 14 × 15 alpha-lattice design. All freeze tests comprised two replications. In addition, six inbred lines from the seed (Lo7, Lo90, Lo115, Lo117, Lo176, and Lo191) and the pollen (Lo225, Lo282, Lo298, Lo310, Lo348, and Lo351) parent pool, respectively, and Puma–SK were assessed for REC in the freeze test in a single climate chamber with four replications. Each test unit in the freeze test contained five plants per line and testcross entry. A typical temperature profile in the controlled platform is shown in Figure S1.
Field trials were carried out at one Russian location, Lipezk (52°37′N, 39°36′E, 160 m. a. s. l.) and three Canadian locations, Minto (49°24′N, 100°01′W, 487 m. a. s. l.), Portage la Prairie (49°58′N, 98°17′W, 262 m. a. s. l.) and Saskatoon (52°8′N, 106°40′W, 481 m. a. s. l.). Daily minimum temperatures from trial sites in the winter seasons are shown in Figure S2. Lines per se and testcrosses were evaluated in alpha-lattice designs (13 × 13, 14 × 15, or 15 × 15) with two replications in each year, except for Lipezk in 2012/2013 where testcrosses were evaluated in three replications. One plot comprised 50–70 plants in Russia and 80–100 plants in Canada. Phenotypic data were collected on survival after winter (SAW) and development after winter (DAW) 2 weeks after snow melt in April or May. SAW was measured as the percentage of plants that survived the winter in each plot. DAW was assessed as a score with a range from 1 to 9 where 1 corresponds to a plot with severely damaged plants and 9 corresponds to a plot with completely healthy and vital plants as described for the scoring of REC.
Phenotypic data analysis
Lines per se and testcrosses were analyzed separately. All phenotypic analyses were performed using the ASReml-R package (Butler et al. 2009). In the controlled platform, the combination of year and temperature was defined as an individual environment. Accordingly, the tests in 2011/2012 and in 2013/2014 and both tests at different minimum temperatures in 2012/2013 were treated as individual environments. In the field trials, an environment is defined as a location–year combination. Phenotypic data analyses for controlled experiments and field trials were performed for individual environments and across environments. Distribution of residuals was inspected by means of residual diagnostic plots and observations were identified as outliers and removed from the data set when their standardized residuals exceeded the threefold standard deviation.
In the freeze test in 2013/2014 and in the field trials, lattice analyses were carried out for each individual environment. As the freeze tests in 2011/2012 and 2012/2013 were carried out in two freezers and several series during the winter, freezer and series were included as factors in the model. Consequently, models were adapted to the respective experimental design in individual environments in the controlled platform and analysis across environments was performed in a two-stage approach. In the first stage, adjusted means for individual environments were obtained for each genotype by assuming genotype as fixed effect. In the second stage, adjusted means were calculated across environments based on adjusted means obtained from the first stage including genotype as fixed effect and environment and genotype × environment as random effects. Adjusted means from the first stage were weighted according to method 1 of Möhring and Piepho (2009).
The combined analysis in the field trials was performed across all environments that exhibited significant genotypic variance and a repeatability >0.10, based on the following model:
where y ijkm is the trait observation, µ is the overall mean, g i and l j are the effects of genotype i and environment j, respectively, gl ij is the interaction effect of genotype i with environment j, r jk is the effect of replication k nested in environment j, b jkm is the effect of incomplete block m nested in replication k nested in environment j, and e ijkm is the residual error. For the estimation of variance components, all effects were assumed random. Adjusted means across environments were obtained by fitting genotype as fixed effect. Broad-sense heritabilities were estimated according to Holland et al. (2003).
Phenotypic correlations between lines per se and testcrosses and between phenotyping platforms were calculated based on adjusted entry means across environments using Pearson`s correlation coefficient. Genotypic correlations between lines per se and testcrosses were obtained from a bivariate model using adjusted entry means from common individual environments and fitting genotype and environment as random terms.
Genotypic data and genetic linkage mapping
DNA was extracted from 263 F3 and 260 F4 plants from the Lo157 × Puma–SK population and analyzed with a subset of 384 SNPs from the Illumina iSelect Rye5k SNP array (Haseneyer et al. 2011). SNPs with more than 20% missing values were excluded from the analyses. After quality checks, 211 F3 and F4 lines and 180 SNPs were available for further analyses. Genetic analysis of polymorphisms in four candidate genes from the frost-responsive network (ScCbf9, ScCbf12, Vrn–R1, and ScMybs3) was carried out by cleaved amplified polymorphic site (CAPS) marker assays or by sequencing. Primers for ScCbf9, ScCbf12, and Vrn–R1 were adopted from Li et al. (2011b). Primers for ScMybs3 were designed based on information from the homoeologous gene in rice (Lu et al. 2002). In addition, two putatively novel ScCbf genes were discovered by blasting the first signal motif contained in Cbf genes described by Skinner et al. (2005) against the rye whole-genome sequence resource of the reference inbred line Lo7 (Bauer et al. 2017) using the ViroBLAST Web server (Deng et al. 2007). According to their closest homologs in Triticum aestivum and Triticum monococcum to which they were identical by 95 and 93%, these Cbf genes were designated as ScCbf1 and ScCbf18. Their sequences were submitted to GenBank under accession numbers KY780081 and KY780082. Based on sequence contigs of Lo7 and 11 other sequenced rye lines (Bauer et al. 2017), SNPs were identified for genotyping. PCR was carried out in 20 µl reaction volumes containing 30 ng DNA, 150 nM of each primer, 0.2 nM of each dNTP, 1× Paq DNA polymerase reaction buffer, and 1.0 U Paq DNA Polymerase (Stratagene, Europe). The primer sequences and details on PCR conditions are listed in Table S3. Genetic maps were established with the software JoinMap 4.1 (Van Ooijen 2006) using the maximum-likelihood algorithm and Haldane’s mapping function (Haldane 1919). One SNP in each of the five candidate genes was used to integrate ScCbf9, ScCbf12, ScCbf18, Vrn–R1, and ScMybs3 in the genetic linkage map of the F4 generation.
From the F5 generation, 200 plants were analyzed with a 16k custom Illumina Infinium SNP array (Illumina Inc., San Diego, CA, USA). Monomorphic SNPs and SNPs with more than 10% missing or 10% heterozygous genotype calls were excluded from the analyses. After quality checks and SNP filtering, 192 F5 lines and 2950 SNPs were available for further analyses. For mapping of SNPs in seven candidate genes, ScCbf1, ScCbf9, ScCbf11, ScCbf12, ScCbf18, ScDhn3, and ScMybs3, KASP (Kompetitive Allele Specific PCR) marker assays were applied (LGC Genomics, Hoddesdon, UK). The F5 linkage map contained 2346 SNPs including nine SNPs in the seven candidate genes. Cosegregating markers were removed from the F5 map resulting in 1050 markers for further analyses.
QTL mapping
The QTL analysis was carried out based on the F4 linkage map. Marker data from F3 plants were associated with adjusted means of F4 lines and F3 testcrosses in individual environments and with adjusted means from combined analyses. Marker data from F4 plants were associated with adjusted means of F5 lines and F4 testcrosses in individual environments. A summary of the data sets is given in Table S2. QTL analyses were performed by composite interval mapping, including only additive effects or both additive and dominance effects. The LOD threshold for each data set was determined with a permutation test based on 1000 reshuffles according to Churchill and Doerge (1994). A LOD threshold corresponding to a genome-wise p value of 0.30 was chosen to declare a putative QTL as significant (Schön et al. 2010). The additive effects at the QTL and the proportion of phenotypic variance explained by individual QTL (partial R 2) were estimated by fitting all QTL simultaneously in a multiple regression model. The QTL support interval was determined as the chromosomal region surrounding a QTL peak plus/minus a LOD fall-off of 1.0. QTL detected in different environments were declared as congruent when their support intervals overlapped and additive effects were of the same sign. All analyses were performed with the software PlabMQTL version 0.9 (Utz 2011). QTL detected in the combined analysis were denominated following the recommendations of the Catalogue of Wheat Gene Symbols (McIntosh et al. 2013).
Evaluation of prediction models for frost tolerance
Based on the data set of the F5 generation comprising 1050 markers, we compared the predictive ability of a QTL-based model with genome-wide prediction approaches. Adjusted means from the combined analysis across environments were used as input data for all models. QTL mapping was carried out as described above using PlabMQTL version 0.9 (Utz 2011). Genomic best linear unbiased prediction (GBLUP) was performed using the R package synbreed (Wimmer et al. 2012). Marker genotypes were coded according to the number of minor alleles with 0 and 2 for the homozygous and 1 for the heterozygous genotypes. Coding of marker genotypes and imputation of missing marker data by flanking markers in the linkage map were carried out using the software package BEAGLE 3.3 (Browning and Browning 2009). For GBLUP models, a genetic relationship matrix was constructed according to Habier et al. (2007). In addition to the standard GBLUP model, a GBLUP model including a SNP of the mapped candidate gene ScCbf12 from the Fr–R2 locus on 5R as fixed effect (denoted as GBLUP+Fr–R2) was calculated. In addition, the variable selection method LASSO (Tibshirani 1996) was applied using the R package glmnet (Friedman et al. 2010). Predictive ability of the QTL-based and the three GP models was assessed in a fivefold cross-validation (CV) with 20 replications. The predictive ability of the QTL model was calculated from the CV as the square root of the adjusted phenotypic variance (R 2adj. ) explained in the test set using PlabMQTL version 0.9 (Utz 2011). The predictive ability of GP models was estimated by the Pearson’s correlation coefficient of observed versus predicted phenotypes for each test set. The mean predictive ability and its standard deviation were calculated based on 100 CV runs for all models. The same partitioning of individuals into estimation and test set was applied in QTL and GP models.
Evaluation of population structure
Population structure of the diverse panel of 122 accessions was investigated by principle coordinate analyses (PCoA) using genetically mapped SNPs from the Rye600k array (Bauer et al. 2017). PCoA was performed for the whole genome (66,561 SNPs), for Fr–R2 (179 SNPs) and Vrn–R1 (212 SNPs), and for both loci together (391 SNPs) based on modified Rogers’ distances between accessions using the R package ape (Paradis et al. 2004). To avoid bias arising from the higher degree of heterozygosity in genetic resources compared to the inbred lines from the seed and pollen parent pools, pseudo-S0 genotypes were used instead of original inbred line genotypes. Pseudo-S0 genotypes were obtained separately for each pool by combining pairs of haplotypes from two randomly sampled individuals of the same pool as described in Meyer et al. (2016). Haplotype phasing was performed with BEAGLE 4.0 (Browning and Browning 2007).
Results
Phenotypic variation for frost tolerance
The level of frost tolerance of six lines from each of both elite breeding pools and the highly frost tolerant Puma–SK was assessed in a freeze test. Adjusted means for recovery after freezing (REC) ranged from 1.29 to 8.64 (Fig. 1). Elite lines from the pollen parent pool exceeded the frost tolerance level of elite lines from the seed parent pool significantly (p < 0.05). Significant differences (p < 0.05) for REC were also observed between inbred lines within each of the pools. The highest REC score was obtained for Puma–SK with about two times the maximum observed score of the lines in the European breeding pools.
Quantitative genetic analysis of frost tolerance in the Lo157 × Puma–SK mapping population
Analysis of frost response
F4 and F5 generations of the biparental mapping population Lo157 × Puma–SK were phenotyped for frost tolerance during 3 years in two phenotyping platforms, a controlled freeze test and field trials in Russia and Canada (Table S2). In the freeze test, the REC progeny mean of lines per se was not significantly different from the parental mean (Table 1). Testcross performance for REC significantly (p < 0.05) exceeded line per se performance in combined analyses across environments (Table 1) and in individual environments (Table S4). The only exception was in the winter of 2011/2012 where testcrosses were tested at 2 °C lower temperatures than lines per se. The genotypic variance component for REC was highly significant (p < 0.01) for lines per se and testcrosses in the combined analysis across environments and in single environments. The heritability of REC of the lines per se and testcrosses was 0.79 and 0.87, respectively. Phenotypic and genotypic correlations between line per se and testcross performance for REC were strong and highly significant (p < 0.01) with 0.86 and 0.98, respectively.
In the field trials, long frost periods occurred in all environments with minimum air temperatures of −10 to −38 °C (Figure S2). Testcross performance for DAW and SAW significantly (p < 0.05) exceeded line per se performance in combined analyses across environments (Table 1) and in single environments, except in Lipezk in 2011/2012 (Table S4). In the combined analysis across environments, the genotypic variance components for DAW and SAW were significant (p < 0.05) except for DAW in the testcrosses (Table 1). The variance components for genotype × environment interaction greatly exceeded the magnitude of the genotypic variance components for DAW and SAW in lines per se and testcrosses. Heritability estimates for DAW and SAW were 0.38 and 0.49 for lines per se and 0.15 and 0.26 for testcrosses, respectively (Table 1). The phenotypic correlation between line per se and testcross performance was significant (p < 0.01) for DAW (r p = 0.26) but not for SAW (r p = 0.03); genotypic correlations were not significant. Phenotypic correlations between the traits DAW and SAW assessed in the field and REC assessed in the freeze tests were highly significant (p < 0.01) and ranged between 0.31 and 0.50 for lines per se and testcrosses (Fig. 2).
Phenotypic correlations between freeze test and field in the Lo157 × Puma–SK population based on adjusted means across environments: a Recovery after winter (REC) and development after winter (DAW) for lines per se b REC and survival after winter (SAW) for lines per se c REC and DAW for testcrosses and d REC and SAW for testcrosses. Genotypes are colored according to their allele at the Fr–R2 locus: Lo157 allele, red squares ; Puma–SK allele, orange circles; heterozygous, brown triangles
Genetic linkage maps and mapping of candidate genes
Linkage maps for QTL mapping were constructed from the F4 generation and contained 158 SNPs including five SNPs in candidate genes from the frost-responsive network (ScCbf9, ScCbf12, ScCbf18, ScMybs3, and Vrn–R1). The two candidate genes ScCbf9 and ScCbf12 were closely linked and mapped on 5RL (Table S5; Fig. 3). As a cluster of Cbf genes, including Cbf9 and Cbf12, was identified as candidate genes for the Fr–2 locus in wheat and barley (Båga et al. 2007; Pasquariello et al. 2014), the genomic region including ScCbf9 and ScCbf12 is referred to as the Fr–R2 locus in the following. The vernalization gene Vrn–R1 (originally designated Sp1 in rye; Plaschke et al. 1993) was mapped 16 cM distal to both Cbf genes. ScCbf18 and ScMybs3 were located on chromosomes 6R and 1R, respectively. The seven chromosomes were represented by 8 (7R) to 30 (1R) markers (Table S6). The total map length was 1171 cM with an average distance between loci of 7.9 cM (cosegregating markers treated as one locus). Significantly distorted segregation (p < 0.01) in favor of the elite line Lo157 was observed at 30% of the markers. Distorted segregation is frequently observed in rye and may be even more severe in crosses involving self-incompatible genetic material such as the open-pollinated cv. Puma (Erath et al. 2016; Hackauf et al. 2009; Korzun et al. 1998).
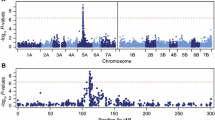
QTL analysis in the Lo157 × Puma–SK population. LOD score profiles are shown from the combined analysis across environments along the linkage maps of the seven chromosomes (1R–7R). Recovery after freezing (REC), development after winter (DAW) and survival after winter (SAW) are indicated in dark blue for lines per se and in light red for testcrosses. Horizontal dashed lines represent the LOD score threshold assessed by permutation in individual data sets. Vertical dashed lines indicate the position of mapped candidate genes
QTL analysis
In the combined analysis across environments, QTL for frost tolerance as measured by the traits REC, DAW, and SAW were detected on 4R (QRec.tum–4R), 5R (QRec.tum–5R, QDaw.tum–5R, QSaw.tum–5R), and 7R (QDaw.tum–7R, QSaw.tum–7R) (Table 2). A high proportion of phenotypic variance was explained by the QTL on 5R for all traits. The highest R 2 values were found for QRec.tum–5R in lines per se (66.7%) and in testcrosses (65.3%). These QTL were consistently detected in single environments in both phenotyping platforms (Table S7). The peak of QRec.tum–5R coincided with the map position of the two genes ScCbf9 and ScCbf12 and accordingly with the Fr–R2 locus (Fig. 3). In the lines per se, the peaks of QDaw.tum–5R and QSaw.tum–5R were slightly shifted distal from the Fr–R2 towards the vernalization locus Vrn–R1. These shifted QTL explained a smaller proportion of the phenotypic variance (8.5 and 9.6%, respectively) than QRec.tum–5R. At the QTL on 5R (QRec.tum–5R, QDaw.tum–5R, QSaw.tum–5R), Puma–SK contributed the allele that increased frost tolerance. QRec.tum–4R, QSaw.tum-5R, QDaw.tum–7R, and QSaw.tum–7R were detected in lines per se, but not in testcrosses. QRec.tum–4R explained 8.5% of the phenotypic variance. QDaw.tum–7R and QSaw.tum–7R explained 11.8 and 14.2% of the phenotypic variance, respectively. At these three QTL, the frost tolerance allele was contributed by Lo157. QRec.tum–4R and QDaw.tum–4R were detected in one and two individual environments, respectively. QDaw.tum–7R and QSaw.tum–7R were detected in one and three individual environments, respectively. Additional QTL were found in single environments on all chromosomes, except on 6R (Table S7). They explained 6.5–12.1% of the phenotypic variance. All QTL results reported here were obtained from a model assuming additive effects, since dominance effects were not significant in the full model.
Evaluation of prediction models for frost tolerance
We compared the predictive ability of a QTL-based model and whole-genome prediction approaches using a dense marker data set from the F5 generation with 1050 SNPs (Table S8). Compared to the results from the F4 generation, QRec.tum–4R, QRec.tum-5R, QDaw.tum–5R, and QSaw.tum–5R were detected again, but not QDaw.tum–7R and QSaw.tum–7R. Among the three traits, the highest predictive abilities for the QTL-based and for the GP models were obtained for REC, ranging from 0.84 to 0.87 in the lines per se and similar values in the testcrosses (Table 3). Lower predictability was observed for DAW and SAW assessed in the field trials, ranging from 0.10 to 0.35 in lines per se and 0.05 to 0.26 in testcrosses, respectively. Predictive ability was not assessed for DAW in testcrosses as their genotypic variance component was not significant and heritability was very low. The comparison of different models revealed that the GP models GBLUP, GBLUP+Fr–R2, and LASSO mostly outperformed the QTL-based model. Differences between GP models were smallest for REC, but larger for the field platform. With the exception of SAW in the testcross data set, predictive abilities of LASSO and GBLUP+Fr–R2 slightly outperformed the standard GBLUP. Including the mapped SNP in ScCbf9 instead of the SNP in ScCbf12 as fixed effect in the GBLUP+Fr–R2 model resulted in comparable predictive ability. Including epistatic effects did not improve the predictive ability of the GBLUP model (results not shown).
Population structure in elite breeding pools and genetic resources
The large effect of the Puma–SK allele at the Fr-R2 locus and the significantly superior frost tolerance of Puma–SK compared to elite breeding lines suggest that valuable alleles in genetic resources could be exploited for improvement of frost tolerance in the Central European breeding pools. On a whole-genome level, the three groups of seed and pollen parent pool and genetic resources formed separate clusters in PCoA with a clear separation of the elite breeding pools and an intermediate position of the genetic resources (Figure S3a). There was no clear differentiation among the three groups when only SNPs from the Fr–R2 locus, the Vrn–R1 locus, or both loci were considered (Figure S3b–d). For the Fr–R2 and/or the Vrn–R1 locus, the first two coordinates explained up to 29.3 and 14.4% of the genotypic variation, respectively.
Discussion
Frost tolerance is an important trait in geographic regions which are affected by severe and long periods at sub-zero winter temperatures. In these regions, the cultivation of winter rye is often preferred over other cereals due to its high level of frost tolerance. This study lays the foundation for the development of genome-based breeding strategies by investigating the genetic architecture of frost tolerance in rye.
Genomic regions controlling frost tolerance in winter rye
In the Lo157 × Puma–SK population, consistent QTL were detected on chromosomes 4R, 5R and 7R. A QTL on 5R coinciding with the Fr–R2 locus explained the largest proportion of phenotypic variance for REC and DAW. It was detected in extreme frost stress environments and in the controlled freeze test. The one common feature of those winter environments and the freeze test was a rather fast drop of temperatures until freezing (Figures S1, S2, S4). Both the variation in the threshold induction temperature at which a plant starts to cold acclimate and the initial rate of cold acclimation have been associated with the Fr–2 locus in cereals (Båga et al. 2007; Campoli et al. 2009; Fowler 2008; Knox et al. 2008). In preparation for the freeze tests, plants were allowed to acclimate for 7 weeks, representing optimal conditions for the acquisition of a high level of frost tolerance. Thus, the large variation explained by QRec.tum–5R (Table 2) likely represents variation in the initial rate of cold acclimation rather than variation in threshold induction temperatures. By contrast, in the field, it was not clear to what extent variation in threshold induction temperatures or in the rate of cold acclimation led to the detection of QDaw.tum–5R and QSaw.tum–5R. Interestingly, the QTL peaks of QDaw.tum–5R and QSaw.tum–5R in lines per se in the combined analysis were slightly shifted towards Vrn–R1. Highly significant marker effects on frost tolerance were also detected on chromosomes 5A and 5B in winter wheat, which could not be clearly assigned to the Vrn–1 nor the Fr–2 locus (Case et al. 2014; Snape et al. 2001; Zhao et al. 2013). It cannot be concluded whether the peaks of QDaw.tum–5R and QSaw.tum–5R were shifted distal to the Fr-R2 locus due to joint effects on frost tolerance of the Fr–R2 locus and Vrn–R1 or due to the lower heritabilities in the field trials resulting in lower mapping precision. In wheat and barley, copy number variation (CNV) at both the Fr–R2 and Vrn–R1 locus was associated with enhanced frost tolerance (Knox et al. 2010; Zhu et al. 2014) and CNV of Cbf genes explained up to 24.3% of genotypic variance for frost tolerance in winter wheat (Würschum et al. 2017). Preliminary read depth analyses using the sequence resources in Bauer et al. (2017) indicated CNV of Cbf genes in the Fr–R2 locus in rye. Additional research is ongoing to experimentally validate these results.
In addition to the level of cold acclimation conferred by the Fr–2 locus, phenological processes like the transition from the vegetative to the reproductive stage are involved in the regulation of frost tolerance in cereals. This transition is controlled amongst others by vernalization genes like Vrn–1, Vrn–2, and Vrn–3 (Yan et al. 2003; 2004; 2006). The chromosomal locations of QRec.tum–4R, QDaw.tum–7R, and QSaw.tum–7R are not syntenic to genomic regions harboring Vrn–2 and Vrn–3 in other Triticeae (Dubcovsky et al. 1998; Yan et al. 2006); however, a rye homolog of the early heading date 3 (ehd3) gene identified in rice (Matsubara et al. 2011) might be a potential candidate for QDaw.tum–7R and QSaw.tum–7R. In contrast to QDaw.tum–5R and QSaw.tum–5R, QDaw.tum–7R and QSaw.tum–7R were only detected in environments with a slow decrease of temperatures in autumn resulting in a longer adaptation phase and with milder frost periods (Figure S4). The Lo157 allele conferring frost tolerance at QDaw.tum–7R and QSaw.tum–7R was obviously sufficient, and the Puma–SK allele at QDaw.tum–5R and QSaw.tum–5R was not essential for the survival and development after winter in these lower stress environments. The fact that the tolerance to frost is influenced by additional factors like drought or light stress (Gray et al. 1997; Thomashow 1999) may explain the observed dependence of QTL effects on environmental conditions in this study. The effects of QTL for abiotic stress are known to depend on specific environmental conditions as it was shown in maize for different drought and heat scenarios (Millet et al. 2016). Analyzing QTL effects for tolerance to abiotic stresses like frost in conjunction with other environmental conditions would allow a more detailed understanding of the underlying mechanisms of stress tolerance and enable the identification of candidate genes decisive for adaptation to specific target environments.
Frost tolerance in lines per se and testcrosses
We assessed three different traits as indicators for frost tolerance. Compared to SAW, which measures the survival rate, DAW is a visual score that integrates both the survival rate as well as the development after winter. As this trait likely captures small effects which are involved in cold adaptation, in the fine-tuning of response to frost and in the recovery after frost periods, it might be the more informative trait for selection purposes. On the other hand, SAW can be counted and has a higher heritability compared to DAW. The correlation with REC in the controlled platform was higher for DAW than for SAW, since REC, similar to DAW, integrates survival and development after frost stress. Moderate correlations between the two phenotyping platforms showed that freeze tests cannot fully substitute laborious phenotyping in field trials. In an artificial freeze test, plants are exposed to freezing temperatures for relatively short periods and experience optimal cold acclimation conditions which rarely occur in the field (Gusta et al. 1997). Since this can result in an overestimation of the frost tolerance level (Gusta et al. 2001), phenotyping in field trials remains indispensable.
All traits in the field and freeze test were assessed on lines per se and on testcrosses. No significant genotypic correlations were observed in the field platform for DAW and SAW. Low genotypic correlations between line per se and testcross performance can occur due to significant non-additive variance (Mihaljevic et al. 2005; Schwegler et al. 2014). In most data sets, testcrosses had significantly higher frost tolerance values than lines per se. Significantly higher REC scores were also observed in the F4 generation compared to the F5 generation, whereas differences between the two minimum temperatures within the F5 generation were not significant. In addition, the six lines from the pollen parent pool exhibited significantly higher REC values than the six lines from the seed parent pool. These findings may be explained by different levels of heterozygosity in the respective lines and suggest a role of dominant gene action in the expression of frost tolerance. Epistatic interactions have also been frequently reported to influence frost tolerance (Galiba et al. 2009; Li et al. 2011a; Wooten et al. 2008), but this may depend on the material under study. In this study, the power for detecting dominance or epistatic effects at QTL was low due to advanced selfing and sample size. The role of non-additive gene action for traits related to frost tolerance warrants further research to allow full exploitation of dominance and heterosis in a population or hybrid breeding scheme. On the other hand, the high genetic correlation between lines per se and testcrosses in the freeze test indicates that REC is predominantly influenced by additive effects. Together with the high heritability in the freeze test, this suggests that effective pre-selection using marker-assisted selection (MAS) based on QRec.tum–5R at the Fr–R2 locus could be performed on lines per se. Testcrosses could then be developed from pre-selected lines and evaluated in field trials.
Selection strategies for frost tolerance in rye
We evaluated the predictive ability of a QTL-based model and different GP models using CV for three frost tolerance traits assessed in the field and a freeze test. For REC, GP models only marginally outperformed the QTL-based model. Together with the high variance explained in QTL analyses, this suggests that REC is strongly affected by the QTL at the Fr–R2 (QRec.tum–5R) locus as opposed to traits assessed in the field. Nevertheless, GP models had higher predictive ability than the QTL-based model for all traits, indicating that frost tolerance in cereals also involves genes with small effects that are not detected in QTL analyses. The superiority of the variable selection method LASSO over the QTL-based model and the standard GBLUP model in most data sets underlines heterogeneous contributions of SNPs to the estimated genomic variance for frost tolerance (Wimmer et al. 2013). Large and small marker effects were also captured through GBLUP+Fr–R2 by fitting the QTL at the Fr–R2 locus as a fixed effect, which yielded predictive abilities similar to LASSO. Similarly, integrating markers from the Fr–A2 locus in wheat as a fixed effect into a GP model improved predictive ability for frost tolerance (Würschum et al. 2017). Using additional flanking markers of QRec.tum–4R, QDaw.tum–7R or QSaw.tum–7R as fixed effects in the GBLUP model increased the predictive ability compared to standard GBLUP only slightly and not consistently across traits (results not shown).
This study showed that pre-selection for frost tolerance can be performed by MAS using markers from Cbf genes in the Fr–R2 locus like ScCbf9 or ScCbf12. The SNP in ScCbf12 displayed a highly significant effect on frost tolerance in the controlled platform and in field trials in a previous candidate-gene-based association study in rye (Li et al. 2011a) and, therefore, represents a reliable marker for selection on frost tolerance. Refined selection and selection in testcrosses should be performed in field trials for DAW and SAW. The use of genomic prediction models like GBLUP+Fr–R2 or LASSO can increase selection intensity and accelerate the breeding progress when expensive field trials in frost stress environments are performed with pre-selected candidates.
Genetic resources for the improvement of frost tolerance
In the past, genetic resources were only occasionally used for rye breeding programs due to difficulties associated with self-incompatibility, low agronomic performance, and low inbreeding tolerance (Geiger and Miedaner 2009). In the Lo157 × Puma–SK population, significantly distorted segregation was observed in multiple genomic regions of the F4 and F5 linkage maps in favor of the elite parent Lo157. This suggests that recessive deleterious alleles resulted in unintended selection of genotypes carrying the Lo157 allele in specific genomic regions during the development of the mapping population. Such conditions may impede QTL detection in higher selfing generations. For instance, the genomic region harboring the QTL on 7R in the F4 linkage map was not covered with markers in the F5 linkage map which prevented the detection of this QTL in the F5 generation. Despite the challenges associated with their use in breeding, genetic resources are appreciated as a rich source of valuable alleles or haplotypes that can improve traits of interest in elite material (McCouch et al. 2013). Marker-based approaches for the targeted introgression of favorable alleles can help to overcome difficulties like linkage drag if flanking markers closely linked to the QTL of interest are available (Collard and Mackill 2008).
The freeze test on lines from the European seed and pollen parent pool and Puma–SK and the large effect of the Puma–SK allele at the Fr-R2 locus in the Lo157 × Puma–SK population revealed the superiority of this genetic resource for frost tolerance and demonstrated a high potential for the improvement of frost tolerance in European breeding pools by genetic resources. The frost resistance loci Fr–R2 and Vrn–R1 as major determinants of frost tolerance are supposed to have a large impact on the observed variation for REC. Whereas the seed and pollen parent pools and genetic resources were clearly separated by PCoA on a whole-genome level, no differentiation between the groups was observed when only the Fr–R2 and/or the Vrn–R1 region were considered. This suggests that no strong differential selection has occurred in these genomic regions in winter rye. Based on these findings and the exceptionally high frost tolerance of Puma–SK, we hypothesize that frost tolerance in European elite lines can be improved beyond the existing level in the seed and pollen parent breeding pools through genomics-assisted breeding approaches.
Conclusions
Winter rye is the most frost tolerant small grain cereal and is well suited for production areas where severe winters occur. In a biparental cross between the European inbred line Lo157 and Puma–SK, three main QTL for frost tolerance were detected that exhibited stable effects. The largest effect was explained by a QTL at the Fr–R2 locus at which the Puma–SK allele increased frost tolerance significantly, demonstrating the potential of genetic resources to improve frost tolerance in elite material. Pre-selection of breeding lines can be performed by MAS based on markers from the Fr–R2 locus. Further selection steps on DAW and SAW should be performed in field trials. Using genomic prediction models like GBLUP+Fr–R2 or LASSO can help to increase selection intensity and the efficiency of phenotyping in remote locations.
Author contribution statement
CCS, EB, PW, and VK conceived the study; MP, AG, MS, BS, BF, and WE conducted experiments; EB, BF, PW, VK, AG, BS, and MS provided materials; WE analyzed the data; CCS and EB supervised the research; WE, EB, and CCS drafted the manuscript; all authors read, edited, and approved the manuscript.
References
Akar T, Francia E, Tondelli A, Rizza F, Stanca AM, Pecchioni N (2009) Marker-assisted characterization of frost tolerance in barley (Hordeum vulgare L.). Plant Breed 128:381–386
Båga M, Chodaparambil SV, Limin AE, Pecar M, Fowler DB, Chibbar RN (2007) Identification of quantitative trait loci and associated candidate genes for low-temperature tolerance in cold-hardy winter wheat. Funct Integr Genom 7:53–68
Bauer E, Schmutzer T, Barilar I, Mascher M, Gundlach H, Martis MM, Twardziok SO, Hackauf B, Gordillo A, Wilde P, Schmidt M, Korzun V, Mayer KFX, Schmid K, Schön C-C, Scholz U (2017) Towards a whole-genome sequence for rye (Secale cereale L.). Plant J 89:853–869
Browning SR, Browning BL (2007) Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet 81:1084–1097
Browning BL, Browning SR (2009) A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet 84:210–223
Butler D, Cullis B, Gilmour A, Gogel B (2009) ASReml-R reference manual. Queensland Department of Primary Industries and Fisheries, Toowoomba
Campoli C, Matus-Cadiz MA, Pozniak CJ, Cattivelli L, Fowler DB (2009) Comparative expression of Cbf genes in the Triticeae under different acclimation induction temperatures. Mol Genet Genom 282:141–152
Case AJ, Skinner DZ, Garland-Campbell KA, Carter AH (2014) Freezing tolerance-associated quantitative trait loci in the Brundage × Coda wheat recombinant inbred line population. Crop Sci 54:982–992
Churchill G, Doerge R (1994) Empirical threshold values for quantitative trait mapping. Genetics 138:963–971
Collard BCY, Mackill DJ (2008) Marker-assisted selection: an approach for precision plant breeding in the twenty-first century. Philos Trans R Soc B 363:557–572
Deng W, Nickle DC, Learn GH, Maust B, Mullins JI (2007) ViroBLAST: a stand-alone BLAST web server for flexible queries of multiple databases and user’s datasets. Bioinformatics 23:2334–2336
Doležel J, Greilhuber J, Lucretti S, Meister A, Lysák MA, Nardi L, Obermayer R (1998) Plant genome size estimation by flow cytometry: inter-laboratory comparison. Ann Bot 82:17–26
Dubcovsky J, Lijavetzky D, Appendino L, Tranquilli G (1998) Comparative RFLP mapping of Triticum monococcum genes controlling vernalization requirement. Theor Appl Genet 97:968–975
Erath W, Bauer E, Kastirr U, Schmidt M, Korzun V, Schmiedchen B, Wilde P, Schön C-C (2016) Oligogenic control of resistance to soil-borne viruses SBCMV and WSSMV in rye (Secale cereale L.). Plant Breed 135:552–559
Fowler DB (2008) Cold acclimation threshold induction temperatures in cereals. Crop Sci 48:1147–1154
Fowler DB, Limin AE (1987) Exploitable genetic-variability for cold tolerance in commercially grown cereals. Can J Plant Sci 67:278
Fowler DB, Limin AE (2004) Interactions among factors regulating phenological development and acclimation rate determine low-temperature tolerance in wheat. Ann Bot 94:717–724
Fowler DB, Siminovitch D, Pomeroy MK (1973) Evaluation of an artificial test for frost hardiness in wheat. Can J Plant Sci 53:53–59
Francia E, Rizza F, Cattivelli L, Stanca AM, Galiba G, Tóth B, Hayes PM, Skinner JS, Pecchioni N (2004) Two loci on chromosome 5H determine low-temperature tolerance in a ‘Nure’ (winter) × ‘Tremois’ (spring) barley map. Theor Appl Genet 108:670–680
Friedman J, Hastie T, Tibshirani R (2010) Regularization paths for generalized linear models via coordinate descent. J Stat Softw 33:1–22
Galiba G, Vágújfalvi A, Li C, Soltész A, Dubcovsky J (2009) Regulatory genes involved in the determination of frost tolerance in temperate cereals. Plant Sci 176:12–19
Geiger H, Miedaner T (2009) Rye breeding. In: Carena MJ (ed) Cereals. Springer, US, pp 157–181
Gray GR, Chauvin LP, Sarhan F, Huner N (1997) Cold acclimation and freezing tolerance (A complex interaction of light and temperature). Plant Physiol 114:467–474
Gusta LV, O’Connor BJ, MacHutcheon MG (1997) The selection of superior winter-hardy genotypes using a prolonged freeze test. Can J Plant Sci 77:15–21
Gusta LV, O’Connor BJ, Gao YP, Jana S (2001) A re-evaluation of controlled freeze-tests and controlled environment hardening conditions to estimate the winter survival potential of hardy winter wheats. Can J Plant Sci 81:241–246
Habier D, Fernando RL, Dekkers JCM (2007) The impact of genetic relationship information on genome-assisted breeding values. Genetics 177:2389–2397
Hackauf B, Rudd S, Van der Voort J, Miedaner T, Wehling P (2009) Comparative mapping of DNA sequences in rye (Secale cereale L.) in relation to the rice genome. Theor Appl Genet 118:371–384
Haldane J (1919) The combination of linkage values and the calculation of distances between the loci of linked factors. J Genet 8:299–309
Haseneyer G, Schmutzer T, Seidel M, Zhou RN, Mascher M, Schön C-C, Taudien S, Scholz U, Stein N, Mayer KFX, Bauer E (2011) From RNA-seq to large-scale genotyping-genomics resources for rye (Secale cereale L.). BMC Plant Biol 11:131
Haussmann B, Parzies H, Presterl T, Susic Z, Miedaner T (2004) Plant genetic resources in crop improvement. Plant Genet Resour 2:3–21
Holland J, Nyquist W, Cervantes-Martínez C (2003) Estimating and interpreting heritability for plant breeding: an update. Plant Breed Rev 22:9–112
Knox AK, Li CX, Vágújfalvi A, Galilba G, Stockinger EJ, Dubcovsky J (2008) Identification of candidate Cbf genes for the frost tolerance locus Fr-A m 2 in Triticum monococcum. Plant Mol Biol 67:257–270
Knox AK, Dhillon T, Cheng HM, Tondelli A, Pecchioni N, Stockinger EJ (2010) Cbf gene copy number variation at Frost Resistance-2 is associated with levels of freezing tolerance in temperate-climate cereals. Theor Appl Genet 121:21–35
Korzun V, Malyshev S, Kartel N, Westermann T, Weber WE, Börner A (1998) A genetic linkage map of rye (Secale cereale L.). Theor Appl Genet 96:203–208
Li Y, Böck A, Haseneyer G, Korzun V, Wilde P, Schön C-C, Ankerst D, Bauer E (2011a) Association analysis of frost tolerance in rye using candidate genes and phenotypic data from controlled, semi-controlled, and field phenotyping platforms. BMC Plant Biol 11:146
Li Y, Haseneyer G, Schön C-C, Ankerst D, Korzun V, Wilde P, Bauer E (2011b) High levels of nucleotide diversity and fast decline of linkage disequilibrium in rye (Secale cereale L.) genes involved in frost response. BMC Plant Biol 11:6
Lu C-A, Ho TD, Ho S-L, Yu S-M (2002) Three novel MYB proteins with one DNA binding repeat mediate sugar and hormone regulation of α-amylase gene expression. Plant Cell 14:1963–1980
Matsubara K, Yamanouchi U, Nonoue Y, Sugimoto K, Wang Z-X, Minobe Y, Yano M (2011) Ehd3, encoding a plant homeodomain finger-containing protein, is a critical promoter of rice flowering. Plant J 66:603–612
McCouch S, Baute GJ, Bradeen J, Bramel P, Bretting PK, Buckler E, Burke JM, Charest D, Cloutier S, Cole G, Dempewolf H, Dingkuhn M, Feuillet C, Gepts P, Grattapaglia D, Guarino L, Jackson S, Knapp S, Langridge P, Lawton-Rauh A, Lijua Q, Lusty C, Michael T, Myles S, Naito K, Nelson RL, Pontarollo R, Richards CM, Rieseberg L, Ross-Ibarra J, Rounsley S, Hamilton RS, Schurr U, Stein N, Tomooka N, van der Knaap E, van Tassel D, Toll J, Valls J, Varshney RK, Ward J, Waugh R, Wenzl P, Zamir D (2013) Agriculture: feeding the future. Nature 499:23–24
McIntosh R, Yamazaki Y, Dubcovsky J, Rogers W, Morris C, Appels R, Xia X (2013) Catalogue of Gene Symbols for Wheat. 12th International Wheat Genetic Symposium. Yokohama, Japan, September, 8–13, 2013
Meyer RS, Choi JY, Sanches M, Plessis A, Flowers JM, Amas J, Dorph K, Barretto A, Gross B, Fuller DQ, Bimpong IK, Ndjiondjop M-N, Hazzouri KM, Gregorio GB, Purugganan MD (2016) Domestication history and geographical adaptation inferred from a SNP map of African rice. Nat Genet 48:1083–1088
Miedaner T (2013) Roggenanbau: Eine erfolgreiche Alternative. DLG-Verlag GmbH, AgrarPraxis kompakt
Mihaljevic R, Schön C-C, Utz HF, Melchinger AE (2005) Correlations and QTL correspondence between line per se and testcross performance for agronomic traits in four populations of European maize. Crop Sci 45:114–122
Millet EJ, Welcker C, Kruijer W, Negro S, Coupel-Ledru A, Nicolas SD, Laborde J, Bauland C, Praud S, Ranc N, Presterl T, Tuberosa R, Bedő Z, Draye X, Usadel B, Charcosset A, Van Eeuwijk F, Tardieu F (2016) Genome-wide analysis of yield in Europe: allelic effects vary with drought and heat scenarios. Plant Physiol 172:749–764
Möhring J, Piepho H-P (2009) Comparison of weighting in two-stage analysis of plant breeding trials. Crop Sci 49:1977–1988
Paradis E, Claude J, Strimmer K (2004) APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20:289–290
Pasquariello M, Barabaschi D, Himmelbach A, Steuernagel B, Ariyadasa R, Stein N, Gandolfi F, Tenedini E, Bernardis I, Tagliafico E, Pecchioni N, Francia E (2014) The barley Frost resistance-H2 locus. Funct Integr Genom 14:85–100
Petoukhov V, Semenov VA (2010) A link between reduced Barents-Kara sea ice and cold winter extremes over northern continents. J Geophys Res (Atmos) 115:D21111
Plaschke J, Börner A, Xie DX, Koebner RMD, Schlegel R, Gale MD (1993) RFLP mapping of genes affecting plant height and growth habit in rye. Theor Appl Genet 85:1049–1054
Pomeroy M, Fowler DB (1973) Use of lethal dose temperature estimates as indices of frost tolerance for wheat cold acclimated under natural and controlled environments. Can J Plant Sci 53:489–494
Schön C-C, Dhillon BS, Utz HF, Melchinger AE (2010) High congruency of QTL positions for heterosis of grain yield in three crosses of maize. Theor Appl Genet 120:321–332
Schwegler DD, Gowda M, Schulz B, Miedaner T, Liu W, Reif JC (2014) Genotypic correlations and QTL correspondence between line per se and testcross performance in sugar beet (Beta vulgaris L.) for the three agronomic traits beet yield, potassium content, and sodium content. Mol Breed 34:205–215
Shebeski L, McGinnis R, Evans L, Zuzens D (1973) Puma, a new cultivar of winter rye. Can J Plant Sci 53:67
Skinner DZ, Mackey B (2009) Freezing tolerance of winter wheat plants frozen in saturated soil. Field Crops Res 113:335–341
Skinner J, Zitzewitz J, Szűcs P, Marquez-Cedillo L, Filichkin T, Amundsen K, Stockinger E, Thomashow M, Chen TH, Hayes P (2005) Structural, functional, and phylogenetic characterization of a large Cbf gene family in barley. Plant Mol Biol 59:533–551
Snape JW, Sarma R, Quarrie SA, Fish L, Galiba G, Sutka J (2001) Mapping genes for flowering time and frost tolerance in cereals using precise genetic stocks. Euphytica 120:309–315
Sorokina SA, Li C, Wettstein JJ, Kvamstø NG (2016) Observed atmospheric coupling between Barents Sea ice and the warm-Arctic cold-Siberian anomaly pattern. J Clim 29:495–511
Thomashow MF (1999) Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol 50:571–599
Tibshirani R (1996) Regression shrinkage and selection via the LASSO. J R Stat Soc Ser B (Methodol) 58:267–288
Tóth B, Francia E, Rizza F, Stanca AM, Galiba G, Pecchioni N (2004) Development of PCR-based markers on chromosome 5H for assisted selection of frost-tolerant genotypes in barley. Mol Breed 14:265–273
Utz HF (2011) PlabMQTL-Software for meta-QTL analysis with composite interval mapping. Version 0.9. Institute of Plant Breeding, Seed Science, and Population Genetics, University of Hohenheim
Van Ooijen JW (2006) JoinMap 4, software for the calculation of genetic linkage maps in experimental populations. Kyazma B V, Wageningen
Wimmer V, Albrecht T, Auinger H-J, Schön C-C (2012) synbreed: a framework for the analysis of genomic prediction data using R. Bioinformatics 28:2086–2087
Wimmer V, Lehermeier C, Albrecht T, Auinger H-J, Wang Y, Schön C-C (2013) Genome-wide prediction of traits with different genetic architecture through efficient variable selection. Genetics 195:573–587
Wooten DR, Livingston DP, Holland JB, Marshall DS, Murphy JP (2008) Quantitative trait loci and epistasis for crown freezing tolerance in the ‘Kanota’ × ‘Ogle’ hexaploid oat mapping population. Crop Sci 48:149–157
Würschum T, Longin CFH, Hahn V, Tucker MR, Leiser WL (2017) Copy number variations of Cbf genes at the Fr-A2 locus are essential components of winter hardiness in wheat. Plant J 89:764–773
Yan L, Loukoianov A, Tranquilli G, Helguera M, Fahima T, Dubcovsky J (2003) Positional cloning of the wheat vernalization gene VRN1. Proc Natl Acad Sci USA 100:6263–6268
Yan L, Loukoianov A, Blechl A, Tranquilli G, Ramakrishna W, SanMiguel P, Bennetzen JL, Echenique V, Dubcovsky J (2004) The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 303:1640–1644
Yan L, Fu D, Li C, Blechl A, Tranquilli G, Bonafede M, Sanchez A, Valarik M, Yasuda S, Dubcovsky J (2006) The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc Natl Acad Sci USA 103:19581–19586
Zhao Y, Gowda M, Würschum T, Longin CFH, Korzun V, Kollers S, Schachschneider R, Zeng J, Fernando R, Dubcovsky J, Reif JC (2013) Dissecting the genetic architecture of frost tolerance in Central European winter wheat. J Exp Bot 64:4453–4460
Zhu J, Pearce S, Burke A, See D, Skinner D, Dubcovsky J, Garland-Campbell K (2014) Copy number and haplotype variation at the VRN-A1 and central FR-A2 loci are associated with frost tolerance in hexaploid wheat. Theor Appl Genet 127:1183–1197
Acknowledgements
This research was funded by the Federal Ministry of Education and Research (BMBF, Germany) within the project RYE SELECT (Grant ID 0315946). The authors are grateful to the field and lab team of KWS Lochow GmbH and to Stefan Schwertfirm and Amalie Fiedler from TUM for the technical assistance. The technical assistance of G. Schellhorn, Plant Sciences Department, University of Saskatchewan is also gratefully acknowledged. The authors thank Prof. Dr. H. Friedrich Utz (University of Hohenheim) for providing an extension of PlabMQTL which enabled the comparison of QTL and GP models.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Ethical standards
The authors declare that this study complies with the current laws of the countries in which the experiments were performed.
Additional information
Communicated by Diane E. Mather.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Erath, W., Bauer, E., Fowler, D.B. et al. Exploring new alleles for frost tolerance in winter rye. Theor Appl Genet 130, 2151–2164 (2017). https://doi.org/10.1007/s00122-017-2948-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-017-2948-7