Abstract
Key message
Breeding for field resistance to common bunt in wheat will need to account for multiple genes and epistatic and QTL by environment interactions. Loci associated with quantitative resistance to common bunt are co-localized with other beneficial traits including plant height and rust resistance.
Abstract
Common bunt, also known as stinking smut, is caused by seed borne fungi Tilletia tritici (Bjerk.) Wint. [syn. Tilletia caries (DC.) Tul.] and Tilletia laevis Kühn [syn. Tilletia foetida (Wallr.) Liro.]. Common bunt is known to cause grain yield and quality losses in wheat due to bunt ball formation and infestation of the grain. The objectives of this research were to identify and map quantitative trait loci (QTL) for common bunt resistance, to study the epistatic interactions between the identified QTL, and investigate the co-localization of bunt resistance with plant height. A population of 261 doubled haploid lines from the cross Carberry/AC Cadillac and checks were genotyped with polymorphic genome wide microsatellite and DArT® markers. The lines were grown in 2011, 2012, and 2013 in separate nurseries for common bunt incidence and height evaluation. AC Cadillac contributed a QTL (QCbt.spa-6D) for common bunt resistance on chromosome 6D at markers XwPt-1695, XwPt-672044, and XwPt-5114. Carberry contributed QTL for bunt resistance on chromosomes 1B (QCbt.spa-1B at XwPt743523) 4B (QCbt.spa-4B at XwPt-744434-Xwmc617), 4D (QCbt.spa-4D at XwPt-9747), 5B (QCbt.spa-5B at XtPt-3719) and 7D (QCbt.spa-7D at Xwmc273). Significant epistatic interactions were identified for percent bunt incidence between QCbt.spa-1B × QCbt.spa-4B and QCbt.spa-1B × QCbt.spa-6D, and QTL by environment interaction between QCbt.spa-1B × QCbt.spa-6D. Plant height QTL were found on chromosomes 4B (QPh.spa-4B) and 6D (QPh.spa-6D) that co-located with bunt resistance QTL. The identification of previously unreported common bunt resistance QTL (on chromosomes 4B, 4D and 7D), and new understanding of QTL × QTL interactions will facilitate marker-assisted breeding for common bunt resistance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Common bunt, also known as stinking smut, is a disease of wheat (Triticum species) that reduces grain yield from the formation of bunt balls that replace the grain with brown black unpleasant smelling spores (Cherewick 1953; Martens et al. 1984). Grain with a detectable odor imparted by bunt is downgraded and devalued by grain buyers. In Western Canada common bunt is caused by the seed-borne fungi Tilletia tritici and T. laevis (Gaudet and Puchalski 1989b). The disease is listed as a Priority 1 disease in the registration testing system promoting efforts to breed for resistance. Bread wheat varieties registered in Canada are expected to have a minimum intermediate resistance reaction to common bunt (web link: http://pgdc.ca/pdfs/wrt/2012-2013%20PRCWRT%20Operating%20Procedures.pdf). Although common bunt can be effectively managed with fungicide seed treatment, utilization of genetic resistance in cultivars is the best option for maximizing economic efficiency, reducing exposure to chemical seed treatments, and minimizing environmental impact. Furthermore, genetic resistance is the only effective measure of bunt control for organic or low input farming systems (Ciucă 2011; Matanguihan et al. 2010).
Assessment of common bunt resistance response in the field can be difficult due to the need to distinguish bunt balls in later stages of plant development, the symptoms sometimes are only expressed on the last spikes formed, and the symptoms often are only expressed in a few of the florets. Additionally, common bunt expression is environmentally dependent, for example disease development is favoured by cool soil temperature.
Marker-assisted breeding can be utilized to overcome limitations of direct assessment of common bunt resistance in the field or growth chambers. The application of marker-assisted breeding requires an understanding of the genetics of sources of resistance. Several studies have been conducted to unravel the genetic control of bunt resistance. Metzger et al. (1979) suggested that bunt resistance is qualitatively controlled and governed by single genes with complete dominance and race specificity, while other researchers have reported incomplete dominance of bunt resistance genes (Holton and Heald 1941; Knox et al. 1998). Gaudet and Puchalski (1989a) documented the quantitative nature of bunt resistance through demonstrating a continuous range in reaction of cultivars. They also revealed the complexity of resistance with the possibility of race non-specific and race specific resistance. In wheat, 16 race specific bunt resistance genes, Bt1 to Bt15, and Btp are reported (Goates 2012; Goates and Bockelman 2012). Major genes for common bunt resistance are located on several chromosomes. Bt1 is located on chromosome 2B (Sears et al. 1960), Bt7 on 2D (Schaller et al. 1960) and Bt10 on the short arm of chromosome 6D (Menzies et al. 2006). Demeke et al. (1996) determined random amplified polymorphic DNA (RAPD) marker UBC primer 196 is closely linked to Bt10 and subsequently Laroche et al. (2000) developed a SCAR (sequence characterized amplified region) marker, FSD_RSA, for marker-assisted selection. Bt10 is effective against all identified common bunt races in western Canada (Gaudet et al. 1993). Three genes are located on chromosome 1B, Bt4, Bt5, and Bt6, and are linked (Schmidt et al. 1969; McIntosh et al. 1998).
The chromosome location of quantitatively inherited bunt resistance has also been determined through quantitative trait loci (QTL) mapping. In Canadian cultivar ‘AC Domain’, two QTL were identified on chromosome 1B (QCbt.crc-1B.1 and QCbt.crc-1B.2) along with a smaller effect QTL (QCbt.crc-7A) on chromosome 7A (Fofana et al. 2008). In the cultivar Blizzard, a QTL on chromosome 1B was found in the same genomic region as the QTL reported in AC Domain (Wang et al. 2009). A QTL in the cultivar McKenzie was located on 7B (Knox et al. 2013). A major bunt resistance QTL on 1BS, and smaller effect QTL on 5B, 7A and 7B were identified in the cultivar Trintella (Dumalasová et al. 2012).
The Bt10 gene is used in Canadian spring wheat breeding programs and was first deployed in cultivars such as AC Vista, Canada Prairie Spring white wheat (DePauw et al. 1998a) and AC Cadillac, hard red spring wheat (DePauw et al. 1998b). AC Cadillac, which expresses the Bt10 phenotype of resistance to common bunt, originates from the cross BW90×3/BW553 where BW553 possesses Bt10 (DePauw et al. 1998b). While this gene is currently effective, reliance on just one major gene for disease resistance presents unnecessary risk to the wheat industry. Fortunately other sources of bunt resistance have been used in Canadian breeding programs, such as resistance from Thatcher and Hope which express intermediate to high levels of race non-specific resistance (Gaudet et al. 1993). Carberry is another Canadian cultivar known to demonstrate a resistant reaction to common bunt based on field bunt nursery evaluations (DePauw et al. 2011). Carberry originates from the cross Alsen/Superb where Superb has a moderately resistant reaction to bunt and derives from the cross Grandin × 2/AC Domain (DePauw et al. 2011; Townley-Smith et al. 2010). AC Domain is moderately resistant to bunt (Fofana et al. 2008) and does not have a source of Bt10 in its ancestry. Alsen also has a moderately resistant reaction to common bunt (Fox and Humphreys 2004). The moderately resistant reaction to bunt of the parents of the resistant Carberry lead us to hypothesize that Carberry has a form of field resistance to common bunt, controlled by multiple genetic factors, that is different from the resistance of AC Cadillac. Given its pedigree, AC Cadillac could also have genes other than Bt10 that contribute to its resistance.
Bt10 is closely linked to the stem rust resistance gene SrCad in AC Cadillac which provides resistance to Ug99 races (Hiebert et al. 2011). Using a population derived from a cross of Carberry by AC Cadillac (Singh et al. 2013), we showed that the 6D QTL, QSr.spa-6D (SrCad derived from AC Cadillac) expresses resistance to North American races of stem rust. Given the effectiveness of Bt10 and its linkage to the very useful stem rust resistance gene SrCad, the Bt10-SrCad combination is a popular choice in breeding. The concern is that the popularity of Bt10 further increases the risk of it being defeated by the pathogen. Therefore to preserve the effectiveness of Bt10 it should be stacked with other sources of resistance. By understanding the nature of resistance in a variety of sources, gene pyramiding can be employed to stack race non-specific resistant genes along with race specific genes to improve durability of resistance. Marker-assisted selection is effectively the only strategy for bunt resistance gene stacking, and improves the flexibility and reliability of selection over field trials.
Further building on the concept of associated genes in breeding as we have discussed with bunt and stem rust resistance is developing an understanding of the relationship of bunt resistance loci with other loci for other traits, such as height. Gaudet et al. (1991) consider the relationship between height and bunt infection to be more complex than simply common bunt reducing height. They hypothesize reduced culm height is associated with bunt susceptibility, but did not demonstrate this genetically. Understanding the association of different traits has the potential to assist breeders in more efficiently selecting traits that are clustered. The Carberry/AC Cadillac population is segregating for plant height, presenting the opportunity of providing insights into the relationship of loci controlling height and bunt susceptibility. AC Cadillac is a taller genotype while Carberry is shorter statured, both with a resistant phenotype to the prevalent Canadian prairie races of common bunt (DePauw et al. 1998b, 2011).
Multigenic quantitative forms of resistance can display environmental interactions and epistatsis that would be useful for the breeder to understand when choosing and assembling resistance. The discovery of loci involved in bunt resistance expression, particularly of partial or quantitative resistance, can also lead to an understanding of the interactions between the loci and their association with other traits. Yang et al. (2008) have developed the software QTLNetwork for determining epistatic and QTL by environment interactions between loci. In wheat, Hao et al. (2011) used QTLNetwork to identify interactions among loci for stripe rust resistance. We have previously reported on genetic factors and their interactions in wheat cultivars AC Cadillac and Carberry for stem rust (Singh et al. 2013) and stripe rust resistance (Singh et al. 2014) using QTLNetwork. Understanding interactions between loci and the identification of gene rich loci will enhance the ability breeders to produce agronomically desirable disease resistant cultivars.
Using a doubled haploid population derived from a cross of Carberry and AC Cadillac, the objectives of this study were: (1) to identify and map QTL associated with field resistance to common bunt, (2) to identify QTL × QTL interactions for bunt resistance, and (3) to determine the relative location of plant height and bunt resistance QTL.
Materials and methods
Plant materials
A doubled haploid population was developed at the Semiarid Prairie Agricultural Research Centre (SPARC) of Agriculture and Agri-Food Canada (AAFC) from a cross of Carberry with AC Cadillac using the maize pollen method described by Knox et al. (2000). A set of 261 lines were evaluated along with the parents.
Disease and agronomic assessment
A common bunt disease nursery was established near Swift Current, SK, on 5 May 2011, 21 April 2012, and 2 May 2013 with materials and methods described by Knox et al. (2013) and Wang et al. (2009). Soil temperatures were recorded at the soil depth of 5 cm at the nearby meteorological site and analysed for the first 2 weeks from planting. Treatments were 261 lines, parents, and checks grown in unreplicated trials of 3 m long rows per treatment at a seeding rate of 100 seeds per row. The bunt susceptible check was Biggar, and the intermediate bunt reaction check was Neepawa. Twenty plots of each parent and Biggar, and ten plots of Neepawa were randomly interspersed throughout the nursery each year. Prior to planting, seeds were inoculated with Tilletia laevis race L16 and T. tritici race T19 (L16 and T19 races together represent the entire bunt virulence spectrum in Canada). Near maturity, incidence was estimated as a percentage of bunted spikes over total spikes in the row based on a visual assessment.
To avoid the potential confounding effects of bunt on height, the Carberry/AC Cadillac population not inoculated with bunt grown in another field nursery also near Swift Current and about 2 km from the bunt nursery was used for height measurements. In the bunt-free nursery plant height was recorded in centimeters using a measuring pole placed on the ground surface with readings taken at the top of the spike excluding awns.
Correlations of bunt incidence between years were calculated using the software package Statistix 7 (Analytical Software v. 7.0). The option for Pearson correlation coefficient and probability were selected.
Molecular genotyping
Extraction of DNA from the parents and 261 lines from the Carberry/AC Cadillac population was done using the Wheat and Barley DNA Extraction in 96-well plates protocol (http://maswheat.ucdavis.edu/PDF/DNA0003.pdf) and were genotyped with modifications to the PCR of SSR markers as described in Singh et al. (2013). Gentotyping with DArT® of the 261 lines and parents was done by Triticarte Pvt. Ltd. Yarralumla, ACT, Australia (www.triticarte.com.au). The DNA was extracted from parents and doubled haploid lines for DArT® analysis according to the protocol published by Triticarte (http://www.triticarte.com.au/pdf/DArT_DNA_isolation.pdf) and as described by Singh et al. (2013).
QTL analysis
Linkage groups were constructed using the software JoinMap® 4.0 with the regression mapping option, and groupings were created using independence LOD (Van Ooijen 2006). The validity of the linkage groups was confirmed with known chromosomal locations of markers determined through the GrainGenes website (http://wheat.pw.usda.gov/GG2/index.shtml). Each linkage group was assigned to the corresponding hexaploid wheat chromosome based on the known genomic positions of the DArT® and SSR markers in the groups. MapQTL6® (Van Ooijen 2009) was used to perform QTL mapping to identify molecular markers significantly associated with common bunt incidence and height. The logarithm of odds (LOD) threshold for significance was obtained by the permutation test option (1000 permutations) within MapQTL®. Genome-wide threshold levels were used to declare significant QTL based at a 5 % significance level. Automatic co-factor detection based on backward elimination as well as manual co-factor selection was used to identify the co-factor markers for Multiple QTL Mapping (MQM). The marker trait associations were further confirmed using the non-parametric rank sum test of Kruskal–Wallis (KW) to determine significant markers in each disease environment. Linkage groups and LOD bars were drawn with Map-Chart v2.2 (Voorrips 2002).
Epistasis analysis
QTLNetwork version 2.1 (Yang et al. 2008) was used to study QTL interactions. Both single-locus effect QTL and epistatic QTL were generated. QTL effects were estimated by the mixed linear model (MLM) approach. The “2D genome scan” option was used to map epistatic QTL with or without single-locus effects. To estimate epistatic effects of the additive × additive (A×A) nature in a doubled haploid population, the “map epistasis” option was used. To control the experimental Type I error rate by the permutation test, critical F values were calculated using the “permutation” option.
Results
In the first 2 weeks from planting, mean soil temperatures ranged from 5.5 to 8.8 °C with a median temperature of 6.9 °C in 2011, 4.2 to 9.7 °C with a median temperature of 7.4 °C in 2012, 2.7 to 13.3 °C with a median temperature of 9.7 °C in 2013.
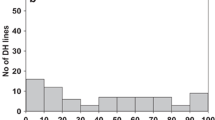
The bunt susceptible control cultivar, Biggar, showed a high level of incidence of common bunt at 57 % in 2011, 64 % in 2012, and 54 % in 2013. The bunt control, Neepawa, that expresses an intermediate level of resistance showed incidence levels of 30 % in 2011, 33 % in 2012, and 28 % in 2013. Each year, AC Cadillac had a similar but numerically lower incidence of bunt than Carberry. AC Cadillac had a mean 3 % bunt incidence in 2011, 5 % in 2012, and 6 % in 2013, while Carberry had 10 % bunt incidence in 2011, 9 % in 2012, and 7 % in 2013. A wide distribution of bunt incidence was observed for the population each year (Fig. 1). The bunt incidence of lines ranged from 0 to 95 % in 2011, 0 to 65 % in 2012, and 0 to 55 % in 2013. In each year, lines segregated from the population with the same or greater incidence of bunt than the susceptible check Biggar. In all the years, the distributions were continuous and similarly shaped being skewed to the right with a preponderance of low-incidence lines.
Frequency distribution of incidence of bunt percentage in the Carberry/AC Cadillac doubled haploid (DH) population. Bunt incidence was measured in common bunt nurseries near Swift Current, Canada against a 1:1 mix of Tilletia laevis race L16 and Tilletia caries race T19 in 2011, 2012 and 2013. Mean common bunt incidence for Carberry was 10 % in 2011, 9 % in 2012, and 7 % in 2013. Mean common bunt incidence for AC Cadillac was 3 % in 2011, 5 % in 2012, and 6 % in 2013. Mean common bunt incidence for the susceptible control, Biggar, was 57 % in 2011, 64 % in 2012, and 54 % in 2013
The mean bunt incidence in 2011 was 15.6 %, in 2012 it was 14.5 %, and in 2013 it was 9.6 %. Out of the 261 lines tested, 122 lines showed 10 % bunt or less in all 3 years and eight lines showed a bunt incidence of 30 % or higher in all 3 years. Some lines consistently expressed intermediate levels of resistance in all 3 years, while other lines were very variable across the 3 years, for example being as resistant as Carberry in one year and nearly as susceptible as Biggar in another year. Correlations of bunt incidence among years were r = 0.64 (p < 0.01) for 2011/2012, r = 0.47 (p < 0.01) for 2011/2013, and r = 0.65 (p < 0.01) for 2012/2013.
The linkage map was constructed using 634 polymorphic DArT and SSR markers. Linkage groups formed were anchored to the 21 wheat chromosomes and spanned 2101.6 cM.
Using MQM, common bunt incidence QTL were identified in the Carberry/AC Cadillac population on chromosomes 1B (QCbt.spa-1B), 4B (QCbt.spa-4B), 4D (QCbt.spa-4D), 6D (QCbt.spa-6D) and 7D (QCbt.spa-7D) (Table 1). Figure 2 shows the interval and relative positions of QTL identified by MQM analysis. QCbt.spa-1B, QCbt.spa-6D and QCbt.spa-7D were identified in more than one environment. Results similar to MQM analysis were obtained using KW analysis: QCbt.spa-1B appeared in 2011 and 2012 (p < 0.001), QCbt.spa-6D and QCbt.spa-7D appeared in all three years (p < 0.001). QCbt.spa-4D expressed in each of the three years (p < 0.01 to < 0.001) with KW analysis. QCbt.spa-5B, which was not identified using MQM mapping, was identified with KW near DArT marker tPt-3719 in 2011 and 2012 (p < 0.01 to <0.001). Carberry contributed to reduced common bunt incidence at QCbt.spa-1B, QCbt.spa-4B, QCbt.spa-4D and QCbt.spa-7D, while AC Cadillac contributed low incidence at QCbt.spa-6D, which was also the largest effect QTL [highest phenotypic value (PV)] (Table 1). Carberry also contributed the lower bunt incidence for QCbt.spa-5B.
Linkage groups of DArT and SSR markers in which QTL for common bunt incidence and plant height were identified with MQM mapping on chromosomes 1B, 4B, 4D, 6D, and 7D in a doubled haploid population derived from Carberry/AC Cadillac. Results are from nurseries grown near Swift Current, Canada in 2011, 2012, and 2013. The QTL intervals of main effects are represented by a bar (LOD 2) and line extending from the bar (LOD 1) for bunt incidence and plant height. Epistatic QTL for bunt incidence generated by QTLNetwork are indicated with a solid circle
With MQM analysis, QTL for plant height were identified on chromosomes 4B (QPh.spa-4B) and 6D (QPh.spa-6D) and each expressed in all the 3 years. The plant height of AC Cadillac was 102.4 cm in 2011, 105.8 cm in 2012, and 117.6 cm in 2013. The plant height of Carberry was 81.0 cm in 2011, 84.6 cm in 2012, and 89.5 cm in 2013.
AC Cadillac contributed to lower plant height at the 6D locus and the QTL explained 3.4–6.5 % of the PV. Carberry contributed to lower plant height at the 4B locus with a much greater contribution to PV of 13–43 %. The plant height QTL, QPh.spa-6D, was located near the bunt resistance QTL, QCbt.spa-6D (Fig. 2). The markers at the peaks of the QTL for bunt resistance (XwPt-5114 and XwPt-1695) and plant height (XwPt-2864 and XwPt-741955) were within 2 cM of each other.
The height QTL, QPh.spa-4B, was located near the QTL for bunt incidence, QCbt.spa-4B, on chromosome 4B. The peak LOD for the QTL for bunt resistance on chromosome 4B spanned two markers, XwPt-744434 and Xwmc617, which lay 27.1 cM apart. Nonetheless, there was overlap of the markers at the peaks of the QTL for bunt resistance (XwPt-744434-Xwmc617) and plant height (XwPt-744434).
QTLNetwork identified the main effect QTL on chromosomes 1B, 4B, 4D and 6D, which was similar to the results of the MQM analysis. The main effects QTL that were additive were detected on chromosomes 4B, 4D and 6D (Fig. 3). The main effect QTL, QCbt.spa-1B, on 1B was not only additive, it showed an epistatic interaction with 4B QTL QCbt.spa-4B (Table 2; Fig. 3). Although QTLNetwork uses the entire map to locate QTL, Fig. 4 is a graphical representation of the interaction using select markers (peak LOD) at the 1B (XwPt-667763) and 4B (XwPt-744434) loci for illustration purposes. The additive effect is seen with reduced incidence of bunt with the contribution of certain alleles at each locus. For example, in 2012 the AC Cadillac alleles at the 1B and 4B loci produced the highest incidence of bunt (32.2 %), whereas the Carberry allele at each of the 1B and 4B loci produced the lowest incidence of bunt (10.1 %). Differences in slopes of the lines in Fig. 4 imply a synergy or epistatic interaction between alleles for bunt resistance between loci with which the markers were associated. In 2013 the effect in reducing the bunt of the 4B AC Cadillac molecular variant with the 1B Carberry molecular variant (8 %) was greater than the 4B plus 1B Carberry molecular variants (9 %) resulting in a crossover interaction for QCbt.spa-1B × QCbt.spa-4B (Fig. 4). Although not declared significant with QTLNetwork as a QTL by environment effect, considering the patterns across environments, the 4B Carberry molecular variant displayed a substantial decrease in bunt incidence in 2011 in the presence of the 1B Carberry molecular variant (7 %) compared to the 1B AC Cadillac molecular variant (21.5 %), a modest decrease in bunt in 2012 (from a high of AC Cadillac 1B with Carberry 4B of 13.7 % to a low of Carberry 1B with Carberry 4B at 10.1 %), and no effect in 2013(AC Cadillac 1B with Carberry 4B of 9 % and Carberry 1B with Carberry 4B at 9 %). However, the 4B Carberry molecular variant dramatically decreased bunt incidence over the 4B AC Cadillac molecular variant in the presence of the 1B molecular variant from AC Cadillac in all years (2011: 37.2 to 21.5 %, 2012: 32.2 to 13.7 %, 2013: 14 to 9 %; Fig. 4).
Main effects QTL with additive effects were detected on chromosomes 4B, 4D and 6D. A main effect QTL with additive plus environment effect was detected on chromosome 1B. Epistatic interaction between QTL on chromosome 1B and 4B and epistatic plus epistatic × environment interaction was detected between QTL on chromosome 1B and 6D
Figure 5 is a graphical representation of the interaction using select (highest LOD) markers at the 1B (XwPt-667763) and 6D (XwPt-1695) loci, again for illustration purposes. A non-crossover epistatic interaction was observed for QCbt.spa-1B × QCbt.spa-6D (Fig. 5). While reduced bunt incidence was observed in all years when the AC Cadillac molecular variant on 6D was present with either 1B molecular variant (e.g. 2.5 % in 2011 in the presence of either AC Cadillac 1B or the Carberry 1B), the same was not true for the Carberry molecular variant. In the presence of the Carberry molecular variant on 6D, lower bunt incidence was observed in conjunction with the chromosome 1B Carberry molecular variant (e.g. in 2011 9.6 % bunt incidence) in contrast with the 1B AC Cadillac molecular variant with the Carberry 6D (e.g. 2011 41.1 %). The epistatic response between the 1B and 6D loci varied sufficiently over the 3 years to be declared a significant QE effect with QTLNetwork.
Discussion
The planting of wheat experiments at the early extreme of what is typical for the Canadian prairie region exposed the bunt inoculated seed to cool soil conditions that did not exceed a median temperature of 10 °C within the first 2 weeks of planting in any of the 3 years. Cool soil conditions at the time of planting favour the expression of common bunt (Goates 1996). The high level of bunt incidence in the bunt susceptible check cultivar Biggar indicated good expression of the disease, confirming that good disease expression was obtained with the high level of bunt incidence in particular lines of the population. Because the two races of Tilletia were used to inoculate the tests represented the spectrum of virulence found on the Canadian prairies, resistance loci revealed by the races have relevance to Canadian conditions.
The low incidence of common bunt expressed by Carberry and AC Cadillac confirmed their resistance to the disease. The segregation of the Carberry/AC Cadillac progeny (Fig. 1) that expressed bunt incidence as high as the susceptible check Biggar indicated those lines were susceptible to bunt. Notably, because of the segregation of susceptible progeny, there is no indication of resistance genes in common between the two parents. The positively skewed and continuous frequency distribution of incidence suggested multiple genes of varying levels of penetrance were segregating. The skewed nature of the distribution of progeny, with a high proportion of lines expressing a low level of incidence to common bunt, indicated the presence of a major gene for resistance. This pattern of segregation is consistent with a contribution of a gene such as Bt10 that is considered to be present in AC Cadillac based on pedigree and phenotype (DePauw et al. 1998b). The similarly skewed shape of the distributions of the population for bunt incidence in response to the different environments across the 3 years of testing indicated a reasonably consistent response to disease. However, subtle differences in the distributions from year to year as indicated by moderate correlations from 0.47 to 0.65 among years and by year-to-year differences in incidence of some genotypes also demonstrated a variable response of resistance to different environmental conditions.
The MQM, KW, and QTLNetwork algorithms produced similar results reinforcing the presence of QTL on chromosomes 1B, 4D and 6D. Support for a QTL on 7D was provided by both MQM and KW and for 4B by MQM and QTLNetwork. Only the KW method identified a significant QTL on chromosome 5B, QCbt.spa-5B, but the occurrence in 2 years provides credibility that the locus effect on bunt incidence is real. The identification with QTL analysis of multiple factors controlling resistance was consistent with the shape of the histogram distributions for bunt incidence.
The QCbt.spa-6D QTL contributed by AC Cadillac and located on chromosome 6D had, as indicated by the phenotypic value, a large effect on bunt incidence which is consistent with the effect of a major gene. As previously mentioned, based on the pedigree of AC Cadillac and its phenotype, the gene on 6D is likely Bt10, further evidence of which comes from its position on chromosome 6D. Menzies et al. (2006) mapped the FSD_RSA marker and the Bt10 bunt resistance to chromosome 6D based on linkage with markers such as Xgwm469. Xgwm469 is linked to the microsatellite marker Xcfd49 (Hiebert et al. 2011) that is about 7 cM from the DArT marker XwPt-1695 at the peak of the QTL we found associated with the major reduction in bunt incidence. Consistent with the major gene effect of QCbt.spa-6D was the appearance of the locus in all 3 years of testing.
All other QTL were contributed by Carberry and although their dissected effect on reducing bunt incidence based on phenotypic value was low to moderate, their cumulative effect provided resistance similar to AC Cadillac. The genetic factor or factors producing the effect of the QTL we identified on chromosome 1B, QCbt.spa-1B, encompassing markers Xbarc128 and Xgwm374, may be the same as those reported by Wang et al. (2009), Fofana et al. (2008) and Dumalasová et al. (2012). QCbt.spa-1B is in the same region as the QTL identified by Wang et al. (2009) that included the same two markers, Xbarc128 and Xgwm374, plus Xgwm264. The markers Xgwm264 and Xgwm374 are consistent with those associated with the QTL Cbt.crc-1B.1 discovered by Fofana et al. (2008), and according to the map of Somers et al. (2004) these markers are in the same vicinity as Xbarc8 which was in the QTL interval determined by Dumalasová et al. (2012). QCbt.spa-1B appearing in only two of the 3 years of testing indicated the locus was affected by environment.
The genetic factor or factors we identified at QTL QCbt.spa-5B on chromosome 5B that produced reduced incidence of bunt and was associated with the DArT marker XtPt-3719 may be the same as those generating the QTL for bunt resistance identified by Dumalasová et al. (2012). XtPt-3719 is found close to marker Xwmc289 on the map of Jighly et al. (2015, Supplementary File 1). According to Somers et al. (2004) Xwmc289 is close to Xgwm408 which is in the interval of the QTL for bunt resistance identified by Dumalasová et al. (2012). The QCbt.spa-5B appears to be influenced by environment with the KW test identifying the locus in only 2 out of 3 years of testing.
In addition to QCbt.spa-1B and QCbt.spa-5B, the QTL QCbt.spa-4B, QCbt.spa-4D and QCbt.spa-7D also contributed to reduced bunt incidence in Carberry in particular years. Although QCbt.spa-7D produced a significant result in only 2012, but the QTL was very close to significant in 2011 and 2012 suggesting the locus produces a consistent but minor effect. This is supported by the modest PV values measured for this locus. The QCbt.spa-4B and QCbt.spa-4D QTL variation over years and modest PV indicated the loci are more influenced by environment than QCbt.spa-7D, at least within the scope of this study. Identification of QTL in one, two, or three environments as well as variation in PV explained by each QTL supports complex inheritance of bunt resistance contributed by Carberry and the role of environment in gene expression.
Sources of race nonspecific bunt resistance have been reported previously (Gaudet and Puchalski 1989a). Fofana et al. (2008) studied the bunt resistance in AC Domain and identified three loci that contributed to the moderately resistant reaction. Dumalasová et al. (2012) reported on bunt resistance in Trintella that segregated for a major gene and three minor QTL. As with our results, in 2 years of testing by Dumalasová et al. (2012), the minor QTL expression varied between years with two of the QTL being expressed in 1 year and a third QTL expressed in the other year.
A novel aspect of the present research is the observation that the bunt resistance in Carberry can also be explained in part by QTL × QTL interactions. There is a lack of information on such epistatic interactions of common bunt resistance loci. The analysis using QTLNetwork provides greater insights into the nature of the relationship of QTL for bunt resistance in the Carberry by AC Cadillac population. By the simple identification of multiple QTL derived from Carberry, the concept of multiple genes contributing to the phenotype is reinforced, but furthermore certain gene combinations were found to be functioning more than additively. In addition to main effects of the QTL at the 1B, 4B, 4D and 6D loci, these loci were involved in interactions that also affected bunt incidence. Interestingly, these interactions were not always simple as demonstrated with the 1B QTL interacting with QTL on both 4B and 6D. The analysis using QTLNetwork showed that certain combinations of alleles could be more effective than others. For example, while AC Cadillac allelic contribution at QCbt.spa-6D and QCbt.spa-1B gave the best bunt control (lowest bunt incidence), the 6D Carberry allele at QCbt.spa-6D interacted with the Carberry allele at QCbt.spa-1B to reduce bunt incidence substantially compared to the interaction of AC Cadillac allele at QCbt.spa-1B, particularly in 2011. Not only were there additive and epistatic components to the variation, the level of effect on bunt incidence of interactions between QTL can vary from year to year. This type of interaction was observed between the 1B and 6D loci which varied enough among years to be declared a significant QTL by environment (QE) interaction with QTLNetwork. The 6D locus showed a dominant epistatic effect of the 6D AC Cadillac major allele, compared to the Carberry 6D allele, with the factor located on 1B. This is the type of epistatic interaction often found with plant disease resistance as described by Sidhu (1984). In other words, in the presence of the 6D allele from AC Cadillac the 1B allele from Carberry showed an attenuated effect. Although we found no previous reports of these types of interactions of QTL for common bunt incidence in wheat, there are reports with other disease systems in wheat. For example, Hao et al. (2011) reported on additive, and additive by environment effects with stripe rust resistance in winter wheat using QTLNetwork. Similarly, Singh et al. (2014) used QTLNetwork to identify epistatic effects between loci within each form of disease for stripe rust severity, stripe rust infection response, and leaf rust severity in spring wheat. In earlier work on wheat they had identified epistatic interactions between loci for stem rust severity, infection response, and seedling infection type (Singh et al. 2013).
The Carberry/AC Cadillac population was segregating for height. We used this opportunity to obtain an understanding of the relationship of short stature of wheat with bunt susceptibility proposed by Gaudet et al. (1991), by performing genetic analysis on the population for height in addition to the analysis of bunt incidence. The potential confounding effect of associating height with resistance from common bunt itself affecting height was avoided by using a second nursery in which the Carberry/AC Cadillac population was growing primarily for rust assessment (Singh et al. 2013, 2014) to collect height data each year. We did not map the direct effect of bunt on plant height. This latter interaction between host and pathogen required much more detailed height measurements, demanding more manpower than was available, so the phenomenon remains to be investigated.
Two loci, one on 6D and one on 4B, were found to be coincident for effects on bunt expression and plant height expression. Although AC Cadillac was the taller cultivar in all 3 years, along with bunt resistance AC Cadillac contributed the lower plant height allele at QPh.spa-6D. This is consistent with AC Cadillac considered to be a short conventional height cultivar. On 4B, the factors for reduced height and bunt resistance were contributed by Carberry. In both instances, bunt resistance was associated with reduced plant height and contrary to the condition proposed by Gaudet et al. (1991). However, in the intervening 24 years it is possible that favourable loci for height and bunt resistance that were at one time in repulsion in breeding germplasm have been selected for recombination to favourable loci being in coupling.
The understanding of relationship of traits at particular loci is valuable in breeding. The trend in breeding has been towards shorter cultivars. Knowing that the reduced plant height allele on 6D from AC Cadillac lies in the same interval as resistance to bunt, and that the reduction in height was stable over environments for the 3 years of trialing will appeal to breeders. This information along with the current knowledge of the association of the locus with Ug99 stem rust resistance (Hiebert et al. 2011; Singh et al. 2013) could contribute to the over-use of Bt10, adding to the urgency of developing other sources of common bunt resistance. Like the 6D plant height factor, the second factor for plant height at QPh.spa-4b was highly expressive and stable over the 3 years of testing. As with the 6D locus, the 4B locus will be appealing for use in breeding programs because of the ability to select for reduced height and bunt resistance at the same time. The common bunt resistance on 4B is interesting as our results from this and a previously published study (Singh et al. 2013) suggest that the bunt resistance lies within a gene rich region. The bunt resistance and plant height LOD peak was at marker XwPt-744434 in the present study, and this marker was also at the peak for resistance to the Ug99 races of stem rust contributed by AC Cadillac (Singh et al. 2013). The stem rust resistance is in repulsion with the bunt resistance, requiring the identification of a line with recombination that would assemble favourable alleles for both stem rust and bunt resistance. Adjacent to the XwPt-744434 was a marker about 20 cM distance with a peak LOD association with a QTL for leaf rust resistance contributed by Carberry (Singh et al. 2014). The relationship of the Carberry QTL for height on chromosome 4B with Rht-B1 is not entirely clear, but consistent with mapping results of Ellis et al. (2002) in which Rht-B1b was located near microsatellite marker Xwmc048. The height QTL from Carberry was located in the 27 cM interval between Xwmc657 and Xwmc617 which encompasses Xwmc048 based on the maps by Somers et al. (2004) and McCartney et al. (2005). This is the same interval in which McCartney et al. (2005) identified a QTL for plant height. Lv et al. (2014) identified a height QTL between Xgwm149 and Xgwm495 which overlaps with the Xwmc657 and Xwmc617 interval according to Somers et al. (2004). Liu et al. (2011) mapped Rht-B1 about 12 cM from Xgwm495. Somers et al. (2004) mapped Rht-B1 outside the Xwmc657 and Xwmc617 interval. Shankar et al. (2008) also appeared to map Rht-B1 outside the Xwmc657 and Xwmc617 interval using Somers’ et al. (2004) map to cross reference common markers. Distances between reported maps varied substantially based on common markers. Future work could include testing the perfect markers developed by Ellis et al. (2002) for Rht-B1. Based our work, the 4B locus possesses factors important in breeding of plant height, and bunt, stem rust, and leaf rust resistance. Also in the region is FHB resistance (Lv et al. 2014) and stagonospora leaf spot resistance (Shankar et al. 2008).
The QCbt.spa-7D QTL on 7D appeared stable expressing bunt resistance in each of the 3 years of testing and coincides with the stem rust infection type QTL, QSr.spa-7D, reported by Singh et al. (2013) using the same Carberry/AC Cadillac population. Unfortunately the favourable alleles for resistance to the two diseases are in repulsion phase linkage necessitating selection within an optimum sized population to obtain favorable recombinants to bring favourable alleles together in coupling.
The identification of co-location of important genes for breeding is positive in providing breeders with information on loci with which to focus selection to have the broadest impact on desirable traits. In some cases more work will have to be done to assemble genes in coupling, such as with the bunt and stem rust resistance on 4B. In some cases such recombinants will already be available in the Carberry/AC Cadillac population. In other cases the gene or genes may have to be introduced into the genetic pool, which may be the case with, for example, stagonospora leaf spot resistance identified on chromosome 4B (Shankar et al. 2008). Breeders will need to give consideration to using markers that flank the interval to capture the entire favourable linkage block.
The availability of a complex of genes for desirable traits, however, could put major disease resistance genes such as Bt10 at risk if the gene is used to the extent that it puts pressure on the pathogen to adapt. Although common bunt resistance provided by Bt10 is effective to all the known races in Western Canada, Goates (2012) reported for the first time a new race, D-18, of T. contraversa with virulence on the combination of Bt9 and Bt10 genes. Fortunately Carberry pulls together a number of minor genes that combined to produce a very effective form of bunt resistance, comparable to the major gene resistance in AC Cadillac. The number of genes involved in the Carberry bunt resistance will present a challenge to breeders to reassemble. Knowledge of the location of the genes will be critical to reassembling the genes in other crosses using markers, and knowledge of relationships of the bunt resistance genetic factors with factors controlling other traits will provide incentive for breeders to select for the gene dense loci. In addition to the fact that the loci from Carberry have additive effects, the variable stability of the loci across environments along with a portion of the variation being explained by epistasis would indicate that as many of the loci as possible should be targeted for inclusion in new cultivars during development to maximize phenotypic expression.
In conclusion, Carberry provides an effective source of common bunt resistance, but we found evidence that the resistance is built on the cumulative effect of at least five loci. Within this study, Carberry’s resistance to common bunt was stable in 3 years of testing, but components of the resistance showed year to year variation, and certain factors interacted epistatically. Additionally, QTL by environment interaction was present. The complex nature of resistance has implications on breeding in that the reassembly of the loci in new cultivars will be challenging. However, the location of some loci contributing to the bunt resistance appears to be rich in other beneficial genes. Thus some bunt resistance loci in coupling with other favourable genes will enhance appeal to use such loci in breeding. The information should ultimately lead to breeders being able to incorporate several beneficial traits simultaneously. Surprisingly, among the beneficial combinations is plant height, with QTL on chromosomes 4B and 6D collocated for plant height and bunt resistance with favourable alleles in coupling in the cultivars Carberry and AC Cadillac. More work is needed to assemble other beneficial loci in coupling, but the research here assists in understanding the traits still needing to be recombined. A concern is the projected over-use of Bt10, especially with it being favourably associated with desirable height and Ug99 stem rust resistance factors. With the information we have presented on Carberry bunt resistance, it should be possible to recombine the resistance with Bt10. The durability of the resistance can only be tested over time, but stacking the Carberry and AC Cadillac resistance should help protect Bt10.
This work validates QTL for common bunt resistance on chromosomes 6D, 1B, and 5B, with the identification of new small effect QTL on 4B, 4D and 7D. Going forward, there is a need for further understanding and validation of each of the identified bunt resistance loci, their interactions, and whether or not other loci are involved from Carberry or AC Cadillac. Work is underway to phenotype and genotype over 800 lines of the Carberry/AC Cadillac population with high throughput genotyping, which will allow fine mapping of the bunt resistant QTL.
Author contribution statement
AS., R.E.K., R.M.D., A.K.S., R.D.C.: conceived and designed experiments. A.S., R.E.K., R.M.D., R.D.C., A.K.S.: managed experiments. R.E.K. and R.M.D.: developed and provided plant lines and doubled haploid population. A.S., R.M.D., R.D.C., R.E.K., A.K.S., S. Kumar, H.L.C.: Performed trials and collected data. A.S., R.E.K., A.K.S., R.M.D.: Analyzed and interpreted data. A.S., R.E.K., A.K.S., R.M.D.: prepared the manuscript.
References
Cherewick WJ (1953) Smut diseases of cultivated plants in Canada. Canada Department of Agriculture, Ottawa
Ciucă M (2011) A preliminary report on the identification of SSR markers for bunt (Tilletia sp.) resistance in wheat. Czech J Genet Plant 47:S142–S145
Demeke T, Laroche A, Gaudet DA (1996) A DNA marker for the Bt-10 common bunt resistance gene in wheat. Genome 39:51–55
DePauw RM, McCaig TN, Knox RE, Clarke JM, Fernandez MR, McLeod JG (1998a) AC Vista hard white spring wheat. Can J Plant Sci 78:617–620
DePauw RM, Thomas JB, Knox RE, Clarke JM, Fernandez MR, McCaig TN, McLeod JG (1998b) AC Cadillac hard red spring wheat. Can J Plant Sci 78:459–462
DePauw RM, Knox RE, McCaig TN, Clarke FR, Clarke JM (2011) Carberry hard red spring wheat. Can J Plant Sci 91:529–534
Dumalasová V, Simmonds J, Bartoš P, Snape J (2012) Location of genes for common bunt resistance in the European winter wheat cv. Trintella. Euphytica 186:257–264
Ellis M, Spielmeyer W, Gale K, Rebetzke G, Richards R (2002) “Perfect” markers for the Rht-B1b and Rht-D1b dwarfing genes in wheat. Theor Appl Genet 105:1038–1042
Fofana B, Humphreys D, Cloutier S, McCartney C, Somers D (2008) Mapping quantitative trait loci controlling common bunt resistance in a doubled haploid population derived from the spring wheat cross RL4452 x AC Domain. Mol Breeding 21:317–325
Fox SL, Humphreys DG (2004) Report on central bread wheat co-operative tests, 2003. In: Fox SL, McCallum KJ (eds) Prairie registration recommending committee for grain, wheat, rye, & triticale subcommittee report. Saskatoon, SK, pp 183–230
Gaudet DA, Puchalski BJ (1989a) Status of bunt resistance in western Canadian spring wheat and triticale Can J. Plant Sci 69:797–804
Gaudet DA, Puchalski BL (1989b) Races of common bunt (Tilletia caries and T. foetida) of wheat in western Canada. Can J Plant Pathol 11:415–418
Gaudet DA, Puchalski BJ, Entz T (1991) Culm height and susceptibility of Canadian winter and spring wheat cultivars to common bunt (Tilletia tritici and T. laevis). Can J Plant Sci 71:677–687
Gaudet DA, Puchalski BJ, Kozub GC, Schaalje GB (1993) Susceptibility and resistance in Canadian spring wheat cultivars to common bunt (Tilletia tritici and T. laevis). Can J Plant Sci 73:1217–1224
Goates BJ (1996) Common bunt and dwarf bunt. In: Wilcoxson RD, Saari EE (eds) Bunt and Smut diseases of wheat: concepts and methods of disease management. CIMMYT, Mexico, pp 12–25
Goates BJ (2012) Identification of new pathogenic races of common bunt and dwarf bunt fungi and evaluation of known races using an expanded set of differential lines. Plant Dis 96:361–369. doi:10.1094/PDIS-04-11-0339
Goates BJ, Bockelman HE (2012) Identification of new sources of high levels of resistance to dwarf bunt and common bunt among winter wheat landraces in the USDA-ARS national small grains collection. Crop Sci 52:2595–2605
Hao Y, Chen Z, Wang Y, Bland D, Buck J, Brown-Guedira G, Johnson J (2011) Characterization of a major QTL for adult plant resistance to stripe rust in US soft red winter wheat. Theor Appl Genet 123:1401–1411
Hiebert CW, Fetch TG, Zegeye T, Thomas JB, Somers DJ, Humphreys DG, McCallum BD, Cloutier S, Singh D, Knott DR (2011) Genetics and mapping of seedling resistance to Ug99 stem rust in Canadian wheat cultivars ‘Peace’ and ‘AC Cadillac’. Theor Appl Genet 122:143–149
Holton CS, Heald FD (1941) Bunt or stinking smut of wheat: a world problem. Burgess Publishing Co, Minneapolis
Jighly A, Joukhadar R, Alagu M (2015) SimpleMap: A pipeline to streamline high-density linkage map construction. The Plant Genome 8:1 (Supplementary File 1)
Knox RE, Fernandez MR, Brûlé AL, DePauw RM (1998) Inheritance of common bunt resistance in androgenetically derived doubled haploid and random inbred populations of wheat. Crop Sci 38:1119–1124
Knox RE, Clarke JM, DePauw RM (2000) Dicamba and growth condition effects on doubled haploid production in durum wheat crossed with maize. Plant Breed 119:289–298
Knox RE, Campbell HL, DePauw RM, Gaudet D, Puchalski B, Clarke FC (2013) DNA markers for resistance to common bunt in ‘McKenzie’ wheat. Can J Plant Pathol 35:328–337
Laroche A, Demeke T, Gaudet DA, Puchalski B, Frick M, McKenzie R (2000) Development of a PCR marker for rapid identification of the Bt-10 gene for common bunt resistance in wheat. Genome 43:217–223
Liu G, Xu S, Ni Z, Xie C, Qin D, Li J, Lu L, Zhang J, Peng H, Sun Q (2011) Molecular dissection of plant height QTLs using recombinant inbred lines from hybrids between common wheat (Triticum aestivum L.) and spelt wheat (Triticum spelta L.). Chin Sci Bull 56:1897–1903. doi:10.1007/s11434-011-4506-z
Lv C, Song Y, Gao L, Yao Q, Zhou R, Xu R, Jia J (2014) Integration of QTL detection and marker assisted selection for improving resistance to Fusarium head blight and important agronomic traits in wheat. Crop J 2:70–78
Martens J, Seaman WWL, Atkinson TH (1984) Diseases of Field Crops in Canada. Canadian Phytopath. Soc, Ottawa
Matanguihan JB, Murphy KM, Jones SS (2010) Control of common bunt in organic wheat. Plant Dis 95:92–103
McCartney CA, Somers DJ, Humphreys DG, Lukow O, Ames N, Noll J, Cloutier S, McCallum BD (2005) Mapping quantitative trait loci controlling agronomic traits in the spring wheat cross RL4452 × ‘AC Domain’. Genome 48:870–883
McIntosh RA, Hart GE, Devos KM, Gale MD, Rogers WJ (1998) Catalogue of gene symbols for wheat. In: Slinkard (ed) proceedings 9th international wheat genetics symposium. Vol 5. Saskatoon
Menzies JG, Knox RE, Popovic Z, Procunier JD (2006) Common bunt resistance gene Bt10 located on wheat chromosome 6D. Can J Plant Sci 86:1409–1412
Metzger RJ, Schaller CW, Rohde CR (1979) Inheritance of resistance to common bunt in wheat, C.I. 7090. Crop Sci 19:309–312
Schaller CW, Holton CS, Kendrick EL (1960) Inheritance of the second factor for resistance to bunt, Tilletia caries and T. foetida, in the wheat variety Martin. Agron J 52:280–282
Schmidt J, Morris R, Johnson V (1969) Monosomic analysis for bunt resistance in derivatives of Turkey and Oro wheats. Crop Sci 9:286–288
Sears E, Schaller C, Briggs F (1960) Identification of the chromosome carrying the Martin gene for resistance of wheat to bunt. Can J Genet Cytol 2:262–267
Shankar M, Walker E, Golzar H, Loughman R, Wilson RE, Francki MG (2008) Quantitative trait loci for seedling and adult plant resistance to Stagonospora nodorum in wheat. Phytopathology 98:886–893
Sidhu GS (1984) Parasitic epistasis. Phytopathology 74:382–384
Singh A, Knox RE, DePauw RM, Singh AK, Cuthbert RD, Campbell HL, Singh D, Bhavani S, Fetch T, Clarke F (2013) Identification and mapping in spring wheat of genetic factors controlling stem rust resistance and the study of their epistatic interactions across multiple environments. Theor Appl Genet 126:1951–1964
Singh A, Knox RE, DePauw RM, Singh AK, Cuthbert RD, Campbell HL, Shorter S, Bhavani S (2014) Stripe rust and leaf rust resistance QTL mapping, epistatic interactions, and co-localization with stem rust resistance loci in spring wheat evaluated over three continents. Theor Appl Genet 127:2465–2477
Somers DJ, Issac P, Edwards K (2004) A high-density microsatellite consensus map for bread wheat (Triticum aestivum L.). Theor Appl Genet 109:1105–1111
Townley-Smith TF, Humphreys DG, Czarnecki E, Lukow OM, McCallum BM, Fetch TG, Gilbert JA, Menzies JG, Brown PD (2010) Superb hard red spring wheat. Can J Plant Sci 90:347–352
Van Ooijen JW (2006) JoinMap® 4, Software for the calculation of genetic linkage maps in experimental populations. Kyazma BV, Wageningen
Van Ooijen JW (2009) MapQTL® 6, Software for the mapping of quantitative trait loci in experimental populations of diploid species. Kyazma BV, Wageningen
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78
Wang S, Knox RE, DePauw RM, Clarke FR, Clarke JM, Thomas JB (2009) Markers to a common bunt resistance gene derived from ‘Blizzard’ wheat (Triticum aestivum L.) and mapped to chromosome arm 1BS. Theor Appl Genet 119:541–553
Yang J, Hu C, Hu H, Yu R, Xia Z, Ye X, Zhu J (2008) QTLNetwork: mapping and visualizing genetic architecture of complex traits in experimental populations. Bioinformatics 24:721–723
Acknowledgments
Financial support was received from Agriculture and Agri-Food Canada, Western Grains Research Foundation, and SeCan. We thank Jason Nimegeers and Herb Cutforth for provision and interpretation of meteorological data. Research support from the SPARC wheat breeding and molecular genetic teams is greatly appreciated.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by S. Dreisigacker.
Rights and permissions
About this article
Cite this article
Singh, A., Knox, R.E., DePauw, R.M. et al. Genetic mapping of common bunt resistance and plant height QTL in wheat. Theor Appl Genet 129, 243–256 (2016). https://doi.org/10.1007/s00122-015-2624-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-015-2624-8