Abstract
Blooming time is one of the most important agronomic traits in almond. Biochemical and molecular events underlying flowering regulation must be understood before methods to stimulate late flowering can be developed. Attempts to elucidate the genetic control of this process have led to the identification of a major gene (Lb) and quantitative trait loci (QTLs) linked to observed phenotypic differences, but although this gene and these QTLs have been placed on the Prunus reference genetic map, their sequences and specific functions remain unknown. The aim of our investigation was to associate these loci with known genes using a candidate gene approach. Two almond cDNAs and eight Prunus expressed sequence tags were selected as candidate genes (CGs) since their sequences were highly identical to those of flowering regulatory genes characterized in other species. The CGs were amplified from both parental lines of the mapping population using specific primers. Sequence comparison revealed DNA polymorphisms between the parental lines, mainly of the single nucleotide type. Polymorphisms were used to develop co-dominant cleaved amplified polymorphic sequence markers or length polymorphisms based on insertion/deletion events for mapping the candidate genes on the Prunus reference map. Ten candidate genes were assigned to six linkage groups in the Prunus genome. The positions of two of these were compatible with the regions where two QTLs for blooming time were detected. One additional candidate was localized close to the position of the Evergrowing gene, which determines a non-deciduous behaviour in peach.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Of the Prunus crop species, almond [P. dulcis (Mill) D. A. Webb; syn. P. amygdalus Batsch] has the earliest blooming time. Its flowers are frequently destroyed by late frosts, which severely compromises nut production. Consequently, the manipulation of late blooming is a major target for almost all almond breeding programmes (Kester and Gradziel 1996).
In almond, as in most woody perennials, the physiology and biochemistry of the flowering process is poorly understood, although some investigations have been directed towards elucidation of the genetic control of the determination of flowering time. Variations in blooming time in almond occur due to the differences in the chilling and heat requirements before blooming (Tabuenca 1972; Tabuenca et al. 1972), thereby resulting in a situation where the blooming date may change from year to year depending on the winter weather conditions. However, the blooming sequence of any specific variety is relatively constant over time.
Blooming time is considered to be inherited quantitatively in the majority of fruit species (Anderson and Seeley 1993), and most of the results obtained in almond also confirm this type of transmission (Grasselly 1978). Kester (1965), however, suggested that in some progenies of the late-blooming budsport Tardy Nonpareil, a single dominant gene could be involved in determining the blooming date. He based his conclusion on the bimodal distribution of blooming dates among the progenies he studied. This segregation pattern was also found by Grassely (1978) and Sócias I Company et al. (1998), who followed the transmission of the trait in several progenies. Thus, blooming time in Tardy Nonpareil seems to be determined by a major gene, denoted Lb (with late blooming being dominant over early blooming), and by several quantitatively inherited modifier genes.
The Lb gene was first mapped using the progeny from a cross between D-3-5 (a Lb/lb heterozygous genotype) and Bertina (a Spanish cultivar of unknown origin) (Ballester et al. 2001) and found to be located on Prunus linkage group 4 (G4). Blooming time has also been studied as a quantitative character in an intraspecific peach F2 population (Dirlewanger et al. 1999), in an interspecific Prunus persica × P. ferganensis backcross (Verde et al. 2002) and in the F2 of the interspecific cross between the almond variety Texas and the peach variety Earlygold (Joobeur 1998). However, the sequence and specific function of Lb and the genes responsible for the quantitative trait loci (QTLs) remain unknown.
One of the possible strategies that can be adopted to identify these genes is the candidate gene (CG) approach. CGs are sequenced genes of known or presumed function that are related with the traits of interest and which could correspond to major loci or QTLs. In this context, genes isolated in model species constitute putative CGs for agronomic species. Most of what we know about flowering has been learned by studying the model plant Arabidopsis thaliana. In Arabidopsis, many genes have been characterized and found to play important roles in flowering regulation (for reviews, see Levy and Dean 1998; Mouradov et al. 2002). These genes can be classified into four groups: flowering-time genes, meristem-identity genes, flower-organ-identity genes and late-acting genes (control of ovule development). Therefore, the question that must be addressed is just how to choose with some degree of reliability which kind of genes should be candidates for Lb and/or QTLs for flowering time in almond. While the obvious strategy would be to look within the flowering-time gene group, the fact can not be ignored that, although flowering in perennial plants may have mechanisms in common with those found in annuals, significant differences must always remain a possibility and similar genes may play different roles in different stages. The difficulty in establishing a perfect match between annuals and perennials with respect to the regulation of flowering results in the genes within the three first categories being putative CGs.
In the CG approach the working hypothesis assumes that a polymorphism within the CG sequence is correlated with the phenotypic variation. An efficient strategy for mapping CGs consists of discovering single nucleotide polymorphisms (SNPs) or insertion/deletion events (INDELS) within their sequence among the parents of a mapping population. A role for the CG in the expression of a trait is supported if there is a correlation between phenotype and the allelic polymorphism—i.e., if they cosegregate in a mapping population. In the absence of such a correlation, the CG is ruled out as being involved in trait variation. CGs may have different origins: (1) when the biochemical and/or physiological pathways related to a trait of interest are well known, genes characterized in model species probably have counterparts in species of agronomic interest; (2) genes identified when comparing plants under different conditions or mutants and wild type, by differential display or by microarray experiments.
The availability of whole genome sequences and expressed sequence tag (EST) databases for important crops is accelerating the process of gene discovery. The Prunus Genome Database (http://www.genome.clemson.edu/) provides access to genomic data of peach and almond and constitutes a very useful source of information for studies on Prunus. To date, this database included 9,984 ESTs from a peach mesocarp cDNA library and 2,794 ESTs from an almond developing seed cDNA library. Prunus EST sequences are also available though the GenBank EST database.
The purpose of the investigation reported here was to understand the molecular basis underlying variations in blooming time in almond. We have localized ten CGs putatively homologous to flowering regulatory genes characterized in other plants on the Prunus reference map (Joobeur et al. 1998). The positions of these CGs were compared with those of Lb gene and QTLs and the results discussed within this framework.
Materials and methods
Plant material and DNA isolation
A F2 mapping population of 75 individuals derived from an almond [Prunus dulcis (Mill) D. A. Webb; syn. P. amygdalus Batsch] ‘Texas’ × peach ‘Earlygold’ (T×E) interspecific cross was used for mapping the candidate genes. Trees of the mapping population are maintained in the field at IRTA-Cabrils. Total DNA was extracted from young leaves collected shortly after blooming following the method described by Viruel et al. (1995). Total RNA was extracted from mature flower buds and pistils according to the method of Gévaudant et al. (1999).
Isolation of an almond cDNA homologous to LEAFY
The amino acid sequence of the Arabidopsis LEAFY gene (Weigel et al. 1992) was aligned with the homologous sequences from Pinus radiata (accession no. AAB68601), Populus balsamifera (accession no. AAB51533), Pisum sativum (accession no. AAC49782), Lycopersycon esculentum (accession no. AAF66099), Eucalyptus globulus (accession no. AAC31359) and Malus × domestica (accession nos. BAB83096 and BAB83097) using clustal w (Thompson et al. 1994). Based on the amino acid similarity, the conserved regions HPFIVTEP and WYVPTKLRQ were identified. Using codehop software (http://www.blocks.fhcrc.org/codehop.html), we designed the degenerate primers 5′-CAYCCNTTYATHGTRACDGAGC-3′ (forward) and 5′-TGNCGGAGCTTGGTNGGNACRTAC-3′ (reverse) to target these regions for use in PCR amplification on almond genomic DNA. PCR conditions were adjusted empirically to obtain mainly fragments of the expected size. The PCR products were analysed on a 2% agarose gel in 1× TBE (45 mM Tris-base, 45 mM boric acid, 1 mM EDTA, pH 8.0), and bands of the expected size were excised and gel-purified with the QIAquick Gel Extraction kit (Qiagen, Courtaboeuf, France). Fragments were cloned into the pCR 2.1 vector (Invitrogen, Carlsbad, Calif.) and sequenced. Sequence analyses revealed that a fragment identical to that of the LEAFY gene had been amplified. Based on the nucleotide sequence of the fragment, specific primers were designed for amplification of the corresponding cDNA. 3′-Rapid amplification of cDNA ends (RACE) was performed using the SMART RACE cDNA Amplification kit (Clontech, Palo Alto, Calif.) according to the manufacturer’s instructions. cDNAs were synthesized from total RNA extracted from almond mature flower buds and adaptor-ligated following the instructions of the kit.
Isolation of an almond cDNA homologous to MADS-box genes
The degenerate primers 5′-AARATHGARATHAARMGGATCGARAA-3′ (forward) and 5′-TCRTAGAGRCGGCCDCKGCTDGAGAA-3′ (reverse) described by Kitahara and Matsumoto (2000) for amplification of MADS-box genes in rose were used for PCR amplification on almond genomic DNA. The primers correspond to the sequences KIEIKRIEN and FSSRGRLYE, two regions from the MADS consensus sequence containing the characteristic amino acids within the class-C floral-identity genes (Theissen et al. 1996). PCR conditions, cloning and sequence analyses were as already described. The amplified genomic fragment revealed sequence homology with MADS-box genes from different species. A corresponding cDNA was amplified by RACE, as described in the preceding section, but with total RNA from pistils.
Prunus ESTs homologous to flowering regulatory genes
A dataset for both the almond and peach EST databases containing information on sequence, assembly and homology data is available to users. Homology data displays the results of most of the significant matches (EXP<1e−9) for each contigue and individual clone when compared against the GenBank nr protein database, using the fastx 3.4 algorithm.
Both of these EST databases were screened and ESTs chosen based on their putative identity reported on the datasets. Sequences of selected ESTs were downloaded and stored. The deduced amino acid sequence of each EST was compared with the GenBank nr protein database but using the blast algorithm. The best match at the overall amino acid sequence level was recorded.
Design of PCR primers
Specific primers designed to target CG fragments were used for amplification of CGs from parental lines of the mapping population. To increase the probability of polymorphism detection between the ‘Texas’ and ‘Earlygold’ genotypes, the following rules were applied when choosing the primers: (1) for sequences going throughout 3′ untranslated regions (UTRs) primers were designed in this gene-specific region; (2) primers were chosen to preferentially amplify genomic regions flanking one or more introns. Putative positions of the introns on peach and almond genes were inferred from homologous genes. Primers were designed using the software primer 3 (Rozen and Skaletsky 2000).
Discovery of polymorphisms between parental lines
Genomic fragments of each CG were PCR amplified from both parental lines and the hybrid under the following conditions: 25 ng of genomic DNA was amplified by PCR in a 25-μl final reaction volume containing 100 mM Tris-HCl (pH 8.3), 500 mM KCl, 2.5 mM MgCl2, 200 μM of each dNTP, 400 nM of each primer and 1.25 U Taq DNA polymerase. The PCR conditions consisted of a preliminary denaturation step of 5 min at 94°C, followed by 30 cycles of 30 s at 94°C (denaturation), 30 s at variable temperatures (annealing) (Table 2) and 2 min at 72°C (extension), with a final extension for 5 min at 72°C. PCR products were analysed by electrophoresis in a 1.5% agarose/1.5% NuSieve (FMC, Rockland, Me.) gels in 1× TBE and then visualized by ethidium bromide staining.
Bands of the expected size were sequenced, either directly from the PCR product or after cloning. For cloning, the PCR product was purified using the QIAquick PCR purification kit (Qiagen) and cloned into the pGEM-T Easy Vector (Promega, Madison, Wis.). Inserts were sequenced using the ABI Prism Big DyeTerminator Cycle Sequencing Ready Reaction kit (PE Applied Biosystems, Foster City, Calif.) and an ABI Prism 377 DNA Sequencer (PE Applied Biosystems). A blast search was conducted on either the nucleotide or the predicted amino acid sequences to confirm homologies. Sequence data was analysed with the software package gcg ver. 9.1 (Genetics Computer Group, University of Wisconsin, Madison, Wis.). ‘Texas’ and ‘Earlygold’ genomic fragments were compared for DNA polymorphisms discovery.
Generation of markers
SNPs or small-scale INDELs creating differential restriction enzyme recognition sites between ‘Texas’ and ‘Earlygold’ were used to developed cleaved amplified polymorphic sequence (CAPS) markers. Genomic fragments of CGs amplified from both parental lines were further digested with restriction endonucleases using the recommended temperatures for 3−4 h, according to the manufacturer’s instructions. CAPS markers were separated on agarose gels, stained and observed as described above.
Large INDEL polymorphisms were detected on the basis of band mobility shifts. For simple sequence repeat (SSR) markers, specific primers were designed flanking the SSR sequence and PCR products were size separated by polyacrylamide gel electrophoresis (PAGE) in a 6% gel containing 7.5 M of urea in 0.5× BE buffer.
Inheritance and linkage analysis for candidate genes
The markers were assayed in the T×E mapping population. Segregation data for each candidate gene was added to the corresponding dataset of the markers (Joobeur et al. 1998). Linkage analysis was performed using the software package mapmaker/exp 3.0 (Lander et al. 1987). Assignment of the CGs to the T×E linkage groups was done with the group command with a LOD≥7.0. The commands try and ripple were then used to assign the position of each CG in the T×E previously established marker order. Recombination units were converted into map distances using the Kosambi (Kosambi 1944) function.
Results
Isolation of almond cDNAs putatively homologous to the LEAFY and MADS-box genes
Using the degenerated primers described in the Materials and methods we were able to isolate two genomic fragments from almond. One of the fragments was highly identical to LEAFY genes while the other showed sequence homology with a conserved region of the MADS-box genes. Sequences of these two fragments were used to design specific primers targeting the coding regions. Primers were then used to extend fragments by RACE.
A 3′-end cDNA was amplified from a floral meristem RACE library and named PrdLFY (Prunus dulcis LEAFY). A second 3′-end cDNA was amplified from a pistil RACE library and named PrdMADS1 (Prunus dulcis MADS1). PrdLFY has a length of 750 bp and encodes a polypeptide 85% identical to the apple LFY homologue (Wada et al. 2002). PrdMADS1 has a length of 937 bp, and its predicted polypeptide was found to be 72% identical to MdMADS10 from apple (Yao et al. 1999) (Table 1). MdMADS10 was reported to be the apple homologue of AGAMOUS-like 11 from Arabidopsis. Sequence identities were determined with the blast algorithm by searches throughout non-redundant databases.
Candidate genes selected for mapping
In total, ten CGs (listed in Table 1) were selected for mapping.
Two of the CGs selected for mapping were the two cDNAs isolated from almond, PrdLFY and PrdMADS1. PrdLFY was chosen on the basis of its sequence homology to LEAFY-like genes, since LEAFY is considered to be a key regulatory gene for flowering in Arabidopsis. In fact, the constitutive expression of LFY genes was shown to be sufficient to promote flower initiation in Arabidopsis as well as in perennial species (Weigel and Nilsson 1995; Peña et al. 2001). AGAMOUS11-like from Arabidopsis has a role in the control of ovule and seed development, and its apple homologue (MdMADS10) is expressed only in the fruit core that develops from ovary tissue and contains the seeds (Yao et al. 1999). This may suggest function conservation. Nevertheless, the putative almond homologue (PrdMADS1) was isolated from mRNAs being expressed in the pistils, which may suggest a putative new function. PrdMADS1 was selected as a CG since a putative function in the control of flowering time could not be ruled out.
Eight of the Prunus ESTs eight were selected as CGs. Each EST was designated by the initials Prd or Prp (for Prunus dulcis or Prunus persica, respectively) followed by the initials of the gene for which it showed sequence homology (names of the Arabidopsis genes were used to facilitate identification). Results of blast searches thought the GenBank nr protein database are given in Table 1; for each CG, the best result is indicated. The ESTs selected as CGs were putatively homologous to either flowering-time genes or to genes known to be involved in the response to light quality (PrpFAR1) or to genes from the gibberellins’ biosynthetic pathway (PrdGA20).
Amplification of CG in the parental lines
Specific primers were designed to amplify CGs from Texas and Earlygold genomic DNA. Primer sequences, size of the cDNA fragment within the primers, size of the amplified geomic fragments and annealing temperatures are given in Table 2 for each CG.
For all CGs only a single product was amplified from genomic DNA of both parents and the F1 hybrid. All ‘Texas’ and ‘Earlygold’ PCR products were sequenced, and the homologies previously reported were confirmed by blast analyses. Amplified fragments ranged in size from 282 bp to 1,260 bp (Table 2). For eight of the ten CGs, PCR fragments amplified from genomic DNA were longer (94–931 bp; Table 2) than the cDNA found within the primer sequences. This indicates the presence of one or more introns in the genomic fragments.
Detection of polymorphisms within the CGs
Sequence analysis of the genomic fragments amplified from both parental lines revealed that most of the polymorphisms involved base substitutions, resulting in the gain or loss of a restriction site rather than length variation. For six of the CGs (PrdMADS1, PrpAP1, PrpAGL2, PrpFAR1, PrdGA20, PrpCO) it was possible to identify SNPs within an enzyme recognition site, thereby enabling their conversion into CAPS markers; the restriction endonucleases used are listed in Table 2. For three of the remaining CGs, PrdLFY, PrpFT and PrdTFL, we detected length polymorphisms in the intronic region on the basis of band mobility shifts. The INDELS identified were used to map these three CGs. PrpAP2 also showed a length polymorphism caused by an SSR located in an intronic region.
Assignment of CGs to linkage groups
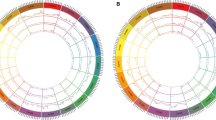
The ten candidate genes were assigned to six of the eight linkage groups in the ‘Texas’ × ’Earlygold’ map. Both the map distances from the origin of the linkage group of the T×E map (Joobeur et al. 1998) as well as the closest reference markers are given in Table 2. Candidate genes were distributed throughout all of the linkage groups except for linkage groups 4 (G4) and 8 (G8) (Table 2, Fig. 1). PrpCO and PrdMADS1 mapped to G1, PrdGA20 to G2 and PrpAGL2 to G3. PrdLFY, PrpAP1 and PrdTFL all mapped to G5, PrpAP2 and PrpFT mapped to G6 and PrpFAR1 mapped to G7.
Linkage groups of the ‘Texas’ × ’Earlygold’ Prunus reference map with the approximate position of the Lb and Evg genes (grey boxes) and the QTLs for blooming time as found by Joobeur (1998) (two thin vertical lines), Dirlewanger et al. (1999) (single thin vertical line) and Verde et al. (2002) (thin and wide vertical lines). The position of the highest LOD for each QTL is indicated with a horizontal line. Names and positions of the candidate genes are in boxes
Discussion
Once linkage mapping identifies a QTL, there are relatively few options for determining the identity of the gene or genes that are actually responsible for variation in the phenotype. Map-based cloning is one possible strategy that can be adopted to accomplish this goal; however, it is difficult to apply the the case of species with a long life cycle, such as fruit trees. The placement of CGs onto QTL maps have proven useful for finding associations between genes involved in relevant pathways and QTLs in fruit trees (Frewen et al. 2000; Etienne et al. 2002). As a first approach to identify floral CGs, we used degenerate primers to target conserved regions of genes identified in model plants as playing important roles in the control of flowering time (LEAFY, APETALA1, TERMINAL FLOWER, AGAMOUS). Using this strategy two almond cDNAs putatively homologous to LEAFY and AGAMOUS-LIKE 11 (AGL 11) genes were isolated, but attempts to optimize conditions and isolate other genes (APETALA1 and TERMINAL FLOWER) were unsuccessful (data not shown).
Studies of comparative mapping lead to the conclusion that, at the genome level, the Prunus genus can be treated as a single genetic entity (Dirlewanger et al. 2004). Because of the high synteny observed, the availability of a Prunus EST database became an excellent alternative strategy for identifying floral CGs for almond. After screening this database, we selected eight ESTs that showed high sequence homology with genes known to be involved in the control of flowering in other species. The two almond cDNAs putatively homologous to LEAFY and AGL 11 genes as well as the eight Prunus ESTs were considered CGs for the control of flowering time in almond and placed in the Prunus genetic map. All ten CGs were polymorphic between the parental lines, which allowed their segregation analysis in the T×E population. One important factor for this success was the high level of polymorphism of this progeny due to its interspecific nature. The advantage of using this population was also demonstrated by Etienne et al. (2002) who used it to map 12 of 18 CGs related to the variability of sugar and organic acid contents in peach. According to these authors, the same CGs were monomorphic in a intraspecific peach population also used for the analysis of these traits. The selection of primers for the amplification of sequences containing 3′UTRs or intronic regions also contributed to the attaining of a high level of polymorphism. The polymorphisms detected in most (60%) of these sequences were SNPs, while the remainder were indels, one of which corresponded to a microsatellite region. For all the SNPs it was possible to develop CAPS markers. Although CAPS have been used successfully for this purpose (Tragoonrung et al. 1992; Gilpin et al. 1997), some authors (Plomion et al. 1999; McCallum et al. 2001) reported their inefficiency when compared to single-stranded conformation polymorphisms (SSCPs). We found CAPS to be very useful and efficient for mapping CGs. The initial investment in sequencing all of the PCR products and obtaining their restriction maps was later a clear advantage since it avoided testing a battery of restriction enzymes, which is costly in both time and money. Moreover, CAPS are technically easier than SSCPs since after digestion the PCR products are resolved in a normal agarose gel.
Joobeur (1998) found QTLs affecting blooming time in four positions (G1, G4, G6 and G7) of the Prunus map using the TxE progeny (Fig. 1). More QTLs for this character have been described in other Prunus populations that were comparable to the positions on the TxE map based on anchor markers: two QTLs (G2 and G7) were detected by Dirlewanger et al. (1999) and one by Verde et al. (2002) in G4. It is interesting that some of these QTLs, although they originate from different genetic backgrounds involving at least three species (almond, peach and P. ferganensis), are located at similar map positions (G4 and G7). This suggests that the same genes may affect blooming time in different species. A QTL found by Joobeur (1998) and another one found by Verde et al. (2002) are located in the neighbourhood of the Lb gene, which indicates that the allele of ‘Tardy Nonpareil’ may be a macromutation (Tanksley 1993) of a gene that produces major effects compared to those of the alleles detected in the other peach progenies.
Despite the wide distribution of our ten CGs throughout the Prunus genetic map, no significant association was found with the Lb gene. In addition to Lb, 27 other major genes have been studied in different populations of various species and integrated into the TxE map (Dirlewanger et al. 2004). One of these genes is Evergrowing (Evg). Evergrowing peach is a mutant that exhibits a different growing winter pattern from that of wild-type deciduous peach. The terminal apices continue to grow in winter until killed by low temperatures, whereas the lateral buds become dormant. Moreover, the flower buds require fewer chilling hours, and evergrowing trees bloom earlier than deciduous trees (Wang et al. 2002a). This gene was first mapped using the progeny from a cross between a deciduous peach (‘Empress op op dwarf’) and an evergrowing peach (P.I. 442380). The microsatellite marker pchgms12 was found to be tightly linked (1 cM) to Evg (Wang et al. 2002a, b). This marker is polymorphic in TxE and maps on G1 at a distance 0.6 cM from the PrdMADS1 CG (W. Howad and P. Arús, unpublished results) (Fig. 1). Considering that Evg influences both cold hardiness and dormancy, it is interesting to speculate about its putative identity as a MADS-box gene. It is known that besides the role of MADS-box genes in floral organ and fruit development they also are likely to be involved in many other aspects of plant development. Therefore, the identity of PrdMADS1 should be confirmed and its function accessed.
With respect to the QTLs, two associations were observed, one in G7 with PrpFAR1 and another in G6 with PrpAP2 (Fig. 1, Table 2).
APETALA2 belongs to the group of meristem-identity genes. In Arabidopsis it promotes the establishment of the floral meristem in collaboration with Apetala1, Leafy and Cauliflower1. During flower development, AP2 is essential for the determination of sepals and petals (Bowman 1993). The co-location we found between the CG PrpAP2 and the QTL for blooming time in the G6 has a limited significance. The highest LOD for the QTL is located at the top of the linkage group, far away from the location of the CG. The role of a putative almond APETALA2 homologue for the control of blooming time is difficult to envisage. Nevertheless, a putative role in almond flowering is likely. To answer whether flowering in almond may involve molecular mechanisms similar to those described in annuals and if these putative orthologues are in fact involved in the process, PrpAP2 must be further characterized.
In contrast, the co-location of the CG PrpFAR1 with the QTLs in G7 seems to be more significant, since the CG is located relatively close to the maximum LOD of one of the QTLs. In Arabidopsis, FAR1 encodes a nuclear protein specific to phytocrome A signalling (Hudson et al. 1999). The Phytocrome A gene belongs to the flowering-time gene group and is involved in the control of flowering time through the photoperiod pathway. Since almond is a perennial species and flower initiation occurs during the year preceding blooming, it is not obvious that similar regulations are involved in the control of flowering time. Nevertheless, almond also has a cold requirement during dormancy of flower buds that resembles vernalization in annuals. Photoperiod regulates dormancy break, which for perennials is probably the most important factor regulating flowering time. Based on these assumptions, further work on the characterization of this putative almond FAR1 homologue would be interesting.
The co-localization of a candidate gene with a QTL does not provide a strong enough basis upon which to draw a conclusion with respect to the responsibility of this CG in phenotypic variation (Pflieger et al. 2001). Validation of the implicated CGs is therefore required. Association mapping provides high QTL resolution and the power to incorporate a wide range of alleles and, therefore, may be very useful for CG validation. Alternatively, physiological analyses may provide evidence for, or against, the role of the CGs. Such analyses involve measuring CG expression at the mRNA level (by quantitative reverse transcription-PCR or Northern blotting) and at the protein level (by Western blotting) as well as the determination of the CG activity (e.g. enzyme activity). If the CG is a member of a multigene family, expression analyses may only provide an estimation of the collective expression of the whole family. For the associations found in this work it will be possible and worthwhile to analyse the expression patterns of the CGs, at both temporal and spatial level.
We report here the map location of ten CGs putatively involved in flower development in almond. Co-segregation analysis between CGs and molecular markers for blooming showed associations between two of the CGs (PrpFAR1 and PrpAp2) and QTLs controlling blooming time. A third candidate gene (PrdMADS1) was located very close to the position of the peach Evergrowing gene. Further characterization of the CGs will be required in order to validate associations. Moreover, the isolation and characterization of CGs may contribute to understanding the molecular basis of flowering initiation and development in almond.
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Anderson JL, Seeley SD (1993) Bloom delay in deciduous fruits. Hortic Rev 15:97–144
Ballester J, Socias I Company R, Arús P, de Vicente MC (2001) Genetic mapping of a major gene delaying blooming time in almond. Plant Breed 120:268–270
Bowman JL, Alvarez J, Weigel D, Meyerowitz EM, Smyth DR (1993) Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development 119:721–743
Dirlewanger E, Moing A, Rothan C, Svanella L, Pronier V, Guye A, Plomion C, Monet R (1999) Mapping QTLs controlling fruit quality in peach (Prunus persica (L.) Batch). Theor Appl Genet 98:18–31
Dirlewanger E, Graziano E, Joobeur T, Garriga-Caldere F, Cosson P, Howad W, Arus P (2004) Comparative mapping and marker-assisted selection in Rosaceae fruit crops. Proc Natl Acad Sci USA 101(26):9891–9896
Etienne C, Rothan C, Moing A, Plomion C, Bodenes C, Svanella-Dumas L, Cosson P, Pronier V, Monet R, Dirlewanger E (2002) Candidate genes and QTLs for sugar and organic acid content in peach [Prunus persica (L.) Batsch]. Theor Appl Genet 105:145–159
Frewen BE, Chen THH, Howe GT, Davis J, Rohde A, Boerjan W, Bradshaw HD (2000) Quantitative trait loci and candidate gene mapping of bud set and bud flush in Populus. Genetics 154:837–845
Gévaudant F, Samson I, Guilliot A, Pétel G (1999) An improved method for isolating polyphenol-free RNA from woody plan tissues. J Trace Microprobe Tech 17:445–450
Gilpin BJ, McCallum JA, Frew TJ, Timmerman-Vaughan GM (1997) A linkage map of the pea (Pisum sativum L) genome containing cloned sequences of known function and expressed sequence tags (ESTs). Theor Appl Genet 95:1289–1299
Grassely C (1978) Observations sur l’utilisation d’un mutant d’amandier á floraison tardive dans un programme d’hybridation. Ann Amelior PLant 28:685–695
Hudson M, Ringli C, Boylan MT, Quail PH (1999) The FAR1 locus encodes a novel nuclear protein specific to phytochrome A signaling. Genes Dev 13:2017–2027
Joobeur T (1998) Construccíon de un mapa de marcadores moleculares y análisis genético de caracteres agronónicos en Prunus. PhD thesis, Universtat de Lleida
Joobeur T, Viruel MA, de Vicente MC, Jáuregui B, Ballester J, Dettori MT, Verde I, Truco MJ, Messeguer R, Batlle I, Quarta R, Dirlewanger E, Arús P (1998) Construction of a saturated linkage map in Prunus using an almond × peach F2 progeny. Theor Appl Genet 97:1034–1041
Kester DE (1965) Inheritance of time of bloom in certain progenies of almond. Proc Am Soc Hortic Sci 87:214–221
Kester DE, Gradziel TM (1996) Almonds. In: Jarnick J, Moore JN (eds) Fruit breeding, vol. 3. Wiley, New York, pp 1–97
Kitahara K, Matsumoto S (2000) Rose MADS-box genes “MASAKO C1 and D1” homologous to class C floral identity genes. Plant Sci 151:121–134
Kosambi DD (1944) The estimation of map distances from recombination values. Ann Eugen 12:172–175
Kusaba S, Honda C, Kano-Murakami Y (2001) Isolation and expression analysis of gibberellin 20-oxidase homologous gene in apple. J Exp Bot 52:375–637
Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L (1987) mapmaker: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181
Levy YY, Dean C (1998) Control of flowering time. Curr Opin Plant Biol 1:49–54
McCallum J, Leite D, Pither-Joyce M, Havey MJ (2001) Expressed sequence markers for genetic analysis of bulb onion (Allium cepa L). Theor Appl Genet 103:979–991
Mouradov A, Cremer F, Coupland G (2002) Control of flowering time: interacting pathways as a basis for diversity. Plant Cell 14:S111–s130
Peña L, Martin-Trillo M, Juárez J, Pina JA, Navarro L, Martínez-Zapater JM (2001) Constitutive expression of Arabidopsis LEAFY or APETALA1 genes in citrus reduces their generation time. Nat Biotechnol 19: 263–267
Pflieger S, Lefebvre V, Causse M (2001) The candidate gene approach in plant genetics: a review. Mol Breed 7:275–291
Plomion C, Hurme P, Frigerio JM, Ridolfi M, Pot D, Pionneau C, Avila C, Gallardo F, David H, Neutelings G, Campbell M, Canovas FM, Savolainen O, Kremer A (1999) Developing SSCP markers in two Pinus species. Mol Breed 5:21–31
Rounsley SD, Ditta GS, Yanofsky MF (1995) Diverse roles for MADS box genes in Arabidopsis development. Plant Cell 7:1259–1269
Rozen S, Skaletsky HJ (2000) primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S (eds) bioinformatics methods and protocols: methods in molecular biology. Humana Press, Totowa, pp 365–386
Sócias I Company R, Felipe AJ, Gómez Aparisi J (1998) Genetics of late blooming in almond. Acta Hortic 484:261–266
Sung SK, Yu GH, An G (1999) Characterization of MdMADS2, a member of the SQUAMOSA subfamily of genes, in apple. Plant Physiol 120:969–978
Tabuenca MC (1972) Necessidades de frío invernal en almendro. An Estac Esper Aula Dei 11:325–329
Tabuenca MC, Mut M, Herrero J (1972) Influencia de la temperatura en la época de floracion de almendro. An Estac Esper Aula Dei 11:378–395
Tanksley SD (1993) Mapping Polygenes. Annu Rev Genet 27:205–233
Theissen G, Kim JT, Saedler H (1996) Classification and phylogeny of the MADS-box multigene family suggest defined roles of MADS-box gene subfamilies in the morphological evolution of eukaryotes. J Mol Evol 43:484–516
Thompson JD, Higgins DG, Gibson TJ (1994) clustal w: improving the sensitivity of progressive multiple sequence alignment through sequence weighing, position-specific gaps penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Tragoonrung S, Kanazin V, Hayes PM, Blake TK (1992) Sequence-tagged-site-facilitated PCR for barley genome mapping. Theor Appl Genet 84:1002–1008
Verde I, Quarta R, Cedrola C, Dettori MT (2002) QTL analysis of agronomic traits in a BC1 peach population. Acta Hortic 592:291–297
Viruel MA, Messeguer R, de Vicente MC, Garcia-Mas J, Puigdomènech P, Vargas F, Arús P (1995) A linkage map with RFLP and isozyme markers for almond. Theor Appl Genet 91:964–971
Wada M, Cao QF, Kotoda N, Soejima J, Masuda T (2002) Apple has two orthologues of FLORICAULA/LEAFY involved in flowering. Plant Mol Biol 49:567–577
Wang Y, Georgi L, Reighard GL, Scorza R, Abbott AG (2002a) Genetic Mapping of the evergrowing gene in peach [ Prunus persica (L) Batsch]. J Hered 93:352–358
Wang Y, Georgi L, Zhebentyayeva N, Reighard GL, Scorza R, Abbott AG (2002b) High throughput targeted SSR marker development in peach (Prunus persica). Genome 45:319–328
Weigel D, Nilsson O (1995) A developmental switch sufficient for flower initiation in diverse plants. Nature 377:495–500
Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowits EM (1992) LEAFY controls floral meristem identity in Arabidopsis. Cell 69:843–859
Yao J, Dong Y, Kvarnheden A, Morris B (1999) Seven MADS-box genes in apple are expressed in different parts of the fruit. J Am Soc Hortic Sci 124:8–13
Acknowledgements
We are grateful to W. Howard for access to unpublished information on the position of pchgms12 and to A. Monfort and W. Howard for fruitful discussions. We also acknowledge the financial support from Fundação para a Ciência e a Tecnologia (Portugal) and Fundo Social Europeu, in the framework III Quadro Comunitário de Apoio for fellowship BD1187/00 awarded to C. Silva and for the research project Sapiens 33499/99.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by E. Guiderdoni
Rights and permissions
About this article
Cite this article
Silva, C., Garcia-Mas, J., Sánchez, A.M. et al. Looking into flowering time in almond (Prunus dulcis (Mill) D. A. Webb): the candidate gene approach. Theor Appl Genet 110, 959–968 (2005). https://doi.org/10.1007/s00122-004-1918-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-004-1918-z