Abstract
The electrocardiogram (ECG) is a widely disseminated method for detecting heart diseases due to its lower cost than other tests. But, some steps are important for detecting cardiac arrhythmias in ECG signals, which are: preprocessing, segmentation, feature extraction and classification. In this work, we assess how four non-morphological feature extraction methods provide useful ECG classification. Moreover, we propose an innovation in the configuration of the structural co-occurrence matrix (SCM), by combining it with the Fourier transform to extract the main frequencies of the signal. We tested theses methods on four well-known classifiers used in the literature and compare the results with six classical feature extraction methods. Moreover, we followed high standard protocols for developing expert systems for clinical usage. The database chosen for evaluation is the MIT-BIH arrhythmia database. We increased the identification of heart dysrhythmia by 2%, representing an advance on reports on the literature. The developed system is 1.3% more reliable than the current best approach reported, being \(10^6\) times faster, as well. The HOS with naive Bayes classified pathologies in 22 patients with 94.3% of accuracy. We perceived that SCM–Fourier is 1.5% more accurate than the SCM or Fourier standalone. The feature extractor proposed in this paper compress 97% of the useful information to provide a reliable arrhythmia classification.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Heart diseases accounted up to 17.5 million deaths in 2012 [29]. The electrocardiogram (ECG) is the first choice for heart diagnosis, because it is noninvasive, easy to apply and low cost [19]. However, it is extremely necessary that a medical specialist diagnose this examination properly, even so, its imprecise characteristics might lead to wrong diagnosis. So, medical area could benefit from computing tools for ECG processing [41].
The electrocardiogram maps the electrical activity associated with contractions of the heart muscle. Such contractions emanating polarization and depolarization of cardiac tissue are called events or waves, which are the main P, Q, R, S and T are also considered intervals of these events such as QT and TS, which have the Q and T, S and T, respectively, or also QRS, known as the QRS complex [17]. Figure 1 shows the characteristic ECG waves in a standard cardiac cycle.
Heart rhythm arrhythmia is caused by changes how the electrical impulse passes through the myocardial tissue [16]. Therefore, from a clinical perspective, it is important to the identify hearth arrhythmia with confidence.
Researches have been studying tools for ECG analysis throughout the years, such as preprocessing, segmentation, feature extraction and automatic classification [24]. For such analyses, parameters such as time, frequency and morphology are important and consider standards, i.e., classical techniques. For example, Acharya et al. [1] used these measures, on an RR interval, to separate different kinds of heart disorders. However, not all relevant information might be available by only using timing and shaping standards; therefore, feature extraction tolls might aid clinical diagnosis of and ECG.
Among the various extraction techniques for ECG found on the literature, the majority use discrete wavelet transform [5, 14, 44, 48,49,50], independent components analysis [48], analysis of linear discriminants [44], RR interval features [21, 48,49,50]. This main focus of this papers is the analysis of nonstandards feature extractors, which were demonstrated to be suitable in others signal processing applications [34, 39].
The Fourier transform is mainly used to identify different frequency components in a signal [31]. It can also be used to highlight the most relevant sine and cosine waves in the signal, such as exhibited in Pławiak [36], who proposed an way to identify heart health based on ECG signals. The algorithm of Goertzel [12] is a numerical way to compute the Fourier coefficients. Another important technique for feature extraction is the higher-order statistics (HOS) [28]. Thus, HOS may be a suitable method to extract features of the ECG signal, especially because Marques et al. [26] have used this kind of features to describe nonlinear relationships in heart’s signals. The structural co-occurrence matrix (SCM) is a feature extraction technique based on frequency occurrences in a know structure. Souza et al. [45] used the SCM to diagnose Parkinson’s disease. Moreover, Peixoto and Filho [34] proposed in a way that combines SCM and Fourier to identify strokes in tomography brain images.
This work aims to assess how Goertzel, HOS, SCM and the fusion SCM–Fourier would perform on heart arrhythmia identification, by evaluating their performance on several classifiers. Besides that, we replicate in this work the approach proposed by Peixoto and Filho [34], and it has never been applied in signals to the best of our knowledge. Four well-known classifiers are used to access the performance of theses methods: The naive Bayes, the optimum-path forest (OPF), the multilayer perceptron (MLP) and the support vector machine (SVM). Moreover, the standard ANSI/AAMI/ISO EC57, 1998-R2008, i.e., the subject-oriented paradigm, is used on the MIT-BIH arrhythmia database in order to provide a reliable to clinical analysis. Our results are also compared to six other feature extraction methods [8, 14, 44, 48,49,50].
In short, our contributions are:
Report a contrast between the most reliable and feasible approaches for heart arrhythmia identification with ECG;
Evaluate how the combination of SCM and Fourier will behave in ECG signals, since it has only been applied in tomography brain images;
Highlight how one could benefit by using such system in real clinical case scenario.
This paper is organized as follows: In Sect. 2, it is presented an overview of related work, which concerns to feature extraction and heart arrhythmia identification; in Sect. 3, the proposed tools are detailed; in Sect. 4 are explained the statistical procedures followed to produce reliable results; then, in Sect. 5 our findings are explained and compared to recent reports on the literature.
2 Background on ECG and Arrhythmia Detection
2.1 Classical Methods for ECG Feature Extraction
Chazal et al. [8] exhibited a method based on the time gaps of RR set of an QRS. This is often called as morphological features. The authors proposed to use 155 features entirely based on time measures of the QRS complex. Güler and Übeylı [14] used the discrete wavelet transform (DWT). Their methods are focused on the coefficients related to details of the first and fourth levels to generate a feature vector of length 19.
The DWT also has been used to generate features for ECG in Song et al. [44]. The authors used the coefficients of different levels to generate 144 features based on DWT and RR morphology. Still in frequency analyses, Yu and Chen [49] provided a set of 21 features, based on morphology and frequency domain.
Yu and Chou [50] used morphological features and independent component analysis (ICA) to generate a feature vector. The 31 most significant features of the RR interval are selected to be used. In the same direction, Ye et al. [48] combine DWT, ICA and PC a set of 96 values plus 4 morphological measures composes the feature vector, years ahead. Afkhami et al. [2] combined similar features with Gaussian mixture models.
It is worth noting that all feature extraction proposed in this work relies on algorithms that automatically identify the QRS complex, i.e. ,signal segmentation. This is not the focus of our work, since other researches have dedicated their efforts to develop algorithms for such purposes. Researches are pushing toward a major breakthrough in heat diagnosis systems. For this reason, it is important to follow standardized procedures. Further explanations are about experimental setting and that shall be taken into consideration to design a proper clinical and reliable system that could be compared to other methods of the literature.
2.2 Related Approaches for Arrhythmia Classification
Morphological features are created by point-to-point measures in the signal. They can be the simplest way to generate features from the QRS complex, but the main disadvantage is that these types of algorithms strongly rely on proper QRS segmentation, which means slightly displacement of R points might lead to a unrealistic morphology. In addition, if the sampling frequency (\(f_\mathrm{s}\)) of the acquisition system is not properly calibrated, this method could not perform as expected. This is the same disadvantage of the feature extractor proposed in Song et al. [44].
ICA methods have been used in time domain and frequency domain [48, 50]. Luz et al. [24] emphasized that ECG signal is composed by a group of smaller signals, which can be separated by ICA. These signals are correlated with dysrhythmia, but the drawback is that it is not easy to set the order of the ICA. Ye et al. [48] proposed an feasible way to achieve this at the cost of a higher-dimensional feature vector, leading this problem into “the curse of dimensionality theorem.”
Afkhami et al. [2] proposed to combine a set of features extraction methods, such as morphological and Gaussian mixture models (GMM), reaching over 90% of accuracy. Therefore, the authors lack to explain their methodology, making their work inappropriate for a fair evaluation.
Still in the context of feature extraction, deep learning (DL) might be used to describe a ECG signal (Rahhal et al. [37]). But, a DL approach is extremely susceptible to overfitting if not proper designed or if not enough data are given.
3 The Proposed Tools
3.1 Proposed Feature Extraction Methods for ECG
The Fourier transform is a widely used tool in practical and academic applications, feasible to be computationally used through the FFT algorithm. Through this transform, any function that can be expressed is a composition of sine-based functions [10]. Among a variety of applications for FFT, there is a highlight for the extraction of characteristics for pattern recognition [13]. The Goertzel [12] algorithm is a another way to compute the amplitude of Fourier transform’s harmonics, in a numerically more efficient way. This might be more suitable for embedded applications, overcoming sampling frequency problems reported in the approaches of Chazal et al. [8] and Song et al. [44].
The Kurtosis is used to measure how a distribution differs from a Gaussian one [9], which is the foundation of higher-order statistics (HOS) [7]. Antoni [4] emphasized this method ability to characterize non-stationary signals. In addition, skewness can be used to describe transition on the signal [27]. Due to its characteristics, this method has been applied successfully on signals with high variability [42]. It has demonstrated to be robust to Gaussian-type noise [20, 28]. This makes this method advantages over all classical methods presented before, since it combines non-stationary analyses, noise invariance with only much lower dimension (four features), compared to Ye et al. [48].
The kurtosis is related to high-frequency transition in the QRS. The skewness indicates a non-Gaussian patterns in the probability density function of a signal, representing a feature to describe smooth transitions during the QRS onset and offset.
The structural co-occurrence matrix (SCM) was recently proposed by Ramalho et al. [38] to analyze how two signals are interconnected. In this method, these two signals generate a 2-D histogram, i.e., a co-occurrence matrix. Report of SCM is mainly found for image classification [34, 43]. Most of applications of SCM are medical diagnosis; for example, Rebouças Filho et al. [40] used a slightly variation of the SCM for automatic detection of skin lesions on images, proposed in Peixoto and Filho [34]. It works the same as the SCM aforementioned; however, one of the input signals is the raw signal reconstructed by using the Fourier harmonics that have an intensity of 1% of the main DC [40]. This configuration has only been applied in images before, never in signals. It could be an improvement in feature extraction robustness, since it would combine statistical features from time and frequency domain filtering.
3.2 The Chosen Machine Learning Methods
The Bayesian classifier is a supervised technique, based on statistics and Fisher‘s rule [11] to classify samples. It uses estimations of co-variance matrices to generate discriminate functions and probability density functions of each class. A commonly found report of this method is the naive Bayes classifiers, which is based on the assumption that the features are uncorrelated, generating a diagonal co-variance matrix [32].
Support vector machines (SVMs) are based on the statistical learning theory proposed by Vapnik [46]. This is considered a large margin classifiers, and its goal is to place a hyperplan that maximizes distances between two classes. It was first proposed to solve binary problem, but one-versus-all structures are adopted for multiclasses. It also uses kernels to map the input features to a higher-dimensional output space, such as the radian basis function (RBF) [47].
The multilayer perceptron (MLP) is a nonlinear classifiers, designed to model how a signal flows through a human brain [15]. Its basic units are the perceptrons, which are connected with each other, layer by layer, through a weights matrix. This is often referred as the “vanilla neural network,” since it is the classical approach in front to deep learning.
The optimal path forest (OPF) is a classifier that mixes features from computational graphs and distance-based classifiers [33]. Its improvement is based on a optimization strategy to order the graph, making the faster to train and to predict than other distance-based classifiers. This classifier has been used for heart arrhythmia classification [23].
4 Methodology
We assess how HOS, SCM, Goertzel and Fourier perform, focusing on ECG signal feature extraction for arrhythmia detection task. We follow the recommendations of ANSI/AAMI [3] to attend to clinical standards and used the dataset division (DS1 and DS2) suggested by Chazal et al. [8]. In this way, we compare with the aforementioned classifiers, as the same adopted by Luz et al. [23] who follow the same medical recommendation.
According to Chazal et al. [8], using the heartbeats from a patient for both the training and the testing adds bias toward the system. So, the authors proposed to split the MIT-BIH dataset into two sets, DS1 and DS2, balancing among the samples.
In Sect. 4.1, we explain the database and the AAMI standards. Followed by Sect. 4.2, the segmentation and feature extraction steps are explained and the classifiers settings in Sect. 4.3. At last, we present the metrics adopted in Sect. 4.4.
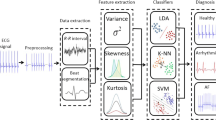
A general overview of our entire process is exhibited in Fig. 2. In (1), the raw data are read using the library provided by in MIT-BIH, which is filtered to reduce noised (2). Then, the QRS complex segmentation is made by using a gold standard algorithm, in which the feature extraction (3) and (4) classification are performed, respectively.
Steps used to identify heart disorders. In (1), a filtering is used under the raw ECG signal for noise removal (2). Then, it is passed to the QRS waves segmentation algorithm (3). The feature vectors are created for each QRS complex in (4). Finally, in (5), the feature vectors are fed into the classifiers to train an detect heart arrhythmia
4.1 The Heart Arrhythmia Database and Clinical Standards
The database adopted in our experiments is the MIT-BIH arrhythmia,Footnote 1 which contains a 48 half-hour ECG signal and records sampled at 360 Hz, of 44 patients. [30, 35]. This is the most used database that researches use to study heart arrhythmia. The Association for the Advancement of Medical Instrumentation (AAMI) published a standard ANSI/AAMI EC57:1998/(R)2008 [3] that recommends to group the heartbeat medical annotations into five classes, as shown in Table 1.
The column AAMI classes, in Table 1, are the suggestion of ANSI/AAMI [3] for grouping the heat diseases found in the MIT-BIH database. The description of each disease is presented in the last column of this table. Llamedo and Martínez [22] proposed to turn this 5-class problem into a 3-class problem by grouping F and Q together and add them to the VEB class, due to the unbalancing number of samples.
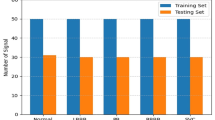
The AAMI standards also recommend to split records, i.e., patients, into two database of patients. One should be used for training (DS1) and other for testing the system (DS2). This is exhibited in Table 2. Thus, the system will be developed using data that represents real clinical cases, which means, patients that are used to design the system are different than the ones used to evaluate it. In addition, Chazal et al. [8] went one step further and proposed to balance both sets, by grouping the patients exhibited in Table 2.
As emphasized by Luz et al. [23], the heartbeats of a patient will never be in both DS1 and DS2, which means a patient should be only used for training or testing the system. This procedure might increase the reliability of the methods to arrhythmia detection.
Luz et al. [24] reported the main works published in the literature are grouped following the intra-patient scheme, where heartbeats of the same patients can be seen in both training and testing sets. Luz et al. [24] are strict to say these results do not contribute to the develop a system, from a clinical point of view, since the values are not similar in a real-life scenario regarding the accuracy rates reported.
4.2 Segmentation and Feature Extraction Steps
The R point the ECG signal is found using the algorithm proposed by Madeiro et al. [25]. And, the algorithm outputs the Q and S point as well, which is the entire QRS complex. But, we adjusted the algorithm to return 64 samples, as proposed in Yu and Chen [49], in order to center the R peak at each signal. Therefore, the segmented QRS corresponds to 2.8ms. There have been already efforts to develop algorithms for QRS segmentation, so this is not the main goal of our work.
We used the QRS segmented to extract features, with the methods presented in the previous section, and we compare them against the classical feature excitation exhibited in Luz et al. [23]. Tables 3 and 4 summarize all features extractors used in this work, showing the number of samples and features of each one.
4.3 Classifiers Settings
It is worth noting that the main purpose of our work is not to identify which is the most suitable classifier, but rather is to access how different feature extractors would perform on a set of classifiers. Among the methods, we use the naive Bayes, SVM, the MLP and OPF.
Eight datasets are generated with the five methods, combined with the recommendation of 5 and 3 classes. We equally preprocessed all sets. The DS1 set was standardized with to zero mean and unit variance, and the DS2 was transformed using the weights from the former process.
A tenfold cross-validation was employed for model selection and hyperparameter tuning, with a random search with 50 candidates [6]. For the SVM, the settings are: RBF kernel, \( \gamma \) ranging between \([2^{-15}, 2^{33}]\) and C\([2^{-5}, 2^{15}]\). For the MLP: Adam optimizer [18], one hidden layer and neurons range between 2 and 500. For the naive Bayes, the Gaussian probability density function was used, and for OPF, the Euclidean distance was used.
4.4 Evaluation Metrics
The metrics used to analyze the performance of all methods are accuracy, sensitivity, specificity and harmonic means. These values are calculated from the confusion matrix, wherein true positives (TPs) are the number of samples which are correctly identified as a heartbeat without arrhythmia, i.e. normal. False negatives (FNs) express normal beats that are misclassified as arrhythmic, while false positives (FPs) indicate how many arrhythmic samples are classified as normal. And, true negatives (TNs) are disorder heartbeats that are correctly classified as arrhythmic. For more information about how the metrics are calculated for the heart arrhythmia problem, refer to Luz et al. [23].
Accuracy (Acc) is the rate of correct classifications without taking into account the difference between the heartbeats analyzed of the others,
N AAMI class accounts up to 89.46% of samples in this database, and this will be used as the baseline for our analyses, which means a system with accuracy below 90% is considered ineffective.
Specificity (Sp) highlights true-negative samples. This is a metric to measure showing how cardiac arrhythmia, or a single kind of disease, is being identified. Higher values of Sp are desired since a low Sp means the system is labeling patients with a disease as normal, and this means the patient might not receive the proper treatment in time.
The sensitivity (Se) highlights true-positive samples, which accounts for disease’s samples labeled as normal, if the system is labeling normal patients with diseases and this could potently lead to unnecessary medical costs that trigger mental stress over the patients.
All experiments were run in a PC Intel i7 of 3.1 GHz, 8Gb of RAM, on a Linux Ubuntu 18.04. The timing metrics for feature extraction, training and testing are accounted. We coded the feature extractors in MATLAB and the classifiers in the Python.
5 Results, Discussion and Comparisons
In this section, our results will be highlighted in front of works on the literature, comparing and explaining advantages and disadvantages. Table 5 highlights how the classifiers perform on the five feature extraction methods. The dataset used it the one with 5-class version of [3]. The reports of Luz et al. [23] are shown as well.
Against the results of Luz et al. [23], we compare to HOS–naive Bayes, since it has been reached over 94% of Acc, and in Luz et al. [23] best approach, SVM reached 92.2%, using the features set of Chazal et al. [8]. Taking care into detecting only the negative classes, i.e., heart arrhythmia, our approach seems to be more effective, since V-class reaches 87.8% of sensitivity, against 48% in Luz et al. [23]. The same goes for the other classes. On the other hand, classes Q and F have too little samples, compared to normal. This naturally adds a step into hardness to understand the hidden pattern between infusion beats and unknown beats..
The SCM–Fourier extractor performed better with naive Bayes, achieving 92.4% of accuracy. It performed lower than the baseline of 90% with OPF and SVM. According to Peixoto and Filho [34], 1–5% would have most of the features important for the structural space of SCM; therefore, the results still behind the simplistic features of HOS.
For ECG description, the feature extractor HOS provided better results. Therefore, HOS might be understand as an way to calculate dissimilarities between a input signal and a pure Gaussian reference. Among all classifiers, the average accuracy of HOS is over 90%, higher than SCM, Goertzel and Fourier.
During our experiments, we realize that Chazal et al. [8] used features based on time references, to measure morphology’s of an ECG signal. The authors were strict in design and algorithm completely based on the ANSI/AAMI [3] dataset, which have unique setting such as the sample frequency. Their parameters are based on this type of signal, while our approach tends to be generalized to other types of ECG, specially because HOS is intrinsic to a probability distribution of a signal. In addition, HOS gives out 4 features, in contrast to [8] Chazal et al. who proposed 155.
In what concerns the feature sets created by SCM and SCM–Fourier, MLP and Bayes performed over 90% of accuracy, and its sensitivity reached 99.2%. For a clinical usage, this indicates this method tends to provide less false-positive predictions for diseases. It is similar on SVM, which reached over 99% of sensitivity. But, abstracting the filter design, the SCM–Fourier with the signal reconstructed with 1% of DC components performed 1.5% higher than using the SCM standalone.
Fourier and Goertzel did not show to be suitable, because in any of the classifier presented results compared to HOS or SCM. Both methods are based on frequency domain, and a key parameter is to have a resolution good enough to proper identify harmonics in a signal. The ECG signal of MIT-BIH database is sampled at a lower frequency, which certainly decreases the resolution in the frequency spectrum. Methods such as decimation could tackle the problem of resolution for frequency domain spectra in the ECG signal.
The results on all feature extraction methods, for the 3-class version of ANSI/AAMI [3] dataset, are presented in Table 6. The results are similar to the 5-class version of the dataset, by combining the feature-set HOS and classifier Naive Bayes. The results are 94% of Acc, 99% Se and 99% Sp. And it is also better than that of the approach of [23] on the same set. It is another evidence that this method could be suitable for clinical diagnosis of healthy conditions and arrhythmia diseases.
However, the feature extractor SCM and Goertzel performed worse in the 3-class set, with 87% and 71% of Acc, respectively. The accuracy improvements are due to the fact that F-class and Q-class represent less than 1% off all samples, and by grouping them together, there were no misclassifications between these two. We believe the 3-class dataset is more realistic for medical applications, since the patalogies represented in classes F and Q tend to be rare in front of other.
In what concerns to time performance, the results are shown in Table 7. Luz et al. [23] do not report measures of time for feature extraction, only for training and testing. We considered this time, since a real system to aid decision ought to be composed by all theses steps, so this time could not be unconsidered. In our findings, the system delivery higher than Luz et al. [23] (+ 1.3%) is also \(10^6\) times more computationally efficient, with the combination of HOS–naive Bayes. We measured the total time for a full heartbeat prediction, which is about \(1.74E^{-4}\) s (Fig. 3).
We aim to design a system that can be also conservative in providing a diagnosis, which means no Acc below 90% would be considered. From a clinical perspective, a system with as lower false-positive rate as possible is desired. On that matter, we present in Figs. 4 and 5, which are bar graph of the results found by Luz et al. [23]. As aforementioned, the highlight goes to HOS, which achieved over 2% of Acc in the best results of the literature.
HOS seems to be presenting better results than other feature extraction methods. In Fig. 3, we exhibit an example of a patient’s histogram of heartbeats. The distribution tends to be Gaussian, and these differences are measured in skewness and kurtosis.
5.1 Comparison to Other Approaches
We presented before improvements of the features extractors proposed in this paper over the ones reported by over the classical ones reported by Luz et al. [23]. In Fig. 6, the most recent works are compared to ours. We compiled here the works reported between 2016 and 2018, which follows the ANSI/AAMI [3] medical standards and the database split of Chazal et al. [8]
By taking a look at the graph, Rahhal et al. [37] accounted for the best results, achieving over 97%. But, they exclude all classes, except SVEB and VEB diseases. By doing this, they are designing a system focused in recognizing two different pathologies. From the clinical point of view, their system would not recognize normal heart condition, which is the most common class in this type of problem. There are fundamental differences between our goals and the ones of Rahhal et al. [37]. Moreover, the approach of Rahhal et al. [37] is based on deep learning, when ours is based on Gaussian classifier combined with higher-order statistics. By estimating skewness and kurtosis, our difference from Rahhal et al. [37] results is about − 3%. To use a neural network, a lot of concerns arises, such as number of neurons, parameters and hidden layer.
Therefore in a embedded application, a naive Bayes classifyer with statistical measures would be more feasible than computational expensive deep neural networks. This is the same reason that our system is more feasible to reproduce than the one proposed by Afkhami et al. [2] that use an ensemble of 100 decision trees, with a higher feature vector
We could not give a fair comparison for the system‘s metric, since none of the works discussed here calculated this metric.
6 Conclusions
The feature extraction methods proposed in this paper are promising tools to help heart arrhythmia detection on heartbeats, since by using non-classical approaches we achieved 2% more accurately than state-of-the-art ones which is based on morphology and statistical measures, as exhibited in Luz et al. [23].
There are strong advantages of using HOS and SCM–Fourier over morphological feature extraction methods for ECG, not only that leads to higher accuracy, but also reducing false diseases diagnosis on patients. We perceived that a feature extractor that is based only in a frequency domain is not suitable for this kind of task, but combining Fourier with SCM the results are promising and should be more investigated in future works, since it is more 1.5% than SCM and Fourier standalone.
Moreover, the system classified QRS complex of an ECG signal \(10^6\) times faster than reports of Luz et al. [23]. Being also more simple and feasible for embedded applications, it uses only four-dimensional feature vector, which is about 97% smaller than the one proposed Chazal et al. [8].
To summarize our achievements, we designed a system that is:
2% more accurate than reports on the literature;
designed to embedded devices, being \(10^6\) times faster than others reports and also compress 97% of the useful features.
References
U.R. Acharya, V.K. Sudarshan, J.E.W. Koh, R.J. Martis, J.H. Tan, S.L. Oh, A. Muhammad, Y. Hagiwara, M.R.K. Mookiah, K.P. Chua, K.C. Chua, R.S. Tan, Application of higher-order spectra for the characterization of coronary artery disease using electrocardiogram signals. Biomed. Signal Process. Control 31, 31–43 (2017). ISSN 1746-8094
R.G. Afkhami, G. Azarnia, M.A. Tinati, Cardiac arrhythmia classification using statistical and mixture modeling features of ECG signals. Pattern Recognit. Lett. 70, 45–51 (2016). ISSN 0167-8655
ANSI/AAMI, Testing and Reporting Performance Results of Cardiac Rhythm and ST Segment Measurement Algorithms. Technical report, Association for the Advancement of Medical Instrumentation (American National Standards Institute, Inc. (ANSI), 2008)
J. Antoni, The spectral kurtosis: a useful tool for characterising non-stationary signals. Mech. Syst. Signal Process. 20(2), 282–307 (2006)
R. Arvanaghi, S. Daneshvar, H. Seyedarabi, A. Goshvarpour, Classification of cardiac arrhythmias using arterial blood pressure based on discrete wavelet transform. Biomed. Eng. Appl. Basis Commun. 29(05), 1750034 (2017)
J. Bergstra, Y. Bengio, Random search for hyper-parameter optimization. J. Mach. Learn. Res. 13, 281–305 (2012). ISSN 1532-4435
V. Capdevielle, C. Serviere, J. Lacoume, Blind separation of wide-band sources: application to rotating machine signals, in European Signal Processing Conference, 1996 (EUSIPCO 1996), 8th edn. (IEEE, 1996), pp. 1–4
P. Chazal, M. O’Dwyer, R.B. Reilly, Automatic classification of heartbeats using ECG morphology and heartbeat interval features. IEEE Trans. Biomed. Eng. 51(7), 1196–1206 (2004). ISSN 0018-9294
R. Dwyer, Detection of non-Gaussian signals by frequency domain kurtosis estimation. in Acoustics, Speech, and Signal Processing, IEEE International Conference on ICASSP’83, vol 8 (IEEE, 1983), pp. 607–610
O.K. Ersoy, A comparative review of real and complex Fourier-related transforms. Proc. IEEE 82(3), 429–447 (1994)
R.A. Fisher, On the mathematical foundations of theoretical statistics. Philos. Trans. R. Soc. Lond. A 222(594–604), 309–368 (1922)
G. Goertzel, An algorithm for the evaluation of finite trigonometric series. Am. Math. Mon. 65, 34–35 (1958)
R.C. Gonzalez, R.E. Woods, S.L. Eddins et al., Digital Image Processing Using MATLAB, vol. 624 (Pearson/Prentice-Hall, Upper Saddle River, 2004)
I. Güler, E.D. Übeylı, Ecg beat classifier designed by combined neural network model. Pattern Recognit. 38(2), 199–208 (2005). ISSN 0031-3203
S. Haykin, Neural Networks: Principles and Practice. Bookman (2001)
B. Hemmeryckx, Y. Feng, L. Frederix, M. Lox, S. Trenson, R. Vreeken, H.R. Lu, D. Gallacher, Y. Ni, H.R. Lijnen, Evaluation of cardiac arrhythmic risks using a rabbit model of left ventricular systolic dysfunction. Eur. J. Pharmacol. 832, 145–155 (2018). ISSN 0014-2999
G. Israel, S. Ayelet, E. Yair, D.R. Zwas, L. Chaim, K. Andre, Temporal changes in electrocardiographic frontal QRS-T angle and survival in patients with heart failure. PLoS ONE 13(3), e0194520 (2018)
D.P. Kingma, J.Ba. Adam, A method for stochastic optimization. CoRR, arXiv:1412.6980 (2014)
K. Kirti, Accurate interpretation of the 12-lead ECG electrode placement: a systematic review. Health Educ. J. 73(5), 610–623 (2014). ISSN 0017-8969
F. Li, G. Meng, L. Ye, P. Chen, Wavelet transform-based higher-order statistics for fault diagnosis in rolling element bearings. J. Vib. Control 14(11), 1691–1709 (2008)
C.-C. Lin, C.-M. Yang, Heartbeat classification using normalized RR intervals and morphological features. Math. Probl. Eng. 2014, 1–11 (2014). https://doi.org/10.1155/2014/712474
M. Llamedo, J.P. Martínez, Heartbeat classification using feature selection driven by database generalization criteria. IEEE Trans. Biomed. Eng. 58(3), 616–625 (2011)
E.J.S. Luz, T.M. Nunes, V.H.C. Albuquerque, J.P. Papa, D. Menotti, Ecg arrhythmia classification based on optimum-path forest. Expert Syst. Appl. 40(9), 3561–3573 (2013). ISSN 0957-4174
E.J.S. Luz, W.R. Schwartz, G. Cámara-Chávez, D. Menotti, ECG-based heartbeat classification for arrhythmia detection: a survey. Comput. Methods Progr. Biomed. 127, 144–164 (2016). ISSN 0169-2607
J.P.V. Madeiro, P.C. Cortez, J.A.L. Marques, C.R.V. Seisdedos, R.M.R. Sobrinho Carlos, An innovative approach of QRS segmentation based on first-derivative, Hilbert and wavelet transforms. Med. Eng. Phys. 34(9), 1236–1246 (2012)
J.A.L. Marques, P.C. Cortez, J.P.V. Madeiro, V.H.C. de Albuquerque, S.J. Fong, F.S. Schlindwein, Nonlinear characterization and complexity analysis of cardiotocographic examinations using entropy measures. J. Supercomput. (2018). https://doi.org/10.1007/s11227-018-2570-8. ISSN 1573-0484
H.R. Martin, F. Honarvar, Application of statistical moments to bearing failure detection. Appl. Acoust. 44(1), 67–77 (1995)
J.M. Mendel, Tutorial on higher-order statistics (spectra) in signal processing and system theory: theoretical results and some applications. Proc. IEEE 79(3), 278–305 (1991). ISSN 0018-9219
S. Mendis, Global Status Report on Noncommunicable Diseases 2014 (World Health Organization, Geneva, 2014)
G.B. Moody, R.G. Mark, The impact of the MIT-BIH arrhythmia database. IEEE Eng. Med. Biol. Mag. 20(3), 45–50 (2001)
K. Najarian, R. Splinter, Biomedical Signal and Image Processing (CRC Press, Boca Raton, 2005)
N.M. Nasrabadi, Pattern recognition and machine learning. J. Electron. Imaging 16(4), 049901 (2007)
J.P. Papa, A.X. Falcao, C.T.N. Suzuki, Supervised pattern classification based on optimum-path forest. Int. J. Imaging Syst. Technol. 19(2), 120–131 (2009)
S.A. Peixoto, Rebouças Filho, Neurologist-level classification of stroke using a structural co-occurrence matrix based on the frequency domain. Comput. Electr. Eng. 71, 398–407 (2018). ISSN 0045-7906
PhysioBank PhysioToolkit, Physionet: components of a new research resource for complex physiologic signals. Circulation v101 i23, e215–e220 (2000)
P. Pławiak, Novel methodology of cardiac health recognition based on ECG signals and evolutionary-neural system. Expert Syst. Appl. 92, 334–349 (2018)
A. Rahhal, M. Mohamad, Y. Bazi, H. AlHichri, N. Alajlan, F. Melgani, R.R. Yager, Deep learning approach for active classification of electrocardiogram signals. Inf. Sci. 345, 340–354 (2016)
G.L.B. Ramalho, D.S. Ferreira, P.P. Rebouças Filho, F.N.S. de Medeiros, Rotation-invariant feature extraction using a structural co-occurrence matrix. Measurement 94, 406–415 (2016)
P.P. Rebouças Filho, N.M.M. Nascimento, I.R. Sousa, C.M.S. Medeiros, V.H.C. Albuquerque, A reliable approach for detection of incipient faults of short-circuits in induction generators using machine learning. Comput. Electr. Eng. 71, 440–451 (2018)
P.P. Rebouças Filho, S.A. Peixoto, R.V. Medeiros da Nóbrega, D.J. Hemanth, A.G. Medeiros, A.K. Sangaiah, V.H.C. de Albuquerque, Automatic histologically-closer classification of skin lesions. Comput. Med. Imaging Graph. 68, 40–54 (2018). https://doi.org/10.1016/j.compmedimag.2018.05.004
R. Saktheeswari, K. Adalarasu, Survey on signal processing techniques for diagnoising cardiovascular diseases, in 2017 International Conference on Innovations in Information, Embedded and Communication Systems (ICIIECS) (2017), pp. 1–4
B. Samanta, K.R. Al-Balushi, Artificial neural network based fault diagnostics of rolling element bearings using time-domain features. Mech. Syst. Signal Process. 17(2), 317–328 (2003)
S.P. Silva, L.B. Marinho, J.S. Almeida, P.P. Rebouças Filho, A novel approach for mobile robot localization in topological maps using classification with reject option from structural co-occurrence matrix, in International Conference on Computer Analysis of Images and Patterns (Springer, Berlin, 2017), pp. 3–15
M.H. Song, J. Lee, S.P. Cho, K.J. Lee, S.K. Yoo, Support vector machine based arrhythmia classification using reduced features. Int. J. Control Autom. Syst. 3(4), 571–579 (2005)
J.W.M. de Souza, S.S.A. Alves, E.S. Rebouças, J.S. Almeida, P.P. Rebouças Filho, A new approach to diagnose parkinson’s disease using a structural cooccurrence matrix for a similarity analysis. Comput. Intell. Neurosci. 2018, 1–8 (2018). https://doi.org/10.1155/2018/7613282
V.N. Vapnik, An overview of statistical learning theory. IEEE Trans. Neural Netw. 10(5), 988–999 (1999)
J. Wang, Q. Chen, Y. Chen, RBF kernel based support vector machine with universal approximation and its application, in eds. by F. Yin, J. Wang, C. Guo, Advances in Neural Networks (Springer, Berlin, 2004), pp. 512–517. ISBN 978-3-540-28647-9
C. Ye, M.T. Coimbra, B.V.K.V. Kumar, Arrhythmia detection and classification using morphological and dynamic features of ECG signals, in Engineering in Medicine and Biology Society (EMBC), 2010 Annual International Conference of the IEEE (IEEE, 2010), pp. 1918–1921
S. Yu, Y. Chen, Electrocardiogram beat classification based on wavelet transformation and probabilistic neural network. Pattern Recognit. Lett. 28(10), 1142–1150 (2007)
S. Yu, K. Chou, Integration of independent component analysis and neural networks for ECG beat classification. Expert Syst. Appl. 34(4), 2841–2846 (2008)
Acknowledgements
We acknowledge the sponsorship from the Coordination for the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—Brasil (CAPES) by providing financial support. This study was financed in part by CAPES—Finance Code 001.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Nascimento, N.M.M., Marinho, L.B., Peixoto, S.A. et al. Heart Arrhythmia Classification Based on Statistical Moments and Structural Co-occurrence. Circuits Syst Signal Process 39, 631–650 (2020). https://doi.org/10.1007/s00034-019-01196-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00034-019-01196-w