Abstract
We used the Roche-454 platform to sequence from normalized cDNA libraries from each of two inbred lines of onion (OH1 and 5225). From approximately 1.6 million reads from each inbred, 27,065 and 33,254 cDNA contigs were assembled from OH1 and 5225, respectively. In total, 3,364 well supported single nucleotide polymorphisms (SNPs) on 1,716 cDNA contigs were identified between these two inbreds. One SNP on each of 1,256 contigs was randomly selected for genotyping. OH1 and 5225 were crossed and 182 gynogenic haploids extracted from hybrid plants were used for SNP mapping. A total of 597 SNPs segregated in the OH1 × 5225 haploid family and a genetic map of ten linkage groups (LOD ≥8) was constructed. Three hundred and thirty-nine of the newly identified SNPs were also mapped using a previously developed segregating family from BYG15-23 × AC43, and 223 common SNPs were used to join the two maps. Because these new SNPs are in expressed regions of the genome and commonly occur among onion germplasms, they will be useful for genetic mapping, gene tagging, marker-aided selection, quality control of seed lots, and fingerprinting of cultivars.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In spite of its economic significance as one of the major vegetable crops worldwide, genetic maps of onion (Allium cepa L.) remain relatively rudimentary. This is due in part to the enormous onion genome, which at 16.3 gigabases per 1C nucleus (Arumuganathan and Earle 1991) makes identification of some molecular markers technically difficult, and the biennial generation time and severe inbreeding depression which slow the development of segregating families. Nevertheless, numerous classes of molecular markers have been developed and mapped in onion, including restriction fragment length polymorphisms (RFLPs), amplified fragment length polymorphisms (AFLPs), simple sequence repeats (SSRs), and single nucleotide polymorphisms (SNPs) (King et al. 1998; Van Heusden et al. 2000; Martin et al. 2005; Baldwin et al. 2012a, b). Molecular markers, such as SSRs and SNPs, are especially useful for onion because they are codominant and efficiently revealed by the polymerase chain reaction. Onion SSRs and SNPs have been used for cultivar identification (Jakše et al. 2005; Mahajan et al. 2009), genetic diversity estimates (Baldwin et al. 2012a), and tagging of chromosome regions affecting economically important traits such as fructan accumulation (Havey et al. 2004; McCallum et al. 2006; Raines et al. 2009), male-sterility restoration (Gökçe et al. 2002), and flavor (Galmarini et al. 2001; McCallum et al. 2007).
Marker-aided selection (MAS) has great potential in onion improvement because of the high cost of harvesting and vernalizing bulbs prior to flowering and completing crosses with insects. High throughput platforms that allow for the genotyping of large numbers of markers across individuals would allow breeders to select plants at an early stage of development and advance to seed production only a fraction of the numbers of plants as compared to classical phenotypic selection. SNPs are the marker of choice for MAS in onion because of codominance, common occurrence among elite germplasms (Martin et al. 2005; Baldwin et al. 2012b), and the availability of commercially available high-throughput genotyping platforms. In this research, we completed transcriptome sequencing of two inbred onion populations, identified SNPs in expressed regions of the onion genome, and extracted gynogenic haploid plants for genetic mapping of these SNPs. Our research has produced a large number of expressed sequences and the most detailed genetic maps based on codominant SNP markers developed to date for onion.
Materials and methods
RNA isolation and cDNA synthesis and normalization
Two long-day onion populations were selected for cDNA synthesis and SNP identification. 5225 is a red onion derived from a cross between North American Spanish and long-storage germplasms, was putatively a doubled haploid derived from the female gametophyte, and was the gift of Seminis Seed Company (Woodland, CA, USA). OH1 is a yellow inbred selected from long-day storage populations and shows relatively high production of gynogenic haploids (Havey and Bohanec 2007). At approximately 6 weeks after planting vernalized bulbs, tissue from leaves, unopened umbels, bulbs, and roots were separately harvested and frozen in liquid nitrogen. RNA extractions, cDNA syntheses, and normalizations were completed by BioS&T (Montreal, Canada). Total RNA was isolated separately from each tissue using the Trizol method (Invitrogen, Carlsbad, CA, USA). RNA concentrations were determined and equal amounts of RNA from each tissue were combined to create an RNA pool for each onion population. These two RNA pools were used for cDNA synthesis using the SMART (Switching Mechanism At 5′ end of RNA Transcript) method (Clontech, Mountain View, CA, USA). Doubled stranded cDNAs were produced by extensions using 5′ cDNA double-stranded adaptor 5′-CAGTGGTATCAACGCAGAGTGGCCATTA CGGCCTAGTT ACGGG-(cDNA)-3′ and 3′-GTCACCATAGTTGCGTCTCACCGGTAAT GCCGGATCAAT GCCC-(cDNA)-5′. The 3′ cDNA double-stranded adaptor was 5′-cDNA-AAAAAAAAAAAAAAAGGCCGCCTCGG CCACTCTGCGTTGATACCACTG-3′ and 3′-cDNA-TTTTTGTGTGTGTTTCCGGCGGA GCCGGTGAGACGCAACTATGGT GAC-5′. The amount and quality of cDNAs were established using agarose-gel electrophoresis and cDNAs over 0.5 kb in size purified from the gel. Normalization of cDNAs was completed by BioS&T using proprietary techniques.
Sequencing of cDNAs and identification of SNPs
The two cDNA libraries were individually barcoded and 2.5 plates (corresponding to 1.25 plate per population) were sequenced at the J. Craig Venter Institute (JCVI) using the Roche 454 FLX platform and protocols as recommended by the manufacturer (Roche, Branford, CN, USA). The 454 reads from 5225 and OH1 were assembled together using the Newbler Assembler (Roche). Well supported SNPs were identified between these two inbreds by keeping only single-base polymorphisms (ignoring indels and multi-base polymorphisms) and completing the filtering steps listed in Table 1. Sequences flanking these SNPs (approximately 60 basepairs on each side of SNP) were compared to the most similar rice genomic sequence to eliminate SNPs near introns; we previously demonstrated that 83 % of introns are shared between onion and rice (Martin et al. 2005). Sequences flanking SNPs were then analyzed by proprietary software programs for the Golden Gate (Illumina, La Jolla, CA, USA) and KASPar (LGC Genomics, Beverly, MA, USA) platforms to identify those conducive for genotyping using the respective platform. The assembled contigs from 5225 and OH1 were annotated by the JCVI EUK-autonaming pipeline using databases of plant proteins from Swissprot and TrEmbl, NCBI NR, and UniRef100.
Extraction of haploids and confirmation of ploidy level
Single plants of OH1 and 5225 were crossed, seed was harvested from the OH1 plant, bulbs were produced in Wisconsin USA, and hybrids identified by red-bulb color. Hybrid bulbs were vernalized for 8 weeks at 5 °C and then shipped to Slovenia for gynogenic haploid extraction as previously described (Jakše et al. 2010). Haploid plants were allowed to form bulbs in a greenhouse in Slovenia. These bulbs were sent to the USA and planted in a greenhouse.
Young expanding leaf tissue was harvested, kept on ice, and immediately prepared for flow-cytometric analyses. Suspensions of nuclei were prepared using CyStain PI absolute P kit (Partec, Swedesboro, NJ, USA). Approximately 1 × 5 cm piece of each leaf was harvested and chopped using a sharp razor blade for 60–90 s in 2 ml extraction buffer in a Petri dish. After incubation at 4 °C for 5 min, the buffer was filtered through a 30 μm CellTrics filter (Partec), and centrifuged for 5 min at 200 RCF. Pellets from individual plants were resuspended in 500 μl staining buffer containing propidium iodide and RNase as recommended by the manufacturer. The stained suspension was incubated at 37 °C for 1 h before analysis using a FACSCalibur flow cytometer with 488 nm Argon laser excitation at the Carbone Cancer Center, University of Wisconsin-Madison. At least 1,200 nuclei were counted per sample. Ploidy level was determined by comparing the histogram of each individual with diploid onions OH1 and 5225 using CellQuest Pro software (BD BioSciences, San Jose, CA, USA).
Marker genotyping
One SNP between 5225 and OH1 was identified on each of 1,256 cDNA contigs and was genotyped using the KASPar platform. DNAs used for SNP genotyping were isolated by CsCl purification (Bark and Havey 1995) from pooled leaf tissues from at least 25 plants from a diverse set of onion populations (Table 2) and from 182 haploid progenies extracted from hybrids between OH1 and 5225. We also genotyped SNPs using DNAs from 57 F2-derived F3 progenies from Brigham Yellow Globe 15–23 × Alisa Craig 43 (BYG15-23 × AC43); this family has been previously used for genetic mapping of RFLPs, SNPs, and SSRs (King et al. 1998; Martin et al. 2005). For haploid progenies selected from different F1 plants, homogeneity of errors (Gomez and Gomez 1984, pages 464–467) was established before pooling segregations across families. Goodness-of-fit to the expected segregation ratios and genetic mapping using the regression algorithm and Kosambi function were completed with the JoinMap® software version 3 (Van Ooijen and Voorrips 2001) for each segregating family. JoinMap was also used to reveal synteny between the two genetic maps from OH1 × 5225 and BYG15-23 × AC43. Linkage groups were assigned to chromosomes based on previous assignments using the BYG15-23 × AC43 family (Martin et al. 2005). Map images were drawn using the MapChart software (Voorrips 2002).
Results
Transcriptome sequencing and annotation
Sequencing of 2.5 454-plates (1.25 plates each for 5225 and OH1) yielded over 1 billion bases of expressed onion sequence (Table 3). The numbers of reads and total numbers of bases were similar for the two cDNA libraries. Sequences are available from Genbank’s Sequence Read Archive accessions SRX188612 and SRX188621 for 5225 and OH1, respectively. Approximately 1.6 million reads from each library assembled into 27,065 and 33,254 contigs for OH1 and 5225, respectively, with average contig length of approximately 1.2 kb. Approximately 12 and 20 % of contigs were unique to OH1 or 5225, respectively. Individual Transcriptome Shotgun Assemblies (TSAs) were deposited at DDBJ/EMBL/GenBank under accessions GAAN00000000 and GAAO00000000, with the first versions described in this research as GAAN01000000 and GAAO01000000 for 5225 and OH1, respectively. About 7.5 % of reads from the combined assembly of OH1 and 5225 remained as singletons. Functional annotations of the cDNA contigs from the combined assembly are listed in Supplemental Table 1. The annotations of 14,357 of the 48,459 (30 %) cDNA contigs remained unknown (i.e., no hit to databases).
SNP identification
Initial assemblies revealed 65,675 SNPs between OH1 and 5225 on 13,861 cDNA contigs. We used the criteria listed in Table 1 to select well supported SNPs between OH1 and 5225, yielding 3,364 SNPs on 1,716 cDNA contigs (Supplemental Table 2) and an average of one SNP per 1.7 kb of expressed sequence. For the Illumina genotyping platform, 1,830 SNPs were identified with designability value of 1 (Supplemental Table 3). For the KASPar genotyping platform, 2,285 SNPs were identified on 1,256 cDNA contigs that were conducive to genotyping using this platform (Supplemental Table 4). One SNP from each of the 1,256 cDNA contigs was randomly selected for genotyping using the KASPar platform (Supplemental Table 5). Of these 1,256 primer sets, 930 produced amplicons and revealed the expected SNPs across the diverse onion populations (Table 2, Supplemental Tables 6 and 7). From these successful amplifications, OH1 and 5225 were heterozygous for 9.8 and 19.9 % of the SNPs; in contrast amplicons from doubled haploids 2107 and H6 which appeared heterozygous for only 1.4 and 1.3 % of the SNPs (Supplemental Table 7).
Extraction of haploids and genetic mapping
Over 400 gynogenic haploids were obtained from 25 hybrid plants from the cross OH1 by 5225. Flow cytometry identified 11 diploid plants out of a random sample 96 gynogenic progenies. These diploid plants were homozygous for all SNPs segregating in the OH1 × 5225 family, indicating that they likely arose from spontaneous doubling of a cell from the female gametophyte. One hundred and eighty-two haploids (48, 44, 32, 30, and 28 haploids extracted from five different hybrid plants) were selected for genetic mapping. Of the 930 primer sets that produced amplicons, 178 were excluded because they did not segregate (158) or produced heterozygous genotypes (20), likely due to amplifications from paralogs, among the haploid progenies. Of the 752 remaining amplicons, 522 (69 %) SNPs fit the expected 1:1 ratio at P > 0.001. For the 230 (31 %) SNPs not fitting the expected 1:1 ratio at P > 0.001, 155 were eliminated because of highly skewed segregation ratios at P < 0.00001. Errors were homogeneous (P > 0.01) for the segregating SNP markers across haploids extracted from independent hybrid plants; therefore segregations were pooled across families.
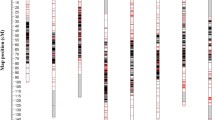
Genetic mapping of 597 SNPs using the haploid progeny DNAs from OH1 × 5225 yielded 10 linkage groups at LOD ≥8.0 (Supplemental Table 8); two linkage groups were assigned to chromosome 4 and two to chromosome 6 based on common markers with the BYG15-23 × AC43 family (described below). SNPs showing significantly (P < 0.001) distorted segregations in the OH1 × 5225 haploid family were concentrated on chromosomes 7 and 8 (Fig. 1; Supplemental Table 8). Many of these distorted markers segregated normally in the BYG15-23 × AC43 family and had essentially the same linkage orders (Fig. 1), indicating that these two genomic regions were likely under selection among haploids from the OH1 × 5225 family. Two small regions on the ends of chromosomes 1 and 6 also showed segregation distortions in the OH1 × 5225 family (Fig. 1).
Genetic maps of single nucleotide polymorphisms (SNPs) segregating in the OH1 × 5225 haploid family (linkage groups on left) and restriction fragment length polymorphisms, simple sequence repeats, and SNPs in the BYG15-23 × AC43 family (linkage groups on right). Genetic distances are in centiMorgans (cM). SNPs segregating in both families are in red and lines correspond to their relative positions in the two maps. Chromosome assignments are based on markers previously assigned to chromosomes using the BYG15-23 × AC43 map (Martin et al. 2005). Blue bars indicate markers showing significant segregation distortion (P < 0.001). Marker names followed by an asterisk and number (*3, *4, and *5) indicate significant distortion from expected ratios at P < 10−3, P < 10−4, and P < 10−5, respectively (color figure online)
A total of 479 (342 phase-known and 137 phase-unknown) markers segregated in the BYG15-23 × AC43 family, of which 339 were new SNPs identified in this research and 140 were previously developed RFLPs, SNPs, or SSRs (Martin et al. 2005), yielding ten linkage groups at LOD ≥4.0 (Supplemental Table 9). Of the SNPs newly identified in this research, 223 also segregated in the haploid families (Supplemental Table 10). The orders of co-segregating SNPs were similar for the OH1 × 5225 and BYG15-23 × AC43 families, although marker orders on chromosomes 2 and 6 were inverted between the two families over relatively short genetic distances (Fig. 1). Common SNPs between these two segregating families allowed for the assignment of linkage groups from OH1 × 5225 to chromosomes (Fig. 1). Eighteen (4 %) markers in the BYG15-23 × AC43 family showed significant (P < 0.001) segregation distortion and many were located on chromosome 5. Again, many of the SNPs in these distorted regions segregated normally in the OH1 × 5225 family and their marker orders were essentially the same (Fig. 1).
Discussion
Due to the extremely large nuclear genome of onion, sequencing of random genomic fragments revealed only 4 % of shot-gun reads showing significant similarities to non-organellar proteins (Jakše et al. 2008). Alternatively, transcriptome sequencing has proven to be an efficient approach to sample lower-copy regions of the onion genome (McCallum et al. 2001). Kuhl et al. (2004) completed 20,000 single pass sequencing reactions from the 5′ end of cDNAs from a normalized library of onion, and these expressed sequences were a good source of SSRs and SNPs for mapping (Martin et al. 2005). Baldwin et al. (2012b) recently reported on transcriptome sequencing of normalized cDNA libraries from doubled haploid and open-pollinated populations of onion. Their results supported transcriptome sequencing as an efficient approach to reveal DNA polymorphisms (SNPs, indels, and cleaved amplified polymorphisms) in expressed regions of the onion genome. We used the same approach as Baldwin et al. (2012b), completing 454 sequencing from two normalized cDNA libraries from two inbred lines (5225 and OH1) of onion. Like Baldwin et al. (2012b), we chose the 454 platform because of longer read lengths in order to aid assembly of random reads. Over 1 billion bases of expressed sequence from onion were generated from our two libraries. All sequences are freely available from Genbank (Sequence read archives SRX188612 and SRX188621 and Transcriptome Shotgun Assemblies GAAN00000000 and GAAO00000000). Numbers of reads from our libraries were twice the number generated by Baldwin et al. (2012b), assembling into over 48,000 contigs (Table 3).
We identified highly confident SNPs by requiring that multiple reads supported variants in OH1 and 5225. Primers flanking 1,256 putative SNPs were synthesized, of which 930 consistently produced amplicons across the diverse set of onion DNAs listed in Table 2, yielding the largest number of SNPs identified to date for onion. We chose to use the KASPar assay to genotype these SNPs (Supplemental Table 6) because this platform is most commonly used by seed companies. Importantly 930 SNPs were present among the evaluated DNAs (Supplemental Table 7), indicating that they will be useful for diversity studies and fingerprinting of onion germplasms. Significant heterozygosity (9.8 and 19.9 %) was revealed within OH1 and 5225, respectively, as expected because onion inbreds retain heterozygosity due to significant inbreeding depression (Jones and Davis 1944). 5225 was chosen as a putative doubled haploid for sequencing and mapping; however this line was heterozygous at many more SNPs than the ~1 % observed in doubled haploids 2107 and H6 (Supplemental Table 7). Heterozygous SNPs in 5225 were concentrated at the ends of linkage groups (Supplemental Table 11), indicating that the cell in the female gametophyte that gave rise to 5225 may have undergone second division restitution (Ramanna and Jacobsen 2003). The approximately 1 % heterozygosity in doubled haploids 2107 and H6 likely arose from amplifications from duplicated regions of the onion genome (King et al. 1998).
We exploited gynogenic haploids for efficient genetic mapping of SNPs. Gynogenic haploids offer numerous advantages for mapping of genetic markers in onion, including rapid development of segregating families and the advantage that each haploid plant represents a gamete. CsCl-purified DNAs from the haploid family can be stored over the long term for use by other research groups for mapping of additional markers and joining of different genetic maps. However, challenges include the difficulty of extracting haploids from many onion populations and doubling chromosome numbers in order to seed propagate progenies (Jakše et al. 2010). The relatively low heritability of gynogenic haploid production is well documented in onion (Bohanec et al. 2003). Because OH1 was selected for high gynogenic haploid production (Havey and Bohanec 2007), we hoped that F1 plants from the cross of OH1 by 5225 would efficiently produce gynogenic haploids showing low segregation distortion. However, this was not observed. Distorted ratios (P < 0.001) were observed for 35 % of the SNPs segregating in the haploid family, as compared to 11 % in the sexually produced F2 family from BYG15-23 × AC43 (Supplemental Tables 8 and 9). Segregation distortion across specific chromosome regions has been commonly observed among haploid progenies (Rivard et al. 1996; Tai et al. 2000).
Flow cytometry identified 11 diploids out of a random sample of 96 gynogenic progenies from OH1 × 5225; this frequency of spontaneous diploid plants from the female gametophyte is close to the previous report of 10 % by Bohanec (2002). The diploids were homozygous for all segregating SNPs indicating that they likely arose from spontaneous doubling of a cell in the female gametophyte, and not from maternal tissue (all polymorphic SNPs would be heterozygous) or from restitution gametes (a proportion of polymorphic SNPs would remain heterozygous).
A total of 713 newly identified SNPs were placed on the genetic map of onion: 597 in the OH1 × 5225 family and 339 in the BYG15-23 × AC43 family. Two hundred and twenty-three SNPs segregated in both families and were used to join the two maps together (Fig. 1) and assign linkage groups to chromosomes (Martin et al. 2005). Numerous aberrantly segregating SNPs (P < 0.001) in the OH1 × 5225 or BYG15-23 × AC43 families segregated normally in the other family, mapped to the same linkage groups, and were largely syntenic (Fig. 1). This observation indicates that different chromosome regions were under selection during the extraction, growth, or propagation of gynogenic haploids from OH1 × 5225, or during selfing to produce the BYG15-23 × AC43 family, resulting in skewed segregation ratios across specific regions.
Significant effort has gone into the development and mapping of molecular markers in onion, including RFLPs (King et al. 1998), randomly amplified DNAs (Bradeen and Havey 1995), AFLPs (Van Heusden et al. 2000), SSRs (Jakše et al. 2005; Martin et al. 2005; McCallum et al. 2008; Baldwin et al. 2012a), and SNPs (Martin et al. 2005; McCallum et al. 2008; Baldwin et al. 2012b). Our research has produced the largest number of robust, commonly occurring SNPs in onion, adding significantly to the 43 and 93 SNPs mapped by Martin et al. (2005) and Baldwin et al. (2012b), respectively. All eight chromosomes of onion are relatively well covered by these newly identified SNPs (Fig. 1). Because these SNPs are in expressed regions of the genome and commonly occur among elite germplasms, they will be useful for the development of high throughput genotyping platforms for gene tagging, marker-aided selection, and fingerprinting of onion.
References
Alan A, Lim M, Mutschler M, Earle ED (2007) Complementary strategies for ploidy manipulations in gynogenic onion (Allium cepa L.). Plant Sci 173:25–31
Arumuganathan K, Earle ED (1991) Nuclear DNA content of some important plant species. Plant Mol Biol Rep 9:208–218
Baldwin S, Pither-Joyce M, Wright K, Chen L, McCallum J (2012a) Development of robust genomic simple sequence repeat markers for estimation of genetic diversity within and among bulb onion (Allium cepa L.) populations. Mol Breed 30:1401–1411
Baldwin S, Revanna R, Thomson S, Pither-Joyce M, Wright K, Crowhurst R, Fiers M, Chen L, MacKnight R, McCallum J (2012b) A toolkit for bulk PCR-based marker design from next-generation sequence data: application for development of a framework linkage map in bulb onion (Allium cepa L.). BMC Genomics 13:637
Bark OH, Havey MJ (1995) Similarities and relationships among open-pollinated populations of the bulb onion as estimated by nuclear RFLPs. Theor Appl Genet 90:607–614
Bohanec B (2002) Doubled haploid onions. In: Rabinowitch HD, Currah L (eds) Allium crop science: recent advances. CAB International, Wallinford, pp 145–157
Bohanec B, Jakše M, Havey MJ (2003) Genetic analyses of gynogenetic haploid production in onion. J Am Soc Hortic Sci 128:571–574
Bradeen JM, Havey MJ (1995) Randomly amplified polymorphic DNA in bulb onion and its use to assess inbred integrity. J Am Soc Hortic Sci 120:752–758
Galmarini CR, Goldman IL, Havey MJ (2001) Genetic analyses of correlated solids, flavor, and health-enhancing traits in onion (Allium cepa L.). Mol Gen Genomics 265:543–551
Gökçe AF, McCallum J, Sato Y, Havey MJ (2002) Molecular tagging of the Ms locus in onion. J Am Soc Hortic Sci 127:576–582
Gomez K, Gomez A (1984) Statistical procedures for agricultural research, 2nd edn. Wiley, New York
Havey MJ, Bohanec B (2007) Onion inbred line ‘B8667 A&B’ and synthetic populations ‘Sapporo-Ki-1 A&B’ and ‘Onion Haploid-1′. HortScience 42:1731–1732
Havey MJ, Galmarini CR, Gökçe AF, Henson C (2004) QTL affecting soluble carbohydrate concentrations in stored onion bulbs and their association with flavor and health-enhancing attributes. Genome 47:463–468
Jakše J, Martin W, McCallum J, Havey MJ (2005) Single nucleotide polymorphisms, indels, and simple sequence repeats for onion cultivar identification. J Am Soc Hortic Sci 130:912–917
Jakše J, Meyer JDF, Suzuki G, McCallum J, Cheung F, Town CD, Havey MJ (2008) Pilot sequencing of onion genomic DNA reveals fragmented transposable elements, low gene densities, and significant gene enrichment after methyl filtration. Mol Genet Genomics 280:287–292
Jakše M, Hirschegger P, Bohanec B, Havey MJ (2010) Evaluation of gynogenic responsiveness of selfed doubled haploid onion lines, chromosome doubling procedure via somatic regeneration, and male fertility. J Am Soc Hortic Sci 135:67–73
Jones HA, Davis G (1944) Inbreeding and heterosis and their relation to the development of new varieties of onions. USDA Tech Bull No. 874
King JJ, Bradeen JM, Bark O, McCallum JA, Havey MJ (1998) A low-density genetic map of onion reveals a role for tandem duplication in the evolution of an extremely large diploid genome. Theor Appl Genet 96:52–62
Kuhl JC, Cheung F, Yuan Q, Martin W, Zewdie Y, McCallum J, Catanach A, Rutherford P, Sink KC, Jenderek M, Prince JP, Town CD, Havey MJ (2004) A unique set of 11,008 onion (Allium cepa) ESTs reveals expressed sequence and genomic differences between monocot orders Asparagales and Poales. Plant Cell 16:114–125
Mahajan V, Jakše J, Havey MJ, Lawande KE (2009) Genetic fingerprinting of Indian onion cultivars using SSR markers. Indian J Hortic 66:62–68
Martin W, McCallum J, Shigyo M, Jakše J, Kuhl JC, Yamane N, Sink KC, Town CD, Havey MJ (2005) Genetic mapping of expressed sequences in onion and in silico comparisons show scant colinearity with rice. Mol Genet Genomics 274:197–204
McCallum J, Leite D, Pither-Joyce M, Havey MJ (2001) Expressed sequence markers for genetic analysis of bulb onion (Allium cepa). Theor Appl Genet 103:979–991
McCallum J, Clarke A, Pither-Joyce M, Shaw M, Butler R, Brash D, Scheffer J, Sims I, van Heusden S, Shigyo M, Havey MJ (2006) Genetic mapping of a major gene affecting onion bulb fructan content. Theor Appl Genet 112:958–967
McCallum J, Pither-Joyce M, Shaw M, Kenel F, Davis S, Butler R, Scheffer J, Jakše J, Havey MJ (2007) Genetic mapping of sulfur assimilation genes reveals a QTL for onion bulb pungency. Theor Appl Genet 114:815–822
McCallum J, Thomson S, Pither-Joyce M, Kenel F, Clarke A, Havey MJ (2008) Genetic diversity analysis and single-nucleotide-polymorphism marker development in cultivated bulb onion based on expressed sequence tag–simple sequence repeat markers. J Am Soc Hortic Sci 133:810–818
Raines S, Henson C, Havey MJ (2009) Genetic analyses of soluble carbohydrate concentrations in onion bulbs. J Am Soc Hortic Sci 134:618–623
Ramanna MS, Jacobsen E (2003) Relevance of sexual polyploidization for crop improvement—a review. Euphytica 133:3–18
Rivard SR, Cappadocia M, Landry BS (1996) A comparison of anther culture derived, selfed, and hybrid progenies of Solanum chacoense. Genome 39:611–620
Tai GCC, Seabrook JEA, Aziz AN (2000) Linkage analysis of anther-derived monoploids showing distorted segregation of molecular markers. Theor Appl Genet 101:126–130
Van Heusden AW, Van Ooijen JW, Vrielink-van-Ginkel R, Verbeek WHJ, Wietsma WA, Kik C (2000) A genetic map of an interspecific cross in Allium based on amplified fragment length polymorphism (AFLP) markers. Theor Appl Genet 100:118–126
Van Ooijen JW, Voorrips RE (2001) JoinMap® 3.0. Software for the calculation of genetic linkage maps. Plant Research International, Wageningen
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78
Acknowledgments
We gratefully acknowledge the support of grant 2008-51180-04875 from the USDA Specialty Crops Research Initiative and scholarship support for JD from the Strategic Scholarships for Frontier Research Network of Thailand’s Commission on Higher Education.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by G. Bryan.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Duangjit, J., Bohanec, B., Chan, A.P. et al. Transcriptome sequencing to produce SNP-based genetic maps of onion. Theor Appl Genet 126, 2093–2101 (2013). https://doi.org/10.1007/s00122-013-2121-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-013-2121-x