Abstract
The location of new genes for resistance to common bunt in wheat is valuable for gene pyramiding in breeding. For this purpose, the genetics of the relatively high level of resistance in the European winter wheat variety Trintella was investigated using a doubled haploid mapping population of a cross between Trintella and the susceptible variety Piko. The population was scored for bunt infection in the field for 2 years following inoculation with a mixture of teliospores of Tilletia tritici and T. laevis. A genetic map consisting of 29 linkage groups was constructed using polymorphic simple sequence repeat markers. This map was used for QTL analysis, and in both years, results indicated that resistance to common bunt could mostly be attributed to a gene on chromosome 1B, near to the centromere and closest to marker Xgwm273 on the short arm. Additionally, in 2008, smaller QTL effects were ascribed to chromosomes 7A and 7B, and another smaller QTL effect to chromosome 5B in 2009 only.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Common bunt caused by Tilletia tritici (Bjerk.) Wint. and T. laevis Kühn occurs worldwide wherever wheat is grown. This disease reduces both the yield and quality of the wheat grain. Chemical treatment of the seed is widely used to control common bunt; however it is costly and may have adverse effects on the environment. Seed treatment with chemicals is also not allowed under organic farming conditions. Genetic resistance has been used in several countries to control common bunt. Successful resistance breeding has been based on sources of resistance effective to local populations of common bunt. A number of resistance sources are available and fifteen (Bt1–Bt15) resistance genes have been registered (Goates 1996). In addition to these genes, ‘alien’ sources of resistance also have been used in breeding, for example, BtZ originating from Agropyron intermedium (Host) P. Beauv. (Goates 1996), or resistance derived from Aegilops cylindrica Host (Galaev et al. 2006), and other grasses (Babayants et al. 2006).
Mapping of resistance genes to specific chromosomes is important for gene pyramiding in breeding. However, only a limited number of genes or QTL for common bunt resistance have been located to date. Bt1 (Sel. 2092) has been located on chromosome 2B (Sears et al. 1960), and Bt4 (CI 1558) and Bt6 (Rio) on chromosome 1B (Schmidt et al. 1969). Bt4 and Bt6 are linked together with a gene for red glume colour (Schmidt et al. 1969). They can be distinguished using cv. Turkey 3055 as a definite source of Bt4, and cv. Rio as the source of Bt6. Bt5 (Hohenheimer) has been reported by Metzger and Schaller as also being on chromosome 1B (McIntosh et al. 1998). Bt7 (Sel. 50077) has been located on chromosome 2D by Metzger (after McIntosh et al. 1998), and Bt10 (PI 178383 × Elgin) on chromosome 6D (Menzies et al. 2006). Recently two papers appeared that attributed common bunt resistance to chromosome 1B (Fofana et al. 2008; Wang et al. 2009). The former authors also mentioned an effect associated with chromosome 7A.
Mapping of bunt resistance genes was previously mainly carried out using monosomic analysis, but more recently molecular markers have been used. The aim of the work described here was to map resistance to common bunt in the resistant cv. Trintella using SSR markers on a doubled haploid (DH) mapping population developed from a cross with the susceptible variety Piko.
Materials and methods
Bunt experiments
A recombinant doubled haploid population was developed by Limagrain UK Ltd, Docking, UK, from F1s between the varieties Trintella and Piko, using the maize cross technique. This population was used in the experiments described below. Trintella (CB-239/VDH-256-81//RPB-48-75A/Moulin) is resistant to common bunt (Dumalasová and Bartoš 2006; Nielsen 2003). Cv. Piko (CWW-3319.5/3*/Kraka//Maris Huntsman/Frühgold/) is susceptible to common bunt. Seed of 88 independent recombinant DH lines and the parental varieties were tested in the field at the Crop Research Institute in Prague-Ruzyně (Czech Republic) for 2 years (2008 and 2009). Field trials had four replicates, each of them represented by one row of 1 m long.
Before sowing, seed was inoculated with a mixture of teliospores of T. tritici and T. laevis originating from three different locations in the Czech Republic. This mixture was also tested on controls consisting of lines/varieties possessing Bt0–Bt13 genes (Goates 1996) obtained by courtesy of Dr. B. J. Goates. In total, 18 varieties grown in the Czech Republic were included to illustrate the levels of infection over 2 years, 2008 and 2009 (Table 1).
Inoculation was done by shaking seed with a surplus of teliospores (the same amount for each sample) in Erlenmayer flasks for 1–2 min. Inoculation was carried out in early October 2007. One half of each inoculated sample was sown on the 15th October 2007, the second half was kept in a refrigerator at about 7 °C for a year and sown on the 13th October in 2008 in order to avoid labour linked to inoculation in 2008.
For each year, two replications were scored by counting healthy and diseased ears, while the other two replications were used to estimate the percentage of bunted ears. Healthy and bunted ears were counted for the third and fourth replicates only in the case of a discrepancy between the two replicates scored by counting.
Mean numbers of 178 and 145 ears were evaluated per replication in 2008 and 2009, respectively.
Mapping and QTL analysis
The genetic map of the Trintella × Piko DH population was developed using DNA extracted from pooled leaf material from five seedlings of each of DH line using the Qiagen DNeasy 96 Plant kit as described in the protocol ″Purification of total DNA from fresh plant tissue″(DNeasy 96 Plant Handbook). For mapping, publicly available simple sequence repeat (SSR) markers were used aiming for a marker density of one marker every 10–20 cM. Primer sets used were from the John Innes Centre (JIC), Norwich, UK (psp), Leibniz Institute of Plant Genetics and Crop Plant Research (IPK) Gatersleben, Germany (gwm/gdm), Wheat Microsatellite Consortium (wmc), Beltsville Agricultural Research Center, USA (barc) and Institut National de la Recherche agronomique INRA, France (cfd/cfa) collections (GrainGenes website http://www.wheat.pw.usda.gov/). Around 650 primer pairs, amplifying markers distributed across the entire wheat genome, were first screened on the parents. Polymorphic markers were selected and targeted to provide good genome coverage using published consensus maps (Somers et al. 2004) and for their ease of scoring. DNA fragments were amplified using PCR and run on 5 % polyacrylamide gels for separation. The silver staining technique was used to visualize fragments (Bassam et al. 1991), and band scoring for different alleles was done by eye but checked at least twice by separate individuals. Additional markers were provided by Syngenta PLC to fill in gaps (Leahy 2008).
Joinmap v3.0 (Van Ooijen and Voorrips 2001) was used for genetic linkage map construction, using the default settings with the Kosambi mapping function. Linkage groups were selected at a minimum LOD of 3 for reliable associations. QTL effects were estimated using the single marker analysis, interval mapping (IM) and composite interval mapping (CIM) functions of QTL Cartographer version 2.5 (Basten et al. 1994). For both IM and CIM the parameters were set to default, as follows: Threshold levels for significance were set using the permutation test to empirically estimate the genome-wide significance threshold; the walk speed was set to 1 cM, the threshold value settings were set to 500 permutations at a significance level of 0.05 for all traits. For CIM the default Model (Model 6 Standard) was used as cofactors in the analysis. Each year was analysed separately and comparisons of QTL locations across years were aligned on the genetic map of the population relative to the respective QTL confidence limits.
Results
Bunt infection levels on the check and parental varieties
The ranges of disease incidence on the check varieties tested were 4.6–74.9 % and 4.1–85.9 % in 2008 and 2009, respectively (Table 1). There was a close correspondence between the infection levels in the 2 years with a highly significant Pearson correlation coefficient of 0.895.
Following infection, the % infected ears was recorded: Bt0-57.2, Bt1-30.6, Bt2-36.6, Bt3-7.3, Bt4-3.6, Bt5-1.6, Bt6-7.5, Bt7-54.6, Bt8-7.8, Bt9-0.0, Bt10-0.0, Bt11-0.0, Bt12-0.0, and Bt13-5.4.
Inoculation of Piko with a mixture of teliospores of T. tritici and T. laevis showed a mean disease incidence of 70.9 % in the 2008 test and 77.1 % in 2009. These data confirmed the susceptibility of Piko to bunt. The resistant variety Trintella displayed mean infection levels of 8.5 and 16.6 % in 2008 and 2009, respectively. Results from the checks and parents of the recombinant population provided confidence that the experimental data on the DH lines would be an accurate estimate of the genetic differences between lines.
Bunt infection levels on the DH lines
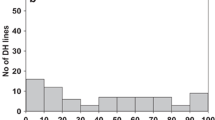
The mean level of infection on the DH lines in 2008 was 49.97 %, with a range from 0.6 to 93.0 %. In 2009 the mean level of infection was 50.05 % with a range from 2.2 to 94.8 %. Thus, the 2 years were quite consistent in mean disease levels. Analysis of variance performed on the field data of the DH population indicated that the DH lines were significantly different in levels of bunt resistance (data not shown), with no significant year effect but a small but significant (P = 2.6 %) interaction, indicating some inconsistencies in performance of some lines over years. Most lines showed a consistent performance over years (Fig. 1) with a few outliers which accounted for the years × lines interaction, probably due to disease escapes. For example, DH10 showed a considerable difference in the infection level over years with a mean disease incidence of 29.3 % in 2008, but much higher at 79.2 % in 2009, indicating that the line most likely escaped infection in 2008 (Fig. 1).
Map development
A genetic map consisting of 29 linkage groups, comprising 144 publically available and 95 proprietary Syngenta polymorphic SSR markers was constructed. Comparisons with published maps and data from GrainGenes allowed these linkage groups to be assigned to the individual 21 wheat chromosomes. Linkage groups could be assigned to all 21 wheat chromosomes, except chromosomes 3A and 3D, with a total map distance of 1379 cM (Leahy 2008). This map was used for the QTL analysis of the field bunt assessment data from 2008 and 2009, using QTL Cartographer.
QTL analysis
The frequency distributions for bunt incidence in the DH population for the two years are shown in Fig. 2. These clearly show bimodality, but with an overlap, suggesting the presence of a single major gene segregating in the cross with the presence of modifying genes of smaller effects. Results of the QTL analysis from using interval and CIM functions in QTL Cartographer (Table 2) confirmed this hypothesis. The field trials of 2008 and 2009 indicated the presence of one major gene for bunt resistance on chromosome 1B with Trintella contributing the resistance allele (Fig. 3). This is effectively a major gene (2009 CIM 1B location at 45 cM, LOD 38), giving an additive effect explaining 29 and 30 % of the bunt infection in 2008 and 2009, respectively.
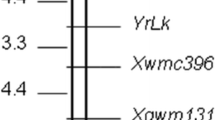
Nevertheless, modifier QTL were also detected, which were different for the 2 years, namely QTL on chromosomes 7A (CIM 7A location 2008 at 44 cM, LOD 2.6; 2-LOD interval 32.7–48.5 cM) near the centromere (nearest marker Xpsp3050), and 7B (CIM 7B 2008 location at 13 M, LOD 2.7; 2-LOD interval 0–23 cM) near the centromere (nearest marker Xgwm43). These were significant in 2008 although also apparent but non-significant in 2009, with Trintella contributing both resistance alleles, both with additive effects of 4 % in 2008. Interestingly, both QTL were near the centromere, and there exists a possibility that they could be homoeoalleles based on their relative positions on the chromosomes, aligned using the consensus maps of Somers et al. (2004) and map data on GrainGenes. There was also one QTL on chromosome 5B (CIM 2009 5B location at 0 cM, LOD 3.4 (at peak); 2-LOD interval 0–19 cM) distal on the long arm (nearest marker Xgwm408) in 2009 (effect also apparent but not significant in 2008), again with the resistance allele coming from Trintella, with an additive effect of 5 % (Table 2). The positions of these three QTL on the respective linkage groups are shown in Fig. 4.
Locations of significant minor bunt resistance QTL (LOD > 2.4) for individual years (upper designation notes the chromosome, lower designation the linkage group). Arrows indicate the location of the peak of the QTL profile. QTL08 and QTL09 relate to QTL detected only in 2008 and 2009, respectively. Map distance in cM are given on the left-hand side of the chromosome, and marker names on the right
Discussion
For our 2008 field trial, most of the resistance to common bunt in this population was attributed to a major gene on chromosome 1B near to the centromere on the short arm, and smaller effects on resistance were ascribed to chromosomes 7A and 7B. In 2009, a major gene was again detected at the same location on chromosome 1B near the molecular marker Xgwm273 and the centromere, and a smaller effect was ascribed to chromosome 5B. As outlined before, the Bt4, Bt5 and Bt6 genes have been described as being on chromosome 1B.
Fofana et al. (2008) used 185 DH spring wheat lines from the cross RL 4452 × AC Domain to locate common bunt resistance in the spring wheat cv. AC Domain. Reaction to common bunt was tested in the field over 2 years. Three QTL were found to be associated with bunt resistance, two of them on chromosome 1B (1BL and 1BS) and the third one on chromosome 7A. They observed a continuous distribution of disease reaction and transgressive segregation for the trait. They defined the results as oligogenic inheritance with additive effects. QTL on chromosome arms 1BL and 1BS explained 29 % of the phenotypic variation, and a QTL located on the long arm of chromosome 7A accounted for 3 % of the phenotypic variation.
Wang et al. (2009) used a DH population from the backcross F1 of 8405-JC3C/Blizzard//2*8405-JC3C to analyze common bunt resistance of the winter wheat cv. Blizzard. Seventy-eight DH lines randomly selected from the 147 lines developed were analyzed. Common bunt resistance in the cv. Blizzard was again located to chromosome 1BS.
In our experiments, chromosome 1B was also found to be the critical chromosome carrying common bunt resistance in cv. Trintella. Another less effective QTL was located on chromosome 7A, a location also described by Fofana et al. (2008); and on 7B or 5B, which were not found by Fofana et al. (2008). However, the gene studied here did not align within QTL confidence limits either with QCbt.crc-1B.1 or QCbt.crc.1B.2 recorded by Fofana et al. (2008), and was located to a separate location between the two genes; their locus on the short arm being much more distal. Similarly, the QTL found here on 7A did not align with the QTL QCbt.crc-7 of Fofana et al. (2008). The locus detected in this study was near the centromere while their locus was distal on the long arm (Somers et al. 2004). Nevertheless, the level of resistance governed by the genes in cvs. AC Domain and Trintella seems to be similar.
Pedigrees of the varieties studied may help to estimate whether the different resistance genes are of the same origin. The pedigree of AC Domain is RL4137/ND499//ND585, and was analyzed in more detail by Fofana et al. (2008). They stated that two lines, Hope (H49-24) and H44-24, derived from the cross Yaroslav Emmer × Marquis and cvs. Regent, Renown and Redman, have been reported as sources of common bunt resistance in Canada. Bunt resistance in them was derived from the cv. Marquis. The high levels of resistance to common bunt in wheat varieties derived from Hope, compared to the parents, was attributed to recombination of genetic factors of the two parents in the form of transgressive segregation. Cv. Red Fife is in the pedigree of cv. Marquis and was derived in Canada from seed from Scotland. Old sources of common bunt resistance used in USA and Canada originate from Europe. Though the places of origin of cvs AC Domain and Trintella are remote from one another, some genetic similarity of bunt resistance in them cannot be therefore excluded. A major difference between our results and those by Fofana et al. (2008) is that they described QTLs of smaller effect, whereas our data indicate the presence of a major dominant gene.
Cv. Blizzard was derived from a cross Utah216C-12-10/Cheyenne/5/PI476212(SM4)/4/Burt/3/Rio/Rex//Nebred, where PI476212 contributed the bunt resistance (Sunderman et al. 1991). The resistance of cv. Blizzard was high in our tests; no infection appeared in the trial where the highest infection was 54.3 % bunted ears (Blažková and Bartoš 2002). Cv. Blizzard has been described as highly resistant not only to common bunt but also to dwarf bunt. None of the European varieties tested by Huber and Buerstmayr (2006) was as highly resistant to dwarf bunt as cv. Blizzard. No data on the dwarf bunt resistance of cv. Trintella was available to us. However, the level of common bunt resistance in Trintella was much lower than in cv. Blizzard. If we presume that the gene in Trintella might be identical to the gene in Blizzard, in spite of the differences described above, then another less effective allele must be present in Trintella but not in Blizzard. In our molecular marker analysis, different molecular markers were used than those by Fofana et al. (2008) and Wang et al. (2009) which complicated the comparison of our results with their results, but some alignment was possible using the consensus maps of Somers et al. (2004).
Wang et al. (2009) stated that further study was required to determine if the gene on 1B in cv. Blizzard was different from Bt4, Bt5 or Bt6. Nevertheless, results by Cichy and Goates (2009) indicated that the gene in Blizzard was Bt12. In the papers by Fofana et al. (2008) and Wang et al. (2009) reasons for difficulties of obtaining comparable results in repeated field tests were described in detail. Similar reasons can be applied to our field trials with the Trintella × Piko DH population, and they probably caused different results regarding QTL with small effects in the two experimental years.
References
Babayants LT, Babayants OV, Baranovskaya VL, Dubinina LA (2006) Tilletia caries and resistance of wheat to this pathogen in Ukraine. Czech J Genet Plant Breed 42(Special Issue):33–36
Bassam BJ, Caetanoanolles G, Greshoff PM (1991) Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal Biochem 196:80–83
Basten CJ, Weir BS, Zeng Z-B (1994) Zmap-a QTL cartographer. Proceedings of the 5th World Congress on Genetics Applied to Livestock Production: Computing Strategies and Software, vol 22. Guelph, pp 65–66
Blažková V, Bartoš P (2002) Virulence pattern of European bunt samples (Tilletia tritici and T. laevis and sources of resistance. Cereal Res Commun 30:335–342
Cichy K, Goates BJ (2009) Evaluation of molecular markers for common bunt resistance genes in diverse wheat genotypes. ASA-CSSA-SSSA International Annual Meetings, 1–5 Nov, Pitsburgh. Available via http://a-c-s.confex.com/crops/2009am/webprogram/Paper54548.html. Accessed February 2010
Dumalasová V, Bartoš P (2006) Resistance of winter wheat cultivars to common bunt, Tilletia tritici (Bjerk.) Wint. and T. laevis Kühn. J Plant Dis Protect 113:159–163
Fofana B, Humphreys DG, McCartney CA, Somers DJ (2008) Mapping quantitative trait loci controlling common bunt resistance in a doubled haploid population derived from the spring wheat cross RL4452 × AC Domain. Mol Breed 21:317–325
Galaev AV, Babayants LT, Sivolap YM (2006) DNA-markers for resistance to common bunt transferred from Aegilops cylindrica Host. to hexaploid wheat. Czech J Genet Plant Breed 42(Specal Issue):62–65
Goates BJ (1996) Common bunt and dwarf bunt. In: Wilcoxson RD, Saari EE (eds) Bunt and smut diseases of wheat: concepts and methods of disease management. CIMMYT, Mexico, pp 12–25
Huber K, Buerstmayr H (2006) Development of methods for bunt resistance breeding for organic farming. Czech J Genet Plant Breed 42(Special Issue):66–71
Leahy F (2008) Inflorescence characteristics which enhance hybrid wheat production. PhD thesis, the University of East Anglia, UK, pp 232
McIntosh RA, Hart GE, Devos KM, Gale MD, Rogers WJ (1998) Catalogue of gene symbols for wheat. Proceedings of the 9th International Wheat Genetics Symposium. Saskatoon, 2–7 August 1998
Menzies JG, Knox RE, Popovic Z, Procunier JD (2006) Common bunt resistance gene Bt10 located on chromosome 6D. Can J Plant Sci 86:1409–1412
Nielsen BJ (2003) Strategies for avoiding seed-borne diseases. 11th Australian Barley Technical Symposium, Conference Proceedings, pp 75–79
Schmidt JW, Morris R, Johnson VA (1969) Monosomic analysis for bunt resistance in derivatives of Turkey and Oro wheats. Crop Sci 9:286–288
Sears ER, Schaller CW, Briggs FN (1960) Identification of the chromosome carrying the Martin gene for resistance of wheat to bunt. Can J Genet Cytol 2:262–267
Somers DJ, Isaac P, Edwards K (2004) A high-density microsatellite consensus map for bread wheat (Triticum aestivum L.). Theor Appl Genet 109:1105–1114
Sunderman DW, Souza DW, Birzer E, Whitmore J (1991) Registration of “Blizzard” wheat. Crop Sci 31:491–492
Van Ooijen JW, Voorrips RE (2001) JoinMap® 3.0, Software for the calculation of genetic linkage maps. Plant Research International, Wageningen
Wang S, Knox RE, DePauw R, Clarke FR, Clarke JM, Thomas JB (2009) Markers to common bunt resistance gene derived from “Blizzard” wheat (Triticum aestivum L.) and mapped to chromosome arm 1BS. Theor Appl Genet 119:541–553
Acknowledgments
We thank Dr. V. Šíp for critical reading of our manuscript and invaluable comments. The research was supported by the Ministry of Agriculture of the Czech Republic, Projects No. MZE 0002700604 and No. QH 71242.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Dumalasová, V., Simmonds, J., Bartoš, P. et al. Location of genes for common bunt resistance in the European winter wheat cv. Trintella. Euphytica 186, 257–264 (2012). https://doi.org/10.1007/s10681-012-0671-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-012-0671-7