Abstract
In this review, we summarize computational and experimental data gathered so far showing that structural disorder is abundant within paramyxoviral nucleoproteins (N) and phosphoproteins (P). In particular, we focus on measles, Nipah, and Hendra viruses and highlight both commonalities and differences with respect to the closely related Sendai virus. The molecular mechanisms that control the disorder-to-order transition undergone by the intrinsically disordered C-terminal domain (NTAIL) of their N proteins upon binding to the C-terminal X domain (XD) of the homologous P proteins are described in detail. By having a significant residual disorder, NTAIL–XD complexes are illustrative examples of “fuzziness”, whose possible functional significance is discussed. Finally, the relevance of N–P interactions as promising targets for innovative antiviral approaches is underscored, and the functional advantages of structural disorder for paramyxoviruses are pinpointed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
The replicative complex of paramyxoviruses
Negative-stranded RNA viruses (NSRVs) are responsible for numerous human and animal diseases. Some NSRV are classified as potential agents of bio-terrorism, and several have been included in the priority pathogen lists of the NIAID (National Institute of Allergy and Infectious Diseases) (http://www.niaid.nih.gov/topics/biodefenserelated/Pages/default.aspxand) and of the CDC (Center for Disease Control and Prevention) (http://www.bt.cdc.gov/agent/agentlist.asp#m). NSRVs embrace two groups: viruses with segmented RNA genomes and viruses with non-segmented RNA genomes. The latter constitute the Mononegavirales order, which includes severe human pathogens such as the Ebola, rabies, mumps, and respiratory syncytial viruses (Fig. 1). This order is characterized by remarkable common features of their replication machinery minimally involving three proteins that dynamically associated with constitute a polymerase complex that uses a protein–RNA structure, the nucleocapsid, as template.
Phylogenetic tree of the Mononegavirales order showing the viruses mentioned in this review. The tree was built using the maximum likelihood method on the L protein sequences aligned by ClustalW. The viral genera (italic) and the viral families (bold) have been classified using the last taxonomy of the Mononegavirales order [235]. MeV measles virus, RDV rinderpest virus, CDV canine distemper virus, NiV Nipah virus, HeV Hendra virus, SeV Sendai virus, hPIV3 human parainfluenza type 3, NDV Newcastle disease virus, MuV mumps virus, PIV5 parainfluenza virus 5, EboV Ebola virus, RSV respiratory syncytial virus, hMPV human metapneumovirus, VSV vesicular stomatitis virus, RabV rabies virus
The Paramyxoviridae family is among the viral families that belong to this order. This family encompasses some of the major and ubiquitous disease-causing viruses in humans and animals. An illustrative example is measles virus (MeV), a Morbillivirus member, which is one of the most infectious viruses ever known [1]. The widespread use of vaccines for MeV has drastically reduced the worldwide incidence of measles infection. Despite this success, measles remains endemic in many developing countries. Measles infections remain a major cause of childhood morbidity and mortality, especially in the African and South East Asian regions [2]. In addition, measles re-emerged also in western countries (USA and Europe) and is still a major concern for human health with 134,200 deaths worldwide in 2015 (http://www.who.int/mediacentre/factsheets/fs286/en/).
Beyond a number of older, well-studied human and animal pathogens, paramyxoviruses, also include newly emerged severe human pathogens, e.g., the Nipah (NiV) and Hendra (HeV) viruses [3]. Although these newly emerged viruses appear to retain many of the genetic and biological properties of other Paramyxoviridae members, they also have a number of unique characteristics that set them aside and have justified their classification within the new Henipavirus genus [1, 3, 4]. Henipaviruses are the only currently recognised zoonotic paramyxoviruses and are responsible for severe encephalitis in human beings, with case fatality rates reaching more than 75% [4, 5]. Fruit bats of the Pteropus genus have been identified as their natural reservoir [5]. Notably, these fruit bats constitute the natural reservoir for an increasing number of new and often deadly zoonotic viruses [6]. HeV came to light in 1994 in the Hendra suburb of Brisbane (Australia) as a new etiologic agent responsible for a sudden outbreak of acute respiratory disease within horses [5]. NiV emerged in 1998 in Malaysia as the etiologic agent of an outbreak of disease in pigs and humans. The first NiV outbreak in Malaysia resulted in 265 human cases of encephalitis and 105 deaths [5]. After the first cases of human infection in 1998 in Malaysia, NiV has regularly re-emerged since 2001 in Bangladesh. The ability of NiV to be transmitted by direct inter-human transmission further extends its potential to cause deadly outbreaks. In addition, the discovery of henipaviruses in other species of bats in West Africa and China underscores the threat that these viruses constitute to human health. Their high pathogenicity, wide host range, and interspecies transmission led to the classification of henipaviruses as biosecurity level 4 (BSL4) pathogens and as potential bio-terrorism agents.
Paramyxoviruses are enveloped viruses with a pleomorphic structure. Their envelope is derived from the plasma membrane of the host cell enriched in two envelope glycoproteins: a fusion protein (F) and a receptor-binding attachment protein (G/H). Beneath the envelope, the viral matrix protein (M) bridges the cytoplasmic tails of F and G/H proteins with the nucleocapsid and is responsible for virus budding. As in all Mononegavirales members, the genome of paramyxoviruses is encapsidated by the nucleoprotein (N) giving rise to a helical nucleocapsid (N–RNA) with a characteristic herringbone-like structure [7,8,9,10,11,12]. Following fusion between the viral envelope and the host cell membrane, the nucleocapsid is released in the cytoplasm. A peculiar feature of Mononegavirales members is that the nucleocapsid, and not naked RNA, is the template used by the RNA-dependent RNA polymerase (RdRp) during both transcription and replication. The RdRp is made of the large protein (L) and the phosphoprotein (P). During RNA synthesis, the N–P interaction ensures the tethering of L onto the N–RNA template. The P protein is, therefore, an essential polymerase cofactor in that it allows the L protein to be recruited onto the nucleocapsid template. The complex formed by the N, P, and L proteins constitutes the viral replicative unit (Fig. 2a). The N, P, and L proteins are necessary and sufficient to sustain the replication of viral RNA in Paramyxoviridae [1, 13]. As detailed below, during the synthesis of viral RNA, the components of the viral replication machinery engage in a complex macromolecular ballet.
Scheme of Paramyxoviridae RNA synthesis and viral genome. a Schematic illustration of the Paramyxoviridae replicative complex during replication of the viral genome or anti-genome. The N protein is drawn with a bilobal shape according to available structural data. It encapsidates both the viral RNA used as a template and the neo-synthetized RNA. The intrinsically disordered protein regions of N and P are symbolized by lines and α helixes by rectangles. The P–L complex forms the RNA-dependent RNA polymerase (RdRp) complex, which cartwheels onto the nucleocapsid complex via the X domain of P (XD). P oligomerizes through its multimerization domain (PMD) and is shown as a tetramer to reflect the prevalence of this oligomeric state in paramyxoviral P proteins. The α-MoRE at the N-terminus of P (PNTD) binds a monomeric N protein and forms the N0–P complex that prevents N self-assembly and binding to cellular RNA. The long disordered N-terminal region of P (PNT) may allow the binding of multiple monomers of N by a single oligomer of P. In the N0–P complex, XD might also bind to NTAIL. The extended conformation of the disordered regions would allow the formation of a tripartite complex between N0, P, and L that may enhance nucleocapsid assembly by bringing N0 near the encapsidation site. b Schematic representation of the genome of Paramyxoviridae. The negative-sense genomic RNA is presented in the 3′–5′ orientation. Below, the genome is shown a schematic representation of the expression gradient of the mRNA as a result of inefficient transcription re-initiation by the polymerase during transcription [1]
The number of nucleotides in the genome of paramyxoviruses is a multiple of six, consistent with the experimentally supported “rule of six”, which posits that only the genomes, whose number of nucleotides is a multiple of six, will be replicated [13,14,15]. This rule reflects the ability of each nucleoprotein to bind six nucleotides (for a review, see Ref. [16]). Deviation from the rule of six is prevented because of structural constraints imposed by the N to RNA binding. The genome organization of henipaviruses is close to that of others paramyxoviruses. Their genome is, however, bigger (18,234 nt for HeV and 18,246 or 18,252 nt for NiV Malaysia and Bangladesh, respectively) than that of typical paramyxovirus members characterized by an average genome length of approximately 15,500 nt [17]. This extra-length of the genome is due to the presence of long non-coding sequences at the 5′ end of each gene and to the particular large size of the P protein of henipaviruses.
Upon intracytoplasmic delivery of the viral ribonucleoprotein complexes, transcription of viral genes occurs using endogenous NTPs as a substrate. The polymerase enters the nucleocapsid at the promoters for transcription and replication located at the 3′ end of the genome. Transcription of the genes is sequential. At each intergenic region (IGR), the polymerase ends and re-initiates at the downstream gene. Gene-end (GE) signal corresponds to the polyadenylation of the upstream gene. Then, the polymerase scans over three nucleotides (3′-GAA-5′ or 3′-GCA-5′) without transcribing them in search of the downstream gene-start (GS) signal to resume transcription. The efficiency with which the polymerase re-initiates transcription decreases with increasing distance from the 3′ end, thereby leading to a gradient in the transcripts, with the most distal genes being the least expressed [1] (Fig. 2b).
At some stage after primary transcription, the polymerase switches to a replication mode to synthesize a full, complementary strand of genome length leaving the IGRs unrecognized. The sole use of this positive-stranded RNA (anti-genome) is to serve as a template intermediate for genome replication [1]. The relative level of transcription versus replication is postulated to be controlled by the intracellular concentration of the N protein. When the concentration of N is high enough to allow encapsidation of the nascent RNA chain, the replicase mode of the polymerase is favored over the transcription mode (see Ref. [18] for review).
As the most abundant viral protein in infected cells, the N protein from Paramyxoviridae members exists in two forms: a soluble, monomeric one (named N0) and a nucleocapsid assembled one (named NNUC) [19, 20]. Once the N protein has been synthesized, a chaperone is necessary to maintain it in the unassembled form in the cytoplasm. The illegitimate self-assembly of N is prevented by the association with the P protein [21, 22]. The resulting N0–P complex is used as the substrate for encapsidating the nascent genomic RNA chain (Fig. 2a). When assembled together with the RNA genome, N forms complexes with both isolated P (to yield NNUC–P) and P bound to L (to yield NNUC–P–L). These complexes are essential to RNA synthesis by the viral polymerase (see Refs. [1, 15, 18, 23, 24] for reviews on transcription and replication).
Albeit in vitro and in the absence of P, L can synthesize short RNA transcripts using naked RNA as a substrate [25, 26]; in infected cells, P is required to allow the recognition of the nucleocapsid template and also to stabilize L. In the case of MeV, NiV, and mumps virus (MuV), the cellular hsp90, in conjunction with P, is required to enable L to fold into a functional, mature form [27, 28]. The L protein carries out most (or even all) enzymatic activities essential for transcription and replication, i.e., nucleotide polymerization, and the maturation of viral mRNA (capping and polyadenylation). Being unstable unless bound to the P protein, L accumulates in low amounts in infected cells and this makes its molecular characterization very challenging [29]. The present knowledge on paramyxoviral L proteins is essentially based on bioinformatics studies. In fact, no functional paramyxoviral polymerase has been purified and biochemically characterized so far. Among Paramyxoviridae members, Rinderpest virus (RDV) and Sendai virus (SeV, a Respirovirus) constitute the only two examples, where L (or L/P) has been partially purified [30, 31], with SeV L possesses a methyltransferase activity in its C-terminal region [31], in agreement with predictions [32]. Within the Mononegavirales order, the two best characterized polymerases are that of respiratory syncytial virus (RSV, a Pneumoviridae member), whose RNA polymerase activity is documented in vitro [33], and that from vesicular stomatitis virus (VSV, a Rhabdoviridae member), whose structure is solved at almost atomic resolution using cryo-electron microscopy (cryo-EM) [34].
Presently, N and P are the best characterized proteins of the replicative complex of paramyxoviruses thanks to the numerous efforts in their molecular characterization in the last two decades. The N–P interaction has attracted a lot of interest not only from a fundamental perspective, but also from a more applied point of view. Indeed, by allowing the recruitment of L onto the nucleocapsid template, the N–P interaction is regarded as a potential target for antiviral approaches.
In the course of the characterization of paramyxoviral N and P proteins, they were found to be enriched in intrinsically disordered protein regions (IDPRs). These IDPRs play key roles in the formation of the tripartite N–P–L complex and enable a broad molecular partnership (for reviews, see Refs. [18, 35,36,37,38,39,40,41,42,43,44]). From seminal observations on MeV P and N proteins [8, 45], subsequent studies have collectively contributed to enlarge the awareness of the abundance and functional importance of structural disorder within paramyxoviruses N and P.
Intrinsically disordered proteins (IDPs) and IDPRs are ubiquitous functional proteins/regions that lack stable (i.e., highly populated) structure in the absence of a partner/ligand under physiological conditions of pH and salinity [46]. The inability of IDPs/IDPRs to fold is encoded by their amino acid sequence. A specific imbalance in the content of hydrophobic versus polar residues in IDP/IDPRs confers them the ability to populate a continuum of conformations ranging from random coils, RC (i.e., completely extended) to pre-molten globules and molten globules (i.e., more compact). The interactions of IDP/IDPRs with partners dictate both their function and the conformational ensemble that they can sample. The folding of IDPs/IDPRs upon binding leads to either stable complexes amenable to crystallization, or, more frequently, to the so-called “fuzzy” complexes [47, 48]), i.e., complexes with significant residual disorder. These peculiar characteristics of IDPs/IDPRs give them a number of advantages over folded proteins, hence their prevalence in hubs in protein interaction networks and cell signaling (for a recent review on IDPs/IDPRs, see Ref. [49]).
In this review, the molecular information that has been gathered so far on the N and P proteins from three illustrative paramyxoviruses, namely, MeV, NiV, and HeV, is described in detail and how structural disorder ensures an efficient replication and transcription of the paramyxoviral genome is emphasized. The implications of induced folding and residual flexibility in molecular partnership, transcription, and replication are discussed. How targeting the N–P interaction can pave the way for new antiviral approaches is then underscored. Finally, the functional implications and potential advantages arising from structural disorder within paramyxoviruses are pointed out.
Structural disorder in paramyxoviral N and P proteins
Structural organization of paramyxoviral P proteins
The P gene of paramyxoviruses manages to encode multiple information in a single gene: it gives rise to a number of different products by means of either overlapping reading frames (C protein) or messenger editing (V and W proteins). The latter is a peculiar process, whereby one or more non-templated nucleotides are inserted co-transcriptionally at a specific site. This insertion results in a shift of the open reading frame leading to a new protein product. Depending on the number of nucleotides inserted at the edition site, the V or W proteins are the end products. The V and W proteins share with P their N-terminal domain and possess each a unique C-terminal domain.
A similar P modular organization is generally observed in paramyxoviruses with, however, a much larger size of the Henipavirus P proteins (707 amino acids in HeV and 709 amino acids in NiV) than those of other paramyxoviruses (507 amino acids in MeV) (Fig. 3a) [4]. Bioinformatics analyses showed that paramyxoviral P proteins consist of an intrinsically disordered N-terminal domain (PNT), and a C-terminal domain (PCT) further subdivided into various regions [50, 51]. The disordered state of MeV, NiV, and HeV PNT is experimentally supported according to multiple biochemical and biophysical approaches. Specifically, PNT domains (1) are highly sensitive to proteolysis, (2) possess an anomalous electrophoretic behavior (i.e., they migrated in SDS-PAGE with an apparently higher molecular mass), (3) possess NOESY and circular dichroism (CD) spectra typical of IDPs, and (4) possess Stokes radii (R S) by far exceeding than those of globular proteins with similar size [45, 50]. The PNT domain of henipaviruses is spectacularly large, consisting of 404 (HeV) or 406 (NiV) residues, and accounts for the extra-length of Henipavirus P proteins. They are disordered not only in isolation but also within the full-length P protein, as inferred from their high sensitivity to proteolysis (Beltrandi, Habchi, Longhi, and Cavalli, unpublished data). The extra-length of Henipavirus PNT domains is consistent with the higher tolerance of disordered regions to insertions or major rearrangements as compared to ordered ones.
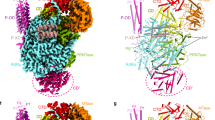
Modular organization of MeV, NiV, and HeV phosphoproteins. Domain organization of P showing two moieties, PNT and PCT. Structured and disordered regions are represented as large or narrow boxes, respectively. PNT N-terminal region of P, PCT C-terminal region of P, PMD P multimerization domain, XD X domain of P adopting a triple α-helical bundle. The α-MoRE at the N-terminal region of P (PNTD), which is partly preconfigured in solution and shown to adopt a stable α-helical conformation upon binding to N0, is shown as a red helix. Available structures and models are shown: MeV and NiV N0–PNTD complexes (PDB codes 5E4V and 4CO6, respectively) [57, 58], MeV PMD: PDB code 4BHV [102], MeV XD: PDB code 1OKS [82], HeV XD: PDB code 4HEO [91], structural model of the PMD trimeric forms of NiV [111] and HeV [110], structure of the tetrameric form in crystals (PDB code 4N5B) [104], and structural model of NiV XD [92]. All structures were drawn using Pymol [236]
All the disorder predictors implemented in the MeDor metaserver predicted the first 40–50 amino acids of MeV, NiV, and HeV P to be order prone [52] (data not shown and [50]). Such an N-terminal module with α-helical folding propensities is conserved amongst Avulavirus, Henipavirus, and Rubulavirus members [51]. Because this N-terminal module has a certain propensity to bind to a partner and to undergo induced folding (i.e., a disorder-to-order transition), it constitutes a so-called α-helical molecular recognition element (α-MoRE) [53,54,55,56]. The involvement of this N-terminal region in binding to N0 has been experimentally demonstrated in the case of NiV and MeV. In these two viruses, the crystal structures of a monomeric, RNA-free form of the N protein in complex with the N-terminal N0-binding region of P (PNTD) were solved and revealed that PNTD adopts an α-helical conformation at the surface of N0 [57, 58] (see also “Structural organization of paramyxoviral N proteins”). Computational approaches showed that paramyxoviral P proteins share a short (11–16 residues) sequence motif within PNTD [59]. The authors proposed that this region is responsible for binding to N0 and conserved in all Mononegavirales P proteins as a result of divergent evolution [59]. Accordingly, a similar N-terminal module was identified and characterized also in the P proteins from RSV [60, 61], human metapneumovirus (hMPV) [61], VSV [62], and in the Ebola virus (EboV) VP35 protein (i.e., the P protein counterpart in Filoviridae) [63]. In these viruses, this module was shown to be globally disordered while containing transient α-helices [63,64,65]. In the case of VSV, EboV, and hMPV, this P motif was shown to fold upon binding to the monomeric form of N [61,62,63].
By analogy with the closely related RDV and human parainfluenza type 3 virus [66, 67] and with rabies virus (RabV) and VSV [68, 69], PNTD might also bind additional partners, such as L and/or SNAP29. In the case of VSV, the N0-binding site of P does not overlap with the L-binding site, thereby enabling a P molecule to be simultaneously engaged in the N0–P complex and in the interaction with L. This tripartite complex could bring N0 in close proximity of the site of RNA synthesis, with transfer of N0 from P to the nascent RNA chain being possibly promoted by the polymerase [70]. Alternatively, as recently proposed by Jamin and Yabukarsky [71], since Mononegavirales P form homo-oligomers, a monomer of P may bind to L, while another monomer may interact with N0, thereby bringing the latter to the encapsidation site. Those studies also shed light on the stoichiometry of the VSV N0–P complex showing that each P monomer within the P dimer [72, 73] can bind up to two client N proteins [70] (Fig. 1).
In accord with the presence of a transiently populated α–MoRE, size exclusion chromatography (SEC), dynamic light scattering (DLS), and far-UV CD studies unveiled that MeV, NiV, and HeV PNT domains are not completely unfolded: they conserve some degree of compactness and residual, fluctuating secondary structure that are typical of a pre-molten globule conformation [45, 50]. Pre-molten globules are characterized by an intermediate conformational state between a random coil and a molten globule and possess a certain degree of residual compactness due to the presence of residual and fluctuating secondary and/or tertiary structures [74, 75]. The extent of residual compactness of the PNT domains varies according to the virus species with the order NiV PNT > HeV PNT > MeV PNT. Whether this different compactness reflects a different degree of pre-configuration of their α-MoRE remains to be established.
The disordered PNT domains contain most of the phosphorylation sites of Paramyxoviridae P proteins [51, 76,77,78]. This observation is in good agreement with the relationship between structural disorder and post-translational modifications and, in particular, phosphorylation [79].
PNT are not the only IDPRs of P. In fact, additional IDPRs exist within PCT, where regions of disorder alternate with ordered regions (Fig. 3a) [45, 50]. Consequently, 70–80% of the residues in P proteins from MeV, NiV, and HeV are disordered. The ordered domains of PCT are the P multimerization domain (PMD) and the C-terminal X domain (XD). PMD ensures oligomerization and XD the binding to the C-terminal domain of N (i.e., NTAIL) [80,81,82,83,84]. PMD and XD are separated by a poorly ordered and flexible linker [51]. In the case of SeV, NMR studies confirmed that the region upstream XD is disordered [85,86,87], while in the case of MeV, a proteolytic cleavage site was mapped therein thus providing (indirect) support for the disordered state of this region [8]. Besides, an additional disordered region (referred to as “spacer”) occurs upstream of the PMD in MeV, NiV, and HeV [50, 51, 88]. Notably, its counterpart in the SeV P protein is ordered [89].
The structures of MeV and HeV XD consist of a bundle of three antiparallel α-helices [82, 83, 90, 91] (Fig. 3a), in line with predictions [50, 51] and spectroscopic studies [92]. They display a large hydrophobic cleft delimited by helices α2 and α3. The structure of NiV XD is expected to adopt a structure similar to that of HeV XD (Fig. 3a) because of high sequence similarity (94%) and common spectroscopic features. Accordingly, a homology-derived structural model of NiV XD was proposed [92]. High-resolution structures X domains of the closely related SeV and MuV were also solved either by nuclear magnetic resonance (NMR) or by X-ray crystallography [86, 93]. Albeit SeV and MeV XD share similar overall structure, the surface created by helices α2 and α3 in SeV is dominated by negatively charged residues [81]. A peculiar feature of MuV XD is that it exists in solution in the form of a molten globule, as judged from CD, NMR, and DLS studies [93]. The observed structure in crystals results from a stabilizing effect brought by a crystallization additive [93]. MuV XD does not interact with the C-terminal, disordered region of N (NTAIL), but rather establishes contacts with the structured NCORE region of N [84]. By analogy with MuV, MeV XD might also bind to a secondary site located on NCORE [94]. This would explain how transcription and replication can be supported by a truncated form of MeVN (N1–439) devoid of NTAIL [94, 95].
Interestingly, while in Paramyxoviridae members, the C-terminal nucleocapsid-binding region of P is prevalently folded, it is disordered in RSV, a Pneumoviridae member [96, 97]. In Rubulavirus P proteins, intrinsic disorder further extends to their X domains. Indeed, these X domains span a structural continuum ranging from largely disordered in solution to a triple α-helical bundle fold [93, 98]. In line with these observations, electron spray ionization mass spectrometry (ESI–MS) studies on MeV XD indicated that this domain is structurally heterogeneous, populating at least two alternative conformations under native conditions [99]. This conclusion is further supported by a recent study, where the structure of a folding intermediate of MeV XD was identified and characterized by means of combined equilibrium and kinetic measurements and replica-averaged metadynamics simulations [100]. The chemical shifts of the ensemble obtained by metadynamics much better agree with those derived by NMR than with those derived from the crystal structure, suggesting that some effects of the dynamics of the ensemble are lost during the crystallization process and that most likely, the protein explores both the intermediate and native state in solution.
As already mentioned, beyond XD, Paramyxoviridae P proteins contain another structured region that is responsible for P multimerization (e.g., PMD). Paramyxoviridae PMDs adopt a coiled-coil structure [50, 51, 96] as experimentally confirmed in the case of SeV [89], RDV [101], MeV [88, 102], MuV [103], NiV [104], human metapneumovirus, and RSV (the two latter are Pneumoviridae members) [96, 105, 106]. The fact that paramyxoviral P proteins studied so far are oligomeric provides support to the so-called “cartwheeling” mechanism proposed for Paramyxoviridae. According to this model, the polymerase complex cartwheels on adjacent N monomers within the nucleocapsid to permit transcription and replication [107]. However, the quaternary organization of P varies among paramyxoviruses as detailed below.
As revealed by X-ray crystallography, SeV [89], MeV [88, 102] RDV [101], human metapneumovirus [105], RSV [96, 108], and MuV [103], PMDs form a tetrameric coiled-coil. A tetrameric organization is also conserved in the P orthologue of EboV, i.e., VP35 [109]. MuV PMD has a unique structural organization in that the tetramer consists of two sets of parallel helices in opposite orientation, i.e., it is a dimer of two antiparallel coiled-coil dimers [103], while all the other paramyxoviral P proteins characterized so far possess a parallel organization. How these differences in the organization can be compatible with a common mechanism of transcription and replication remains to be deciphered. The corresponding PMDs of RabV and VSV form dimers with a different structural arrangement [72, 73].
However, from recent findings, Henipavirus PMDs rather adopt a trimeric organization in solution [110, 111]. This conclusion was supported by several independent biochemical and biophysical approaches (e.g., SEC, SDS-PAGE, cross-linking with suberic acid bis N-hydroxy-succinimide ester, SAB, analytical ultracentrifugation, and SAXS) [110, 111]. However, crystallization [104] or cross-linking experiments carried out with a different cross-linker (glutaraldehyde) [112] NiV PMD assemble into a tetramer. It should be pointed out, however, that the very high cross-linker concentrations used in the study by Salvamani and co-workers may have introduced a bias and could potentially have generated non-specific association. SAXS studies confirmed the trimeric state of HeV PMD not only in isolation but also in the context of the entire PCT region, a finding that extends and strengthens the conclusions based on PMD [110]. The SAXS envelope of HeV PMD is, however, different from that of NiV PMD, e.g., HeV PMD has a much smaller R g and D max. This difference arises from a different orientation of the N-terminal helical region (or “head”) of PMD: the PMD heads of NiV and HeV are in the “up” orientation (i.e., exposed to the solvent) and in the “down” orientation (i.e., packed back against to coiled-coil), respectively (Fig. 4a, b) [110]. In the crystal structure of NiV PMD, the “head” is in the “down” orientation as it is the case for SeV [89]. Because of the high sequence similarity between NiV and HeV PMDs, the structural differences noticed by SAXS were ascribed to their ability to undergo conformational changes resulting in coiled-coil structures of different lengths and compaction, possibly related to the different functions that P plays during the viral cycle [110].
Structures of Henipavirus and MeV P multimerization domains. Structural models of the trimeric coiled-coil PMDs of NiV [111] (a) and HeV [110] (b) embedded into their SAXS-derived ab initio envelopes. c Structural comparison of MeV PMD structures. Left ribbon representations of the crystal structures of the MeV PMD tetramers as observed in the three different MeV PMD forms solved to date. Right superimposition of the three MeV PMD tetramers, with PDB entries 4BHV, 4C5Q, and 3ZDO shown in red, yellow, and green, respectively; data from [102] and reproduced with permission of the International Union of Crystallography (http://journals.iucr.org/). d Structural comparison of NiV PMD crystal structures. Left Ribbon model of the two crystal structures of NiV PMD solved so. Right superimposition of the two NiV PMD tetramers, with PDB entries 4N5B and 4GJW shown in red and green, respectively
What are the reasons for the observed discrepancies between the conformations of NiV PMD in solution and in crystals? The high local protein concentrations and/or the strong inter-molecular interactions forced by crystallization might favor a tetrameric organization at the expenses of a trimeric one. That coiled-coils could adopt different oligomeric states according to the physico-chemical conditions (pH, temperature, etc.) or depending on their intra- or extra-cellular location is well established [113, 114]. For example, the GCN4 leucine-zipper domain can adopt different oligomeric states as a function of the crystallization conditions. Thus, a given amino acid sequence may not dictate a single oligomeric state [115]. Further conflicting experimental evidence is found for SeV PMD that forms trimers in solution [116, 117] and adopts a tetrameric coiled-coil conformation in crystals [89]. The fact that SeV and NiV PMDs would form trimers in solution and tetramers in crystals would likely be a property reflecting their intrinsic ability to adopt different oligomeric states. These different oligomeric states could correspond to different functional forms of P and to the different complexes (i.e., N–P, N0–P, and P–L) that this pivotal protein can form in infected cells. In further support of the ability of P proteins to adopt different oligomeric states, the oligomerization domain of Marburg virus VP35 is a trimer both in crystals (in two different crystal forms) and in solution [118], whereas the corresponding domain in the highly related EboV is a tetramer in solution [109, 118].
Within Paramyxoviridae, structural heterogeneity of PMDs is not unique to henipaviruses and also pertains MeV PMD. Indeed, the comparison of crystallographic structures of MeV PMD reveals surprising structural variations [88, 102]. Although all these structures are tetrameric coiled-coils, they differ both in the quaternary structure and in the degree of disorder within the C-terminal region of the coiled-coil (Fig. 4c). Furthermore, the two crystal structures of NiV PMD (pdb codes 4N5B and 4GJW) display similar polymorphism in their quaternary structure and in their amount of disorder at the C-terminal end (Fig. 4d). The disorder could not be anticipated from the amino acid sequence neither for MeV nor for NiV, an observation that appears to exemplify the limitations of the currently available predictors. These results also show that coiled-coils can exhibit a certain degree of structural freedom, being less rigid than previously thought, in spite of a melting temperature exceeding 80 °C [88, 102, 110, 111]. They also defy to some extent the reliability of predicting the valence of coiled-coils from their amino acid sequence [102].
In conclusion, we can postulate that the ability to form a transcriptase versus a replicase complex would rely on the ability of the PMD region of P protein to adopt different oligomeric states (SeV, NiV, and HeV) and/or to dynamically sample different forms differing in the degree of compaction and in the extent of disorder (MeV). Indeed, such conformational changes might be the basis for the ability of P to form different complexes critical for transcription and replication. Although in the case of the related viruses, such as human parainfluenza virus 3 (hPIV3), RSV, and VSV, mutational studies suggest that P oligomerization is necessary for transcription and replication [119,120,121]; additional studies are required to draw definite conclusions as to whether P oligomerization is indispensable in all paramyxoviruses. Indeed, P oligomerization looks dispensable in RabV [122]. Likewise, additional studies are necessary to unravel the functional impact of varying the P oligomeric state.
Structural organization of paramyxoviral N proteins
Paramyxoviral N proteins are more than 500 amino acids in length. They are responsible for encapsidating the viral RNA into a helical nucleocapsid. This ribonucleoprotein complex has a typical “herringbone”-like structure in electron micrographs [7, 19, 20, 91, 123]. Real-space helical reconstruction of MeV nucleocapsids has been achieved in 2004 by EM [9, 124]. In Paramyxoviridae, each N protein covers precisely six nucleotides, hence explaining the “rule of six”. Once wrapped by the N protein, the RNA genome is protected in a nuclease-resistant form. N is, however, much more than a “simple” structural protein; in that, it renders the viral genome competent for transcription and replication. In fact, only minimal processivity of the polymerase is observed on naked RNA without the transcription or replication of RNA if the genome is not encapsidated by N [25, 26].
In infected cells, N selectively binds to genomic RNAs, thus avoiding the encapsidation of cellular RNA or viral mRNAs. Upon isolated expression in heterologous systems, N enwraps cellular RNAs into nucleocapsid-like structures [10, 16, 19, 123]. Elegant studies that combined real-time NMR and fluorescence spectroscopy provided a detailed kinetic description of the in vitro assembly process of N (both full length and NTAIL-free) into nucleocapsid-like particles upon the addition of six-nucleotide RNA strands [125]. Incidentally, those studies demonstrated that a continuous RNA polymer is not strictly required for N self-assembly. The assembly efficiency strongly depends on the RNA sequence, where genomic 5′ end and poly-A6 sequences trigger assembling efficiently, while poly-U6 RNAs fail to do that. Taking into account the fact that N has to encapsidate the entire genome, these results suggest that successful encapsidation proceeds via the recognition of an initial specific nucleation site [125]. Importantly, these observations enable understanding why properly capped viral messengers are not encapsidated.
Paramyxoviridae N proteins consist of two regions: a globular N-terminal moiety, NCORE, and a disordered C-terminal domain, NTAIL (Fig. 5a) (for reviews, see Refs. [38, 40, 41, 43, 44, 126, 127]). NCORE is well conserved and contains all the determinants required for self-assembly and RNA binding. NTAIL protrudes from NCORE being exposed on the nucleocapsid [7, 128,129,130]; it contains the regions responsible for P binding in both N0–P and NNUC–P complexes [8, 81, 83, 84, 126, 131, 132].
Modular organization of MeV, NiV, and HeV nucleoproteins and structure of the assembled form of MeV N. a Modular organization of N from MeV and henipaviruses consisting of a folded domain, NCORE, and a C-terminal disordered region, NTAIL. The various boxes corresponding to putative or experimentally proven MoREs are shown. The α-MoRE involved in binding to XD (see light blue helix) is highlighted by an arrow. b Schematic illustration of the MeV NCORE region. c Cryo-electron microscopy reconstruction of the MeV nucleocapsid [10, 124]. d Surface representation of the cryo-EM 3D reconstruction of the MeV trypsin-digested, helical nucleocapsid (cut away view). The colour code is the same as in a. The RNA is shown in green. The scale bar corresponds to 50 Å. Reproduced with permission from Ref. [12]. e Schematic representation of three adjacent N protomers with the same colour code as in b and d
High-resolution structural data on Paramyxoviridae N describe both monomeric and assembled forms of N. The crystal structure of the parainfluenza virus 5 (PIV5, a Rubulavirus member) N protein devoid of the disordered NTAIL appendage reveals N:RNA rings that correspond to the turns of the nucleocapsid helix [133]. The RNA is tightly packed between two lobes, referred to as NTD and CTD (for N- and C-terminal domains, respectively), separated by a hinge. The RNA is located on the external face of the N:RNA rings [133]. Each N protomer contacts six nucleotides. Two extended arms, N terminal and C terminal (NTD arm and CTD arm) contact the preceding (Ni−1) and following (Ni+1) protomer, respectively. A very similar arrangement is observed in the case of RSV N:RNA rings, except that each N protomers bind seven nucleotides and that N is comparatively shorter and devoid of the disordered NTAIL region [134].
Gutsche and co-workers first docked a model of MeV N:RNA into the electron density map of MeV nucleocapsids using the structure of RSV N:RNA rings as template [11]. Albeit unresolved, the disordered NTAIL domain was predicted to points towards the interior of the helical nucleocapsid [11]. This orientation was further confirmed by cryo-EM studies [12] that led to near-atomic resolution of NCORE-made nucleocapsids and unveiled the key role of both NTD- and CTD arms in maintaining the cohesion of between neighboring N protomers (Fig. 5). The NTD- and CTD arms also critically rigidify the CTD maintaining the RNA trapped into a closed conformation of NCORE.
Structural comparison between paramyxoviral N proteins in their assembled form (i.e., PIV5 N:RNA rings and MeV nucleocapsid) and in their monomeric, RNA-free form explained the ability of P to prevent both N self-assembly and RNA binding. Indeed, the crystal structures of monomeric, RNA-free forms of both NiV and MeVN devoid of the NTD arm and of NTAIL (N32–383 for NiV and N21–408 for MeV) in complex with the N-terminal N0-binding region of the homologous P (PNTD, P1–50 for NiV and P1–48 for MeV) were also solved [57, 58]. The superimposition of structures of the RNA-free, monomeric N in complex with PNTD and N in its assembled form reveals in N0–PNTD structure the substitution of the α-helix of the NTD arm of the MeV Ni-1 protomer by the helix α1 of PNTD (Pα1) and the overlapping of the loop of the CTD arm of the MeV Ni+1 protomer with helix α2 of PNTD (Pα2) (Fig. 6). In other words, as proposed for NiV [57], Pα1 and Pα2 compete with the NTD arm of the Ni-1 protomer and the CTD arm of the Ni+1 protomer, respectively. Hence, P can sterically prevent the association of adjacent protomers to the growing helical nucleocapsid, thereby providing a structural explanation for its chaperon role vis-à-vis of N. In the case of hMPV, in the N0–P complex, the CTD arm occupies the RNA-binding cleft thus preventing unspecific N–RNA binding by a distinct mechanism [61]. However, in the MeV N0–P complex structure obtained by Guryanov and co-workers [58], the CTD arm does not fold into the RNA-binding site suggesting that this mechanism may not be conserved in the Paramyxoviridae family or that it requires the downstream residues of the MeV CTD arm. The two MeV N structures exhibit another difference: the CTD and NTD domains move relatively to each other by 40° [58] and this results in a modification of the RNA-binding groove. Thus, while in both NiV and MeV N0–PNTD structures, the RNA-binding groove is open, and it is closed in the assembled form of N. Similarly, modeling the open conformation of PIV5 N onto its N:RNA ring structure shows a rotation of its CTD towards the centre of the ring cavity, which results in the exposure of the RNA in the RNA-binding groove that, therefore, becomes accessible for the viral polymerase [133].

Modified from Ref. [58]
Structural illustration of the chaperon role of P. Cartoon representation of the superimposition between MeV N0–PNTD (N0, light blue; PNTD, orange, pdb code 5E4V) [58] and the assembled form of N (gray, PDB code 4UFT) [12]. PNTD (aa 1–48 of P) overlaps with both NTD arm of protomer Ni−1 (yellow) and CTD arm from the Ni+1 protomer (pink). Molecules were aligned using the NTDs.
Besides the differential bending at the hinge between the two lobes, another difference between the two states of N concerns the location of the helix α6, which forms the lower lobe of the RNA-binding cleft. The different conformations adopted by this helix result in a pronounced change in the surface charge distribution. Of note, in both the NiV N0–PNTD and PIV5 N–RNA structures [57, 124], a loop preceding the corresponding helix is not defined in the electron density further supporting the inherent mobility of helix α6. Based on the flexibility of different parts of N in the various N structures, Severin and co-workers suggested that the polymerase may interact only with this flexible loop-helix motif to release the RNA instead of inducing a pronounced CTD rotation that would require more energy [126].
The amino acid sequence of NTAIL domains in Paramyxoviridae members is highly variable and characterized by a compositional bias (i.e., it is enriched in polar and charged residues and depleted in hydrophobic residues) [8, 50, 51, 80]. MeV NTAIL is hypersensitive to proteolysis and cannot be visualized in (cryo)-EM reconstructions of nucleocapsids [8, 11, 124]. All these features are hallmarks of intrinsic structural disorder, and hydrodynamic and spectroscopic analyses did effectively confirm the disordered state of NTAIL in the case of MeV, SeV, and henipaviruses [8, 50, 80, 81]. The presence of a long disordered domain within paramyxoviral N proteins is thus a conserved feature.
EM [8, 9, 11, 124, 135] and solid-state NMR studies [136] revealed a cross-talk between NCORE and NTAIL: both pitch and twist of the helicoidal nucleocapsid change upon removal of the disordered NTAIL domain which results in a more rigid helix.
As already mentioned, no direct structural data on the disordered NTAIL domain could be obtained from EM studies in that this domain was either unresolved [11] or removed by limited proteolysis [12]. By combining NMR and both small-angle X-ray and neutron scattering (SAXS and SANS), a model of the RNA-bound form of full-length MeV N could be obtained (Fig. 7) [130]. This model showed that NTAIL conserves a high conformational freedom also in the context of the nucleocapsid: the first 50 residues of NTAIL point towards the helix interior, and the remainder of the chain sneaks out of the nucleocapsid through the confined interstitial space between successive turns [130]. This model plausibly explains the increased rigidity of MeV nucleocapsids observed upon proteolytical removal of the flexible NTAIL region [8, 9, 12].

Modified from Ref. [130]
Proposed model of the location of NTAIL in intact nucleocapsids. NTAIL (red) escape from the interior of the nucleocapsid through the confined interstitial space between successive turns. NCORE monomers are colored in green and yellow and RNA in blue. a Representation of the conformational sampling of NTAIL from a single N protomer in the capsid. Different copies of NTAIL are shown to indicate the available volume sampling of the chain. Only the first 50 amino acids of NTAIL are shown. b, c Representation of the 13 NTAIL conformers from a single turn of the nucleocapsid shown across (b) or along (c) the axis of the nucleocapsid.
Subsequently, similar NMR studies of nucleocapsids showed that NiV and HeV NTAIL domains too remain disordered with the first 50 disordered amino acids of NTAIL being conformationally restricted. MeV and henipaviruses share, therefore, a conserved nucleocapsid organization: the disordered NTAIL domain is only partly exposed outside of the nucleocapsid and the first 50 residues are packed between turns of the nucleocapsid.
All the studies described above showed that in Paramyxoviridae nucleocapsids, the RNA is hidden within N protein protomers, and must be released, at least partially, to become accessible to the polymerase. This implies a local conformational change of few consecutive N subunits. The disordered NTAIL appendage and/or binding of P are predicted to govern this conformational change. The linear arrangement of N protomers within the nucleocapsid is characterized by a structural polarity in that there is a vacant P-binding site at the 3′ end. It has been proposed that this polarity may favor the recognition of the initiation site and the assembly of the N0–P–L complex [58]. Indeed, the initiation of both transcription and replication occurs at the 3′ end of the genomic RNA [107, 137]. Guryanov and co-workers proposed that the primary interactions would occur between the RNA polymerase complex, L–P, and the nucleocapsid through the low-affinity NTAIL–XD interaction (see the next section). This transient interaction would allow one-dimensional diffusion of the L–P complex along the nucleocapsid. Proper positioning on the complex would occur when PNTD binds to a vacant NTD-arm binding site at the 3′ end of the nucleocapsid. Binding of P may trigger uncoiling of the latter, as observed with MuV [138], to ease the release of the genomic RNA 3′ end from the RNA-binding groove.
Molecular mechanisms of the NTAIL–XD interaction
Residual order within NTAIL domains
In the case of MeV NTAIL, in silico analyses highlighted the presence of a putative α-MoRE (residues 488–499) within one (namely, Box2) out of three regions conserved in Morbillivirus members (referred to as Box1, Box2, and Box3) (Fig. 5a). This α-MoRE binds to XD while undergoing an α-helical-induced folding as revealed by spectroscopic, biochemical [80], and structural studies [83]. MD simulations showed that the isolated α-MoRE behaves like a molten globule [139]. Indeed, most of the conformations of the free form of the α-MoRE are more compact than in the bound conformation (the folded state) as shown by the smaller distribution of the radius of gyration (R g) of the α-MoRE in the unbound state. From the analyses of the mobility of paramagnetic spin labels grafted within Box2 and from analysis of the Cα chemical shifts of NTAIL, the α-MoRE of MeV NTAIL appears to be partly pre-folded in the absence of XD [90, 140, 141]. An atomic-resolution ensemble description of α-MoRE of MeV NTAIL was obtained by combining residual dipolar coupling (RDC) measurements and ensemble optimization methods [85, 142]. Those studies unveiled that the α-MoRE exists in a conformational equilibrium between a completely unfolded form (25%) and four conformers containing α-helical segments of 6, 7, 14, and 18 residues each, and accounting for 22, 30, 10, and 13% of the population, respectively [130]. Four aspartic acids or serines located just upstream of the observed helices stabilize all these α-helices through N-capping interactions [143]. Such N-capping stabilization of helices has already been observed in other IDPs, including SeV NTAIL [142], and nicely illustrates how the primary sequence encodes pre-recognition states. The SeV α-MoRE has a similar conformational behavior, although it samples only three helical conformers [126, 142].
In N of henipaviruses, there are four putative MoREs [50] (Fig. 5a). Their structural properties were unraveled through conformational and spectroscopic analyses of truncated forms bearing various combinations of the four predicted MoREs [144]. Two of them (Box1 and Box4, residues 408–422 and 523–532, respectively) possess irregular forms (i.e., I-MoRE), while the other two (Box2, aa 444–464 and Box3, aa 473–493) have α-helical propensities (i.e., they are putative α-MoREs) [144]. It should be noticed that Box2 had been, however, previously predicted to be a β-strand [50]. This discrepancy between experimental determination and prediction could reflect either an intrinsic plasticity of Box2 that would be able to adopt different conformations in a template-dependent manner (as already shown for IDPs in general [145]) or rather an intrinsic limitation of the predictors.
Box3 functionally corresponds to the Box2 of MeV NTAIL, i.e., it is the XD-binding site [91, 92, 144, 146, 147], while Box2 constitutes an additional putative MoRE with respect to MeV (and also SeV). The presence of this extra MoRE emphasizes the plasticity of IDPs/IDPRs that are characterized by their tolerance to insertions/deletions in functionally relevant regions.
For both NiV and HeV, site-directed spin-labeling (SDSL) EPR spectroscopy studies revealed an important conformational heterogeneity within Box3 arising from the presence of multiple helical conformers of different lengths [146]. Correlatively, Box3 appears to be at least transiently populated as an α-helix as shown by the Cα chemical shifts of free forms of HeV and NiV NTAIL. In spite of the high sequence similarity between NiV and HeV NTAIL domains (74%), hydrodynamic and spectroscopic studies pointed out subtle conformational differences [144]. A second short α-helical region is discernible within the Box2 of NiV [91, 147]. Besides, 15N R2 values argue for a higher degree of pre-configuration of NiV Box3 with respect to its HeV counterpart [147]. EPR measurements detect also a population of longer lived interconverting α-helical segments [146].
Partial pre-configuration of the XD-binding region of NTAIL is, therefore, a conserved feature shared by SeV, MeV, and henipaviruses, implying a functional significance. The partial pre-configuration of MoREs facilitates the folding-upon-binding process. The residual structure restrains the conformational space sampled by the IDP. Consequently the number of interconverting conformers in solution is reduced and less energy is demanded for the structural transition to the (partially) folded conformation [56]. Therefore, although disorder-to-order transitions are entropically disfavored, it appears that IDPs can finely tune their affinity towards partners by varying the extent of pre-configuration of their MoREs.
Molecular polymorphism in NTAIL–PXD complexes
X domains from SeV, MeV, and henipaviruses form with NTAIL a 1:1 stoichiometric complex, whose K D is in the μM range [81, 92, 148]. Binding to XD triggers α-helical folding of NTAIL [81,82,83, 92]. This structural transition takes place within a short NTAIL region (Box2, aa 486–504 of MeV N, and Box3, aa 473–493 of Henipavirus N), while the remainder of the chain remains disordered and does not establish stable contacts with XD [90,91,92, 140, 141, 144, 147, 149, 150]. The crystal structure of an MeV chimeric construct in which XD is covalently attached to the α-MoRE of NTAIL was solved at 1.8 Å [83] (Fig. 8). The structure consists in a pseudo-four helix complex in which the α-MoRE of NTAIL is embedded in a parallel orientation within a large hydrophobic cleft created by helices α2 and α3 of XD [83]. Indeed, interacting residues are mainly hydrophobic involving Leu481, Leu484, Ile488, Phe497, Met500, and Ile504 from XD, and Ser491, Ala494, Leu495, Leu498, and Met501 from NTAIL.

Modified from Ref. [151]
Structures of XD and of the interacting MoRE from NTAIL in MeV, NiV, and HeV. a Structure of the MeV Box2/XD complex (PDB code 1T6O [83]). b, c Structural models of the NiV (b) and HeV (c) Box3/XD complexes with XD [92]. b A model of NiV XD [92], while c shows the crystal structure of HeV XD (pdb code 4HEO) [91]. Box3 is shown in a parallel orientation according to Ref. [151]. In all panels, XD is in surface representation and the MoRE in ribbon representation. Hydrophobic, basic (Arg and Lys), and acidic (Asp and Glu) residues are represented in beige, blue, and red, respectively. All the other residues are shown in gray.
Burying of hydrophobic residues of the MeV α-MoRE at the XD surface likely provides the driving force of its induced folding. The structure of the MeV XD/α-MoRE complex could be used as a template to model the structure of the homologous Henipavirus complexes [92] (Fig. 8). Specifically, the more hydrophobic side of the amphipathic α-MoRE of Henipavirus NTAIL could be docked at the hydrophobic surface delimited by helices α2 and α3 of XD [92] (Fig. 8). The two modeled complexes display a rather small interface area as generally observed at the interfaces of complexes involving IDPs [56]. These models were successively validated by SDSL EPR spectroscopy studies [146].
In spite of the lack of direct structural data on Henipavirus NTAIL–XD complexes, NMR studies provided insights into the structure of these complexes and allowed further model refinement using constraints based on chemical shifts [91]. In contrast with MeV, for both NiV and HeV NTAIL domains, the resonances of the α-MoRE vanish upon the addition of the homologous XD, a finding consistent with a highly dynamic complex in which the α-MoRE undergoes α-helical fraying at the surface of XD [91, 92, 147]. The NiV NTAIL–XD complex is slightly tighter than that of HeV [91, 92, 144, 147], a property that is reflected in the ability to document by SEC complex formation in the case of NiV but not in the case of HeV [92]. SDSL EPR and NMR experiments further support Box2 of NiV as an additional interaction site with XD [146, 147].
Analysis of chemical shift perturbations in reciprocal titration studies and of the crystal structure of HeV XD identified residues involved in the interaction [91]. The binding interface is prevalently hydrophobic, but acidic residues line the binding pocket of XD. These residues establish electrostatic interactions with the basic residues of Box3, as highlighted by isothermal titration calorimetry (ITC) studies carried out at different pH values and mutational studies that targeted charged residues of both NTAIL and XD [151]. Thus, the HeV NTAIL/XD complex formation relies on the so-called “electrostatic steering mechanism” [152]. Long-range electrostatic forces would pull NTAIL towards the acidic patch on the surface of XD, to lead to an “electrostatic encounter complex” [153] with NTAIL being loosely anchored at the periphery of the binding site [151]. According to this model, HeV NTAIL would fold after binding, a scenario that is also supported by NMR titration data (see below) [91].
The role of electrostatics in the association of the HeV NTAIL and XD may explain the differential role of Box2 in binding to XD in the two henipaviruses, since Asp457 in HeV NTAIL is non-conservatively replaced by an Asn in NiV NTAIL.
From a combination of mutational and SAXS analyses, it was concluded that the MoRE adopts a parallel orientation at the surface of HeV XD [151], thus eliminating an ambiguity that persisted even after the analysis of chemical shift perturbations.
The conserved parallel orientation of the MoRE at the XD surface in MeV and HeV argues for a functional relevance. Does it govern the relative orientation of the whole P protein with respect to the NTAIL region protruding from the nucleocapsid and consequently favors an optimal positioning of the polymerase onto the nucleocapsid template? Does it favor a forward directionality of the polymerase moving along the nucleocapsid?
The prevalently hydrophobic nature of the interface of MeV, HeV, and, presumably, NiV NTAIL–XD complexes, is in line with the frequent enrichment in hydrophobic residues of binding interfaces of protein complexes involving IDPs [154]. In the case of SeV, the interaction is dominated by charges, thus giving a good example of how selection pressure in the course of evolution shaped the C-terminal domains of N and P to be structurally and functionally analogous in spite of very limited sequence identity [81].
The α-helical conformation of the XD-bound form of MeV and Henipavirus NTAIL can be presaged from the conformational ensemble that they sample as free forms of in solution [83, 90, 91, 130, 147]. This pre-configuration might be taken as indicative of a “folding before binding” mechanism [155, 156]. However, the pre-existence of ordered structures in IDPs does not necessarily commit to such a mechanism. Indeed, quantitative analysis of NMR titration data of the MeV NTAIL–XD-binding reaction [90] unveiled the presence of a weak, non-specific encounter intermediate complex, thus suggesting an induced folding mechanism [157]. In support of this, a molecular dynamic simulation study revealed that MeV NTAIL binding to XD preferentially occurs via a folding after binding mechanism [139]. Given the complexity of the experimental data, which showed that hints of both induced folding and conformational selection, this experimental system was directly interrogated using kinetics [148] (Fig. 9a). In those studies, a fortuitous complex dependence of the macroscopic rate constant k obs was observed that allowed distinguishing the contribution of the binding and folding steps in the reaction kinetics. Furthermore, by performing pseudo-first-order experiments as a function of the concentration of both XD and NTAIL [158], the clear signatures of the “folding after binding” mechanism were detected, indicating that a conformational selection mechanism, while theoretically possible, is most likely too slow to be operative [148].
Molecular mechanisms of MeV NTAIL–XD complex formation. a Kinetic-based model showing the folding after binding mechanism of the MeV NTAIL–XD interaction. NTAIL recognizes XD by first forming a weak encounter complex in a disordered conformation and is then subsequently locked-in by a folding step. Reprinted with permission from Ref. [148]. Copyright 2014 American Chemical Society. b Cartoon representation of the structural model of the NTAIL–XD complex as derived from a combined ESI–IM–MS and modeling approach. The disordered NTAIL and ordered X domain of P are shown in blue and orange, respectively. A typical MS spectrum is shown. Reprinted with permission from Ref. [99]. Copyright 2014 The American Society for Mass Spectrometry. c Schematic illustration showing the location of the NTAIL regulatory sites (stars), as identified by random mutagenesis coupled to split-GFP re-assembly assays, in the NTAIL fuzzy appendage [173]. d Schematic illustration highlighting the self-inhibitory impact of the NTAIL fuzzy appendage on binding to XD [174]. Structures were drawn using Pymol [236]
A folding-upon-binding mechanism also occurs in the case of HeV. Indeed, quantitative analysis of NTAIL peak intensities at various XD titration points revealed that the signal intensity for the residues located at the extremities of the α-MoRE decreases faster. These residues have also a smaller amount of residual helical structure in the unbound form of NTAIL. Such a differential broadening is compatible with the initial binding of XD to a short central helical segment of the α-MoRE, followed by gradual extension of this helix to the upstream and downstream residues [91]. Whether the higher helical sampling by the NiV NTAIL α-MoRE reflects a different folding mechanism remains to be investigated. A folding-upon-binding mechanism is also shared by the SeV NTAIL–XD pair, where a detailed atomic description of the molecular recognition trajectory could be obtained from relaxation dispersion studies [159].
Residual disorder (e.g., fuzziness) in NTAIL–XD complexes
The concept of “fuzziness” was coined by Tompa and Fuxreiter to describe the persistence of a significant residual structural disorder in IDP-target complexes [47]. Static fuzziness describes the sampling of a number of IDP conformations at the surface of the partner and dynamic fuzziness describes the preservation of the disordered state of the extremities of the binding segment. In this latter case, it may be hypothesized that the flexible adjacent chains could serve as baits for partner fishing through non-specific and transient contacts [160, 161]. The abundance of residual structural disorder in macromolecular complexes advocates for a functional role [48].
The NTAIL–XD complexes from paramyxoviruses combine both static and dynamic fuzziness. Indeed, the α-MoRE of Henipavirus NTAIL remains highly dynamic at the surface of XD and presumably samples many sub-conformations [91, 147] (static fuzziness). Concomitantly, a dynamic fuzzy complex is also formed, since the remainder of the NTAIL chain remains flexible within the NTAIL–XD complexes of the three viruses (see also [162]).
A large set of experimental data support the “fuzziness” of these viral complexes. (1) The majority of NTAIL in the three viruses remains disordered in the bound form, since the chemical shifts of most of the peaks are almost unaltered upon the addition of XD [90,91,92, 147, 149]. (2) The NiV NTAIL–XD complex exhibits R S about 1.6 times larger than expected for a compact complex [92]. (3) From EPR data, MeV Box1 and Henipavirus Box1 and Box4 are not involved in complex formation [141, 146]. (4) The intrinsic fluorescence spectroscopy of a Trp residue introduced within Box4 of Henipavirus NTAIL remains unchanged in the presence of bound XD as expected for a high conformational freedom and solvent exposure of the C-terminal region [92]. (5) In a low-resolution model of the MeV NTAIL–XD complex built from SAXS analyses, most of NTAIL (amino acids 401–488) remains disordered within the complex with XD [149]. (6) In the downstream region, the MoRE remains highly flexible in the MeV complex as concluded by combining SDSL EPR spectroscopy and Monte Carlo simulations [163]. (7) Atomistic models of the MeV NTAIL–XD complex built by combining ESI-Ion Mobility-MS and modeling showed different levels of compaction, i.e., high structural heterogeneity [99] (Fig. 9b). In addition, a previously undetected collapsed form of the complex was identified based on a bimodal charge-state distribution consisting of a high-charge component (18+) and a low-charge (11+) component. The former is compatible with an “open” conformation, in which the disordered arms of NTAIL flanking the α-MoRE maintain high accessibility to the solvent. The latter would represent a compact or “closed” conformation in which these flanking arms collapse onto the surface of the folded partner [99]. Structural models the “open” form of the complex were generated using experimental chemical shifts as restraints. Their solvent accessible surface area (SASA) was found to be in a very good agreement with the SASA experimentally determined by ESI–MS SASA. In the models, the inter-molecular interactions are predominantly hydrophobic both in the ordered core of the complex and in the disordered regions.
Fuzziness may confer many functional advantages, including the ability of interacting with alternative partners or on the contrary of establishing simultaneous interactions with different partners. Moreover, non-specific, transient contacts mediated by disordered appendages can promote partner fishing. Fuzzy parts of complexes can be the target of post-translational modification with regulatory activities. In addition, the reduced entropic penalty of the disorder-to-order transition due to fuzziness can enhance affinity. The interaction strength between an IDP and its partner can thus be modulated by tuning the IDP fuzziness.
In line with these expectations, the fuzzy Box3 region of MeV NTAIL serves as a low-affinity binding site for the major inducible heat-shock protein hsp70 [164, 165] that stimulates both viral transcription and replication [166,167,168]. The major hsp70-binding site is, however, located within Box2 [166, 169] and can compete out XD binding to NTAIL [164]. This suggests a model in which hsp70 would enhance viral RNA synthesis by decreasing the stability of NTAIL–XD complexes, thereby promoting P–L cartwheeling on the nucleocapsid [149, 164].
In addition, the major phosphorylation sites of MeV (S479 and S510) and NiV (S451) NTAIL fall in the fuzzy region [170,171,172], and the phosphorylation of MeV N upregulates transcription in minigenome assays [170]. Likewise, a rapid turnover of the phosphorylation of NiV N critically impacts viral RNA synthesis [172].
Finally, fuzzy regions flanking MoREs can also positively or negatively modulate the interactions established by IDPs. In fact, natural dampeners located in the N-terminal fuzzy region of NTAIL have been identified in MeV by random mutagenesis NTAIL [173] (Fig. 9c). Similarly, the fuzzy Box3 region would naturally serve as a dampener, since MeV NTAIL constructs devoid of Box3 display an enhanced interaction with XD. The impact of the long N-terminal fuzzy region of MeV NTAIL on binding to XD was thoroughly investigated though the generation of truncated forms [174] (Fig. 9d). The shortening of the NTAIL N-terminal fuzzy region increases binding to XD in the case of MeV and henipaviruses, and also to hsp70 in the former case. Although binding increases with decreasing hydrodynamic radius of the NTAIL variants, the relationship is not linear. The molecular bases of this non-monotonic behavior remain, however, to be elucidated, with possible charge-related effects having been ruled out. Results obtained by replacing the MeV NTAIL fuzzy region with a highly dissimilar artificial disordered sequence indicate that the inhibitory effect of the fuzzy region is sequence independent. Kinetics experiments that made use of single-site Trp MeV XD variant and of either NTAIL or a peptide mimicking the α-MoRE shed light on the mechanism by which the fuzzy appendage could inhibit binding. In fact, as recalled above, in the case of NTAIL, there is a hyperbolic dependence of the observed rate constant (k obs) on ligand concentration likely reflecting the fact that folding of NTAIL becomes rate-limiting at high reactant concentrations. Conversely, a linear dependence was observed with a truncated synthetic peptide that mimics the isolated MoRE. Thus, the fuzzy appendage of NTAIL may have a direct effect on the folding of the MoRE, possibly by lowering its rate constant of folding [174]. The molecular determinants of these effects, however, remain to be established.
Irrespective of the underlying mechanism, results suggest that the fuzziness of NTAIL provides a means to modulate the strength of interactions that this domain establishes with partners. Taking into account the fact that the NTAIL–XD interaction needs to be tightly regulated (see below), the discovery that the fuzzy region preceding the α-MoRE dampens the interaction provides a conceptual framework to understand why the MoRE in paramyxoviral NTAIL domains is preceded by a rather long disordered arm. It is tempting to speculate that in the course of evolution, the length of this region has been under selective pressure so as to ensure a “balanced” affinity towards XD, i.e., the arm would have been elongated until the optimized dampening level of the α-MoRE–XD interaction was reached.
Functional impact of the NTAIL–XD interaction
The hypothesis that the NTAIL–XD interaction triggers the opening of the nucleocapsid to expose the viral RNA, so that the polymerase can access to it has been dismissed by NMR studies that could not document any major nucleocapsid rearrangement upon addition of HeV XD [91]. In agreement with these findings, when PNT or PCT from MuV is added to purified MuV nucleocapsids, only PNT induces uncoiling [138]. The addition of PCT creates additional densities on the outside of the nucleocapsid. The size of these extra-densities corresponds to the size of the MuV XD-α-MoRE complex suggesting that MuV XD-α-MoRE complexes may be stabilized on the nucleocapsid core, possibly due to additional interactions between PCT and NCORE. Structural comparison between the crystal structure of VSV N:RNA rings bound or not to VSV PCT shows very little discernable differences, with the only differences concerning the P-binding site [175]. This observation thus reinforces the idea that PCT binding to nucleocapsid does not trigger much conformational rearrangement. The opening of the nucleocapsid to grant access to the polymerase would then require either full-length P, the P–L complex, and/or cellular cofactors. Hsp70, a MeV NTAIL partner [164,165,166], appears as a potential candidate. Indeed, hsp70–nucleocapsid complexes of canine distemper virus, a closely related morbillivirus, display an expanded helical diameter and an increased fragility paralleled by an increased sensitivity of the genomic RNA to nuclease degradation suggesting enhanced solvent exposure [176, 177].
The interaction of NTAIL with XD is critical as it can mediate recruitment of the L–P complex onto the nucleocapsid and/or can ensure optimal transcription and replication [36, 37, 39]. The role of Box2 in recruiting the MeV polymerase complex has been challenged by the finding that Box2 is dispensable for MeV transcription and replication in the absence of the upstream NTAIL region [95]. A truncated form of NTAIL (N1–439) is active in transcription and replication of a minigenome and in a recombinant virus [95]. However, the MeV variant suffered from a severe growth defect [95] and the N variant was very inefficient in supporting transcription re-initiation at the N–P junction in minigenome assays (see below) [94]. By analogy with MuV XD, it is conceivable that XD may bind directly to NCORE, thereby ensuring recruitment of L onto the nucleocapsid in the absence of NTAIL.
According to the so-called “cartwheeling” model, the polymerase would progress along the nucleocapsid by dynamically making/breaking contacts between NTAIL and XD to allow transcription and replication to take place. This model implies that the modifications of the N–P affinity would affect the interaction dynamics and, therefore, the processivity of the polymerase. To directly assess the relationship between the binding affinity of the NTAIL–XD pair and the polymerase elongation rate, a mutational study that targeted the MoRE of MeV NTAIL was carried out [178]. Surprisingly, no relationship was found between the binding strength of the NTAIL–XD pair and the ability of NTAIL to undergo XD-induced folding. Concomitantly, MeV mutants bearing those substitutions were generated and the elongation rates within infected cells were measured. Quite unexpectedly, a reduced binding strength had no impact on the elongation rate. It can, however, be speculated that this tolerance of the polymerase to NTAIL substitutions holds only in a certain range of affinities: in spite of a lower affinity, the NTAIL–XD interaction would still allow the recruitment of the polymerase. This absence of effect of the binding strength on the RNA synthesis was, however, challenged by a recent study, where a correlation was found between the XD–NTAIL affinity and the mRNA accumulation rates [94] (see below). The same result was obtained by mutational studies that targeted MeV XD: while the abrogation of the NTAIL–XD interaction renders the polymerase non-functional, a 1.7-fold increase in the affinity of the NTAIL–XD pair is associated with a 1.7-fold reduction in transcript accumulation rate [179]. Thus, the NTAIL–XD interaction appears to tightly control viral polymerase progression along the nucleocapsid template. The corollary of this property is that the NTAIL–XD interaction strength has to be kept into a precise window to ensure efficient transcription and replication. In line with this requirement, mutational studies carried out on MeV NTAIL showed that random substitutions in Box2 lead to a reduction in the binding strength [173]. This finding implies that the Box2 sequence is poorly evolvable, because it has been naturally selected to bind XD in an optimal way. In support of this observation, the amino acid sequence of Box2 is rather well conserved in naturally occurring MeV strains [173]. The amino acid sequence conservation of Box2 is by no means an exception among IDPs, since MoREs tend to be conserved in spite of the higher evolutionary rates of IDPs compared to structured proteins [180]. Random mutagenesis studies identified substitutions within the N-terminal region of Box2 (aa 489–493) as the most critical ones in terms of interaction strength, a finding that could be rationalized using structural data [173]. Furthermore, a critical NTAIL residue (Arg497) that had escaped detection in previous structural studies was unveiled. This led to a finer description of the hydrogen bonding network that stabilizes the NTAIL–XD complex. Finally, regulatory sites located in the fuzzy NTAIL region (i.e., outside the binding interface) were also identified (Fig. 9c). This well illustrates that in spite of their general power and usefulness, structural studies suffer from some limitations that could be nicely overcome by “descriptive random mutagenesis” approach in that it provided additional information on the NTAIL–XD complex. As such, a combinatorial experimental approach is a valuable general approach to characterize complexes involving IDPs/IDPRs.
The molecular mechanisms were subsequently investigated by examining the functional consequences of substituting critical NTAIL positions on the viral polymerase activity. To this end, molecular dynamics (MD) simulations and biochemical and functional studies were combined. Significant conclusions could be made thanks to the use of recombinant viruses and of minigenomes designed to accurately quantify transcription re-initiation after polymerase scanning through each of the five IGRs of MeV [94]. A detailed molecular explanation for the key role of NTAIL 491 and 497 residues in stabilizing the NTAIL–XD complex was achieved [94]. A correlation between NTAIL–XD affinity and the ability of N to re-initiate transcription at a downstream gene unveiled the key role of this protein interaction in transcription re-initiation at each intergenic region [94] (Fig. 10). In conclusion, the NTAIL–XD interaction is proposed to maintain the polymerase anchored to the nucleocapsid during its scanning upon crossing the IGRs and/or in the transcription re-initiation at each intergenic region. A natural selection for an optimal binding strength of NTAIL–XD would have occurred, since both increasing [179] or decreasing [94] the NTAIL–XD affinity reduces the viral growth. This suggests the establishment of an optimal equilibrium between polymerase recruitment, processivity, and transcription re-initiation efficiency. Consistent with this, none of 1,218 non-redundant circulating MeV sequences harbor the substitutions with the most dramatic impact (i.e., R490S, S491L, and R497G), while others with less drastic impact are found [173]. Accordingly, a conserved amino acid residue and fuzzy appendage length code appear to have shaped the interaction between NTAIL and XD in the course of evolution.
Model of MeV transcription re-initiation. 1 The polymerase complex, composed of the L and P proteins, transcribes the genome. 2 After the addition of the poly-A tail and release of the mRNA, the polymerase complex may re-initiate transcription and transcribe the next gene (a) or stop transcribing (b). Whether the polymerase complex detaches from the genome template (b.i) or travels on it until reaching the 5′ end of the genome (b.ii) remains to be determined. The higher is KD of the NTAIL–XD interaction, the less efficient is the re-initiation of transcription, thus leading to a steeper mRNA gradient. The schematic representation of the polymerase complex transcribing the genome is reproduced from Ref. [94]
Although polymerase activity is lost upon deletion of the last 43 residues of MeV NTAIL (comprising the α-MoRE), it is restored when the deletion is expanded to encompass the last 86 residues of NTAIL. This result led Krumm and co-workers to propose that NTAIL may inhibit polymerase access to the template, and by binding the α-MoRE, XD may overcome this inhibition by ordering NTAIL [95]. Therefore, the NTAIL–XD interaction would not only favor the polymerase anchoring and govern the RNA synthesis dynamics, but may also overcome the inhibition effect created by the disordered appendages covering the nucleocapsid.
Interestingly, the VP30 of EboV that serves as an anti-terminator transcription factor displays a similarly tightly regulated interaction with optimal binding to N being associated with optimal RNA synthesis [181]. One of the major evolution constraints to which the polymerase machinery of MeV, and possibly of paramyxoviruses in general, is subjected to is the need for an optimized interaction between the P and N proteins. It remains to be established whether binding of the C-terminus of P to the globular moiety of N (NCORE), as observed in Mononegavirales, has a similar functional role and thus needs to be similarly tightly controlled.
N–P interactions as potential targets for antivirals
If marketed drugs target mostly the active site of enzymes [182] or ligand-binding sites of receptors [183], there is a growing interest in the inhibition of protein–protein interactions (PPIs) for therapeutic purposes. The theoretical advantages of targeting PPIs lie in the limited conservation of protein interfaces, thus increasing the chances of highly specific inhibition. The inhibition of PPIs constitutes a new way to modulate the activity of proteins. A “proof of concept” of the validity of antiviral approaches relying on PPI inhibition has been obtained by targeting the HIV-1 Nef–SH3 binding surface [184].
Although there is no doubt that targeting viral entry and/or the polymerase (complex) is a valuable approach for antiviral strategies [185], a promising way to inhibit the replication of paramyxoviruses could rely on inhibition of the N–P interaction (i.e., the dual NTAIL–XD and N0–PNTD) that is crucially involved in both transcription and replication.
The antiviral activity of peptides targeting the interaction between N0 and PNTD in NiV [57], RSV [60], and RabV [68] demonstrates the relevance of choosing this interaction as a target for the design of antivirals. The fact that both NTAIL–XD and N0–PNTD interactions involve an IDPR adds further promise for antiviral approaches. In fact, many reports showed how valuable are PPIs involving IDPRs as drug discovery targets [186,187,188,189,190,191,192].
The interaction surface between a globular protein and an IDP is reminiscent of that occurring between an enzyme and its substrate or a receptor and its ligand that are often easily targeted by small molecules. Indeed, the binding of IDPs often occur through the accommodation of a MoRE into a groove of the partner. However, there are several differences. (1) The size of the interfaces of IDPs is ~25% smaller than those of ordered complexes (1141 ± 110 Å2 [56] versus 1600 ± 400 Å2 [193]). (2) Interfaces involving IDPs and globular proteins usually involve a single continuous and multiple (often discontinuous) segments [154], respectively. (3) Hydrophobic residues are preferentially exposed at the interface in the case of IDPs (40–90% exposition for IDPs versus only 5–15% for globular proteins [154]) favoring hydrophobic interactions over polar–polar interactions (33 versus 22%, respectively [154, 194]). These features make interfaces involving IDPs more easily “druggable” by small hydrophobic compounds that are usually more cell permeable, a property that constitutes a prerequisite for targeting paramyxoviral replication machineries, and many other validated therapeutic intracellular targets. There are several examples of small molecule drugs that successfully inhibit PPIs relying on IDRs (see Refs. [188, 190] and references therein cited). They include the complex between Mdm2 and p53 [195,196,197], as well as the C-terminal disordered region of PTP1B and the N-terminal disordered domain of the androgen receptor, the two latter being validated therapeutic targets for diabetes/obesity and prostate cancer, respectively [198, 199].
The NTAIL–XD and N0–PNTD interactions are attractive targets as they both imply an IDPR. Furthermore, NTAIL should be easily competed out by small molecule drugs for binding to XD owing to the rather weak binding affinity (KD in the μM range). The KD of N0–PNTD interaction is still unknown in the case of MeV and henipaviruses, but it is expected to be relatively weak, having to be competitively inhibited by both the CTD arm of the Ni+1 protomer and the NTD arm of the Ni-1 protomer to allow the encapsidation of the nascent RNA chain. It is worth to note, however, that in the case of EboV, the binding affinity of the N-terminal region of VP35 towards the N protomer (i.e., a truncated from of NCORE devoid of the N-terminal arm) is surprisingly high (K D in the nM range) [63].
The relatively small area that is buried at the interface of the NTAIL–XD complex in MeV (<450 Å2 [82]) and in Henipavirus (<700 Å2 as predicted in [92]) presages easy destabilization, in line with the admitted correlation between buried interface area and complex stability [200]. The higher total interface area of N0–PNTD complex in NiV and EboV (1440 and ~2600 Å2, respectively) may not be less favorable, since the contacting PNTD is a bipartite helix separated by a kink [57] with each binding interface being individually “druggable”.
The NTAIL–XD interfaces of henipaviruses and MeV combine core hydrophobic contacts and peripheral electrostatics interactions. Known inhibitors preferentially target hydrophobic interfaces such as those found in PPIs involving IDPRs [201]. Mimicry by small inhibitory molecules is a priori facilitated by the fact that the interaction relies on the embedding of an α-helix in a hydrophobic cleft of the structured partner in both NTAIL–XD and N0–PNTD complexes. In addition, any inhibitor of Henipavirus NTAIL–XD should inhibit both viruses, since their NTAIL and XD domains cross-bind to each other [144]. At the same time, the development of strain specific inhibitors remains possible, since the mode by which NTAIL binds to XD in NiV and HeV subtly differs.
Functional advantages of structural disorder within paramyxoviral N and P proteins
In the previous sections, the numerous experimental lines of evidence pointing to the abundance of structural disorder in the N and P proteins of paramyxoviruses have been described. An obvious question is: “which is the functional benefit that partly/largely disordered N and P proteins bring to paramyxoviruses that couldn’t be brought by fully structured replicative proteins?” In other words, which is the “added value” of structural disorder? Below, the unique functional advantages conferred by structural disorder are discussed.
Disorder as a determinant of interactivity and of extension
Structural disorder is known to be a determinant of protein interactivity: the increased plasticity typical of IDPs/IDPRs enables them to engage in a broad molecular partnership [202,203,204]. In line with this, MeV NTAIL binds to multiple partners including the viral P (XD) [8, 80, 82, 149] and M proteins [206], hsp70 [164,165,166], a nuclear export protein [205], peroxiredoxin 1 [207], cell cytoskeleton components [208, 209] the interferon regulatory factor 3 [210, 211], and a yet unidentified protein cell receptor [212, 213]. A likely broader molecular partnership of NiV and HeV NTAILs is to be identified, because they bear an additional MoRE (i.e., Box2). PNT domains also interact with multiple partners, including N [57, 58, 95, 214], cellular proteins [67, 215], and possibly the viral L protein [216].
Coordinated interactions between the polymerase complex and a large surface area of the nucleocapsid are expected to benefit from the presence of IDPRs within both P and N proteins. Strikingly, MeV PNT can extend up to 40 nm (as determined by SAXS, Longhi, and Receveur-Bréchot, unpublished data). Consequently, the L–P complex could simultaneously bind to successive turns of the helical nucleocapsid that is ~6 nm high in compact nucleocapsids [9, 12, 19]. In addition, since paramyxoviral P proteins are multimeric, they could have an even higher extension. That P remains overall extended upon binding to the nucleocapsid has been shown in the case of VSV [70]. The very large size of both PNT and NTAIL (MeV NTAIL can extend up to 13 nm in solution [8]), together with the additional flexibility brought by the “spacer” and “linker” regions of P, could enable N and P to act as scaffolding engines for partner tethering. Such a long-distance scaffolding property fits very well with the postulated assembly between the N0–P encapsidation substrate and the L–P polymerase complex to yield a tripartite N0–P–L complex.
Disorder as a determinant of nucleocapsid polymorphism
As already mentioned, MeV and henipaviruses share a conserved nucleocapsid structural organization, where the first 50 NTAIL residues are located in the interstitial space between successive turns of the nucleocapsid. The inherent flexibility of the NTAIL region sandwiched between successive turns (and hence, it is potential to undergo conformational changes) can be responsible for the observed variations in pitch and twist [10]. In turn, these conformational differences may affect the recognition of the replication and transcription promoters. Two discontinuous elements constitute the replication promoter at the 3′ end of the viral genome. If juxtaposed on two successive helical turns, they are thought to form a functional unit [217]. This model predicts that the changes of the helical conformation of the nucleocapsid, i.e., changes in the number of N monomers (and thus of nucleotides) per turn, may dictate the switch between transcription and replication by the disruption of the replication or the transcription promoter at the expenses of each other. The large conformational flexibility within Paramyxoviridae nucleocapsids observed in EM supports this postulate [19, 124, 176, 177].
Disorder as a powerful solution to modulate binding affinities
As described in detail in this review, in paramyxoviruses, two critical (and dynamic) interactions for transcription and replication, i.e., N0–PNTD and NTAIL–XD, rely on a disordered segment that undergoes α-helical folding-upon-binding to the structured partner. One can thus raise the question as to why these viruses have conserved this peculiar interaction mode.
Structural disorder is known to allow protein interactions to occur with both high specificity and low affinity [75, 218,219,220,221,222,223,224,225]. The uncoupling between affinity and specificity (i.e., the low affinity coupled to decent specificity) is known to arise from the entropic penalty that is associated with the disorder-to-order transition. However, the persistence of residual disorder in IDP complexes (in the form of fuzzy appendages) and a partial pre-configuration of binding motifs prior to binding afford a way to modulate the binding affinity (for a review, see Ref. [226]). Static fuzziness, i.e., dynamic binding of the binding motif at the surface of the partner as in Henipavirus NTAIL–XD complexes, brings an additional means for regulating the interaction strength.
Taking into account that both N0–PNTD and NTAIL–XD interactions need to be dynamically established and broken to ensure RNA synthesis, their interaction strength has to be tightly controlled. The involvement of disordered regions in these complexes provides an exquisite means to modulate the interaction strength: by tuning the extent of pre-configuration of the binding motifs and/or the length of neighboring fuzzy appendages, the virus can achieve an optimal binding strength.
In addition, the extreme allostery that typifies IDPs (i.e., the long-range nature of the effects of substitutions) affords a supplementary layer of regulation. As a result, amino acid substitutions in fuzzy regions located far away from the binding site have the potential to affect binding.
Disorder as a lessener of evolutionary constraints
Encoding structural disorder is much less constrained than encoding order, because IDRs have much fewer intramolecular interactions and are much more tolerant of substitutions or insertions as well illustrated by the extra-length of the Henipavirus PNT region with respect to other paramyxoviruses [50]. Furthermore, disorder reduces the evolutionary constraints exerted on overlapping reading frames, i.e., regions encoding multiple proteins. This is the case of PNT, which also encodes the C protein, and of the “spacer” region preceding PMD that also encodes the C-terminal domain of the V and W proteins [50, 51]. The disordered nature of these regions is in line with the pinpointed relationship between overlapping reading frames and structural disorder [51, 227,228,229,230]. Because disorder is encoded by a much broader sequence space compared to order, encoding disorder in one of the alternative reading frames may represent a strategy by which genes with overlapping coding capacities may lessen evolutionary constraints imposed on the sequence by the overlap, allowing the encoded overlapping protein products to sample a wider sequence space without losing function.
After the first seminal observations that unveiled the abundance of disorder in paramyxoviral N and P proteins [8, 45, 51, 231], several subsequent computational and experimental studies have reported the prevalence of disorder in viral proteins and especially in the proteome of RNA viruses (see Ref. [232] and references therein cited). The high mutation rates of RNA viruses and the need for compacting the genetic information into an as small as possible genome may explain the wide occurrence of disordered regions in viral proteins: disorder would allow buffering the deleterious effects of mutations and would also afford a broad molecular partnership [233, 234]. More generally, the principal advantage conferred by disorder would reside in an expanded coding capacity, where a single gene would (1) encode a single protein product able to exert multiple concomitant biological effects thanks to its promiscuity and (2) would code for multiple products through overlapping reading frames.
References
Lamb RA, Parks GD (2013) Paramyxoviridae. In: Knipe DM, Howley PM (eds) Fields virology. Lippincott Williams & Wilkins, Philadelphia, pp 957–995
Wolfson LJ, Strebel PM, Gacic-Dobo M, Hoekstra EJ, McFarland JW, Hersh BS, Measles I (2007) Has the 2005 measles mortality reduction goal been achieved? A natural history modelling study. Lancet 369:191–200
Eaton BT, Mackenzie JS, Wang LF (2007) Henipaviruses. In: Fields BN, Knipe DM, Howley PM (eds) Fields virology, 5th edn. Lippincott-Raven, Philadelphia, pp 1587–1600
Wang LF, Yu M, Hansson E, Pritchard LI, Shiell B, Michalski WP, Eaton BT (2000) The exceptionally large genome of Hendra virus: support for creation of a new genus within the family Paramyxoviridae. J Virol 74:9972–9979
Eaton BT, Broder CC, Middleton D, Wang LF (2006) Hendra and Nipah viruses: different and dangerous. Nat Rev Microbiol 4:23–35
Drexler JF, Corman VM, Gloza-Rausch F, Seebens A, Annan A, Ipsen A, Kruppa T, Muller MA, Kalko EK, Adu-Sarkodie Y, Oppong S, Drosten C (2009) Henipavirus RNA in African bats. PLoS One 4:e6367
Karlin D, Longhi S, Canard B (2002) Substitution of two residues in the measles virus nucleoprotein results in an impaired self-association. Virology 302:420–432
Longhi S, Receveur-Brechot V, Karlin D, Johansson K, Darbon H, Bhella D, Yeo R, Finet S, Canard B (2003) The C-terminal domain of the measles virus nucleoprotein is intrinsically disordered and folds upon binding to the C-terminal moiety of the phosphoprotein. J Biol Chem 278:18638–18648
Schoehn G, Mavrakis M, Albertini A, Wade R, Hoenger A, Ruigrok RW (2004) The 12 A structure of trypsin-treated measles virus N-RNA. J Mol Biol 339:301–312
Bhella D (2007) Measles virus nucleocapsid structure, conformational flexibility and the rule of six. In: Longhi S (ed) Measles virus nucleoprotein. Nova Publishers Inc., Hauppage
Desfosses A, Goret G, Farias Estrozi L, Ruigrok RW, Gutsche I (2011) Nucleoprotein–RNA orientation in the measles virus nucleocapsid by three-dimensional electron microscopy. J Virol 85:1391–1395
Gutsche I, Desfosses A, Effantin G, Ling WL, Haupt M, Ruigrok RW, Sachse C, Schoehn G (2015) Near-atomic cryo-EM structure of the helical measles virus nucleocapsid. Science 348(6235):704–707
Halpin K, Bankamp B, Harcourt BH, Bellini WJ, Rota PA (2004) Nipah virus conforms to the rule of six in a minigenome replication assay. J Gen Virol 85:701–707
Kolakofsky D, Pelet T, Garcin D, Hausmann S, Curran J, Roux L (1998) Paramyxovirus RNA synthesis and the requirement for hexamer genome length: the rule of six revisited. J Virol 72:891–899
Roux L (2005) Dans le génome des Paramyxovirinae, les promoteurs et leurs activités sont façonnés par la « règle de six. Virologie 9:19–34
Ruigrok RW, Crepin T, Kolakofsky D (2011) Nucleoproteins and nucleocapsids of negative-strand RNA viruses. Curr Opin Microbiol 14:504–510
Chan YP, Chua KB, Koh CL, Lim ME, Lam SK (2001) Complete nucleotide sequences of Nipah virus isolates from Malaysia. J Gen Virol 82:2151–2155
Blocquel D, Bourhis JM, Eléouët JF, Gerlier D, Habchi J, Jamin M, Longhi S, Yabukarski F (2012) Transcription et réplication des Mononégavirales: une machine moléculaire originale. Virologie 16:225–257
Bhella D, Ralph A, Murphy LB, Yeo RP (2002) Significant differences in nucleocapsid morphology within the Paramyxoviridae. J Gen Virol 83:1831–1839
Tan WS, Ong ST, Eshaghi M, Foo SS, Yusoff K (2004) Solubility, immunogenicity and physical properties of the nucleocapsid protein of Nipah virus produced in Escherichia coli. J Med Virol 73:105–112
Huber M, Cattaneo R, Spielhofer P, Orvell C, Norrby E, Messerli M, Perriard JC, Billeter MA (1991) Measles virus phosphoprotein retains the nucleocapsid protein in the cytoplasm. Virology 185:299–308
Spehner D, Drillien R, Howley PM (1997) The assembly of the measles virus nucleoprotein into nucleocapsid-like particles is modulated by the phosphoprotein. Virology 232:260–268
Albertini AAV, Schoehn G, Ruigrok RW (2005) Structures impliquées dans la réplication et la transcription des virus à ARN non segmentés de sens négatif. Virologie 9:83–92
Longhi S, Canard B (1999) Mécanismes de transcription et de réplication des Paramyxoviridae. Virologie 3:227–240
Morin B, Rahmeh AA, Whelan SP (2012) Mechanism of RNA synthesis initiation by the vesicular stomatitis virus polymerase. EMBO J 31:1320–1329
Morin B, Liang B, Gardner E, Ross RA, Whelan SP (2016) An in vitro RNA synthesis assay for rabies virus defines critical ribonucleoprotein interactions for polymerase activity. J Virol 91(1):e01508-16
Bloyet LM, Welsch J, Enchery F, Mathieu C, de Breyne S, Horvat B, Grigorov B, Gerlier D (2016) HSP90 Chaperoning in addition to phosphoprotein required for folding but not for supporting enzymatic activities of measles and Nipah virus L polymerases. J Virol 90:6642–6656
Katoh H, Kubota T, Nakatsu Y, Tahara M, Kidokoro M, Takeda M (2017) Heat shock protein 90 ensures efficient mumps virus replication by assisting with viral polymerase complex formation. J Virol. doi:10.1128/JVI.02220-16
Chattopadhyay S, Banerjee AK (2009) Phosphoprotein, P of human parainfluenza virus type 3 prevents self-association of RNA-dependent RNA polymerase, L. Virology 383:226–236
Gopinath M, Shaila MS (2008) Recombinant L and P protein complex of Rinderpest virus catalyses mRNA synthesis in vitro. Virus Res 135:150–154
Ogino T, Kobayashi M, Iwama M, Mizumoto K (2005) Sendai virus RNA-dependent RNA polymerase L protein catalyzes cap methylation of virus-specific mRNA. J Biol Chem 280:4429–4435
Ferron F, Longhi S, Henrissat B, Canard B (2002) Viral RNA-polymerases—a predicted 2′-O-ribose methyltransferase domain shared by all Mononegavirales. Trends Biochem Sci 27:222–224
Noton SL, Deflube LR, Tremaglio CZ, Fearns R (2012) The respiratory syncytial virus polymerase has multiple RNA synthesis activities at the promoter. PLoS Pathog 8:e1002980
Liang B, Li Z, Jenni S, Rahmeh AA, Morin BM, Grant T, Grigorieff N, Harrison SC, Whelan SP (2015) Structure of the L protein of vesicular stomatitis virus from electron cryomicroscopy. Cell 162:314–327
Bourhis JM, Longhi S (2007) Measles virus nucleoprotein: structural organization and functional role of the intrinsically disordered C-terminal domain. In: Longhi S (ed) Measles virus nucleoprotein. Nova Publishers Inc., Hauppage, pp 1–35
Longhi S (2007) Measles virus nucleoprotein. Nova Publishers Inc., Hauppage, NY
Longhi S (2009) Nucleocapsid structure and function. Curr Top Microbiol Immunol 329:103–128
Longhi S, Oglesbee M (2010) Structural disorder within the measles virus nucleoprotein and phosphoprotein. Protein Pept Lett 17:961–978
Longhi S (2011) Structural disorder within the measles virus nucleoprotein and phosphoprotein: functional implications for transcription and replication. In: Luo M (ed) Negative strand RNA virus. World Scientific Publishing, Singapore, pp 95–125
Habchi J, Longhi S (2012) Structural disorder within paramyxovirus nucleoproteins and phosphoproteins. Mol BioSyst 8:69–81
Habchi J, Mamelli L, Longhi S (2012) Structural disorder within the nucleoprotein and phosphoprotein from measles, Nipah and Hendra viruses. In: Uversky VN, Longhi S (eds) Flexible viruses: structural disorder in viral proteins. Wiley, Hoboken, pp 47–94
Communie G, Ruigrok RW, Jensen MR, Blackledge M (2014) Intrinsically disordered proteins implicated in paramyxoviral replication machinery. Curr Opin Virol 5:72–81
Longhi S (2015) Structural disorder within paramyxoviral nucleoproteins. FEBS Lett 589:2649–2659
Habchi J, Longhi S (2015) Structural disorder within paramyxoviral nucleoproteins and phosphoproteins in their free and bound forms: from predictions to experimental assessment. Int J Mol Sci 16:15688–15726
Karlin D, Longhi S, Receveur V, Canard B (2002) The N-terminal domain of the phosphoprotein of Morbilliviruses belongs to the natively unfolded class of proteins. Virology 296:251–262
Dunker AK, Babu MM, Barbar E, Blackledge M, Bondos SE, Dosztányi Z, Dyson HJ, Forman-Kay J, Fuxreiter M, Gsponer J, Han K-H, Jones DT, Longhi S, Metallo SJ, Nishikawa K, Nussinov R, Obradovic Z, Pappu RV, Rost B, Selenko P, Subramaniam V, Sussman JL, Tompa P, Uversky VN (2013) What’s in a name? Why these proteins are intrinsically disordered. Intrinsically Disord Proteins 1:e24157
Tompa P, Fuxreiter M (2008) Fuzzy complexes: polymorphism and structural disorder in protein–protein interactions. Trends Biochem Sci 33:2–8
Miskei M, Antal C, Fuxreiter M (2016) FuzDB: database of fuzzy complexes, a tool to develop stochastic structure-function relationships for protein complexes and higher-order assemblies. Nucleic Acids Res 45:D228–S235
Habchi J, Tompa P, Longhi S, Uversky VN (2014) Introducing protein intrinsic disorder. Chem Rev 114:6561–6588
Habchi J, Mamelli L, Darbon H, Longhi S (2010) Structural disorder within Henipavirus nucleoprotein and phosphoprotein: from predictions to experimental assessment. PLoS One 5:e11684
Karlin D, Ferron F, Canard B, Longhi S (2003) Structural disorder and modular organization in Paramyxovirinae N and P. J Gen Virol 84:3239–3252
Lieutaud P, Canard B, Longhi S (2008) MeDor: a metaserver for predicting protein disorder. BMC Genom 9:S25
Garner E, Romero P, Dunker AK, Brown C, Obradovic Z (1999) Predicting binding regions within disordered proteins. Genome Inform Ser Workshop Genome Inform 10:41–50
Oldfield CJ, Cheng Y, Cortese MS, Romero P, Uversky VN, Dunker AK (2005) Coupled folding and binding with alpha-helix-forming molecular recognition elements. Biochemistry 44:12454–12470
Mohan A, Oldfield CJ, Radivojac P, Vacic V, Cortese MS, Dunker AK, Uversky VN (2006) Analysis of molecular recognition features (MoRFs). J Mol Biol 362:1043–1059
Vacic V, Oldfield CJ, Mohan A, Radivojac P, Cortese MS, Uversky VN, Dunker AK (2007) Characterization of molecular recognition features, MoRFs, and their binding partners. J Proteome Res 6:2351–2366
Yabukarski F, Lawrence P, Tarbouriech N, Bourhis JM, Delaforge E, Jensen MR, Ruigrok RW, Blackledge M, Volchkov V, Jamin M (2014) Structure of Nipah virus unassembled nucleoprotein in complex with its viral chaperone. Nat Struct Mol Biol 21:754–759
Guryanov SG, Liljeroos L, Kasaragod P, Kajander T, Butcher SJ (2016) Crystal structure of the measles virus nucleoprotein core in complex with an N-terminal region of phosphoprotein. J Virol 90:2849–2857
Karlin D, Belshaw R (2012) Detecting remote sequence homology in disordered proteins: discovery of conserved motifs in the N-termini of Mononegavirales phosphoproteins. PLoS One 7:e31719
Galloux M, Gabiane G, Sourimant J, Richard CA, England P, Moudjou M, Aumont-Nicaise M, Fix J, Rameix-Welti MA, Eleouet JF (2015) Identification and characterization of the binding site of the respiratory syncytial virus phosphoprotein to RNA-free nucleoprotein. J Virol 89:3484–3496
Renner M, Bertinelli M, Leyrat C, Paesen GC, Saraiva de Oliveira LF, Huiskonen JT, Grimes JM (2016) Nucleocapsid assembly in pneumoviruses is regulated by conformational switching of the N protein. Elife 5:e12627
Leyrat C, Jensen MR, Ribeiro EA Jr, Gerard FC, Ruigrok RW, Blackledge M, Jamin M (2011) The N(0)-binding region of the vesicular stomatitis virus phosphoprotein is globally disordered but contains transient alpha-helices. Protein Sci 20:542–556
Leung DW, Borek D, Luthra P, Binning JM, Anantpadma M, Liu G, Harvey IB, Su Z, Endlich-Frazier A, Pan J, Shabman RS, Chiu W, Davey RA, Otwinowski Z, Basler CF, Amarasinghe GK (2015) An intrinsically disordered peptide from Ebola virus VP35 controls viral RNA synthesis by modulating nucleoprotein–RNA interactions. Cell Rep 11:376–389
Leyrat C, Yabukarski F, Tarbouriech N, Ribeiro EA Jr, Jensen MR, Blackledge M, Ruigrok RW, Jamin M (2011) Structure of the vesicular stomatitis virus N(0)–P complex. PLoS Pathog 7:e1002248
Pereira N, Cardone C, Lassoued S, Galloux M, Fix J, Assrir N, Lescop E, Bontems F, Eleouet JF, Sizun C (2017) New insights into structural disorder in human respiratory syncytial virus phosphoprotein and implications for binding of protein partners. J Biol Chem 292:2120–2131
Sweetman DA, Miskin J, Baron MD (2001) Rinderpest virus C and V proteins interact with the major (L) component of the viral polymerase. Virology 281:193–204
Ding B, Zhang G, Yang X, Zhang S, Chen L, Yan Q, Xu M, Banerjee AK, Chen M (2014) Phosphoprotein of human parainfluenza virus type 3 blocks autophagosome–lysosome fusion to increase virus production. Cell Host Microbe 15:564–577
Castel G, Chteoui M, Caignard G, Prehaud C, Mehouas S, Real E, Jallet C, Jacob Y, Ruigrok RW, Tordo N (2009) Peptides that mimic the amino-terminal end of the rabies virus phosphoprotein have antiviral activity. J Virol 83:10808–10820
Rahmeh AA, Morin B, Schenk AD, Liang B, Heinrich BS, Brusic V, Walz T, Whelan SP (2012) Critical phosphoprotein elements that regulate polymerase architecture and function in vesicular stomatitis virus. Proc Natl Acad Sci USA 109:14628–14633
Yabukarski F, Leyrat C, Martinez N, Communie G, Ivanov I, Ribeiro EA Jr, Buisson M, Gerard FC, Bourhis JM, Jensen MR, Bernado P, Blackledge M, Jamin M (2016) Ensemble structure of the highly flexible complex formed between vesicular stomatitis virus unassembled nucleoprotein and its phosphoprotein chaperone. J Mol Biol 428:2671–2694
Jamin M, Yabukarski F (2017) Nonsegmented negative-sense RNA viruses-structural data bring new insights into nucleocapsid assembly. Adv Virus Res 97:143–185
Ding H, Green TJ, Lu S, Luo M (2006) Crystal structure of the oligomerization domain of the phosphoprotein of vesicular stomatitis virus. J Virol 80:2808–2814
Ivanov I, Crepin T, Jamin M, Ruigrok RW (2010) Structure of the dimerization domain of the rabies virus phosphoprotein. J Virol 84:3707–3710
Uversky VN (2002) Natively unfolded proteins: a point where biology waits for physics. Protein Sci 11:739–756
Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, Oldfield CJ, Campen AM, Ratliff CM, Hipps KW, Ausio J, Nissen MS, Reeves R, Kang C, Kissinger CR, Bailey RW, Griswold MD, Chiu W, Garner EC, Obradovic Z (2001) Intrinsically disordered protein. J Mol Graph Model 19:26–59
Fuentes SM, Sun D, Schmitt AP, He B (2010) Phosphorylation of paramyxovirus phosphoprotein and its role in viral gene expression. Future Microbiol 5:9–13
Gerard FC, RibeiroEde A Jr, Leyrat C, Ivanov I, Blondel D, Longhi S, Ruigrok RW, Jamin M (2009) Modular organization of rabies virus phosphoprotein. J Mol Biol 388:978–996
Leyrat C, Gerard FC, de Almeida Ribeiro E, Jr Ivanov I, Ruigrok RW, Jamin M (2010) Structural disorder in proteins of the rhabdoviridae replication complex. Protein Pept Lett 17:979–987
Iakoucheva LM, Radivojac P, Brown CJ, O’Connor TR, Sikes JG, Obradovic Z, Dunker AK (2004) The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res 32:1037–1049
Bourhis J, Johansson K, Receveur-Bréchot V, Oldfield CJ, Dunker AK, Canard B, Longhi S (2004) The C-terminal domain of measles virus nucleoprotein belongs to the class of intrinsically disordered proteins that fold upon binding to their physiological partner. Virus Res 99:157–167
Houben K, Marion D, Tarbouriech N, Ruigrok RW, Blanchard L (2007) Interaction of the C-terminal domains of Sendai virus N and P proteins: comparison of polymerase-nucleocapsid interactions within the paramyxovirus family. J Virol 81:6807–6816
Johansson K, Bourhis JM, Campanacci V, Cambillau C, Canard B, Longhi S (2003) Crystal structure of the measles virus phosphoprotein domain responsible for the induced folding of the C-terminal domain of the nucleoprotein. J Biol Chem 278:44567–44573
Kingston RL, Hamel DJ, Gay LS, Dahlquist FW, Matthews BW (2004) Structural basis for the attachment of a paramyxoviral polymerase to its template. Proc Natl Acad Sci USA 101:8301–8306
Kingston RL, Walter AB, Gay LS (2004) Characterization of nucleocapsid binding by the measles and the mumps virus phosphoprotein. J Virol 78:8630–8640
Bernado P, Blanchard L, Timmins P, Marion D, Ruigrok RW, Blackledge M (2005) A structural model for unfolded proteins from residual dipolar couplings and small-angle X-ray scattering. Proc Natl Acad Sci USA 102:17002–17007
Blanchard L, Tarbouriech N, Blackledge M, Timmins P, Burmeister WP, Ruigrok RW, Marion D (2004) Structure and dynamics of the nucleocapsid-binding domain of the Sendai virus phosphoprotein in solution. Virology 319:201–211
Houben K, Blanchard L, Blackledge M, Marion D (2007) Intrinsic dynamics of the partly unstructured PX domain from the Sendai virus RNA polymerase cofactor P. Biophys J 93:2830–2844
Communie G, Crepin T, Maurin D, Jensen MR, Blackledge M, Ruigrok RW (2013) Structure of the tetramerization domain of measles virus phosphoprotein. J Virol 87:7166–7169
Tarbouriech N, Curran J, Ruigrok RW, Burmeister WP (2000) Tetrameric coiled coil domain of Sendai virus phosphoprotein. Nat Struct Biol 7:777–781
Gely S, Lowry DF, Bernard C, Ringkjobing-Jensen M, Blackledge M, Costanzo S, Darbon H, Daughdrill GW, Longhi S (2010) Solution structure of the C-terminal X domain of the measles virus phosphoprotein and interaction with the intrinsically disordered C-terminal domain of the nucleoprotein. J Mol Recognit 23:435–447
Communie G, Habchi J, Yabukarski F, Blocquel D, Schneider R, Tarbouriech N, Papageorgiou N, Ruigrok RW, Jamin M, Ringkjøbing-Jensen M, Longhi S, Blackledge M (2013) Atomic resolution description of the interaction between the nucleoprotein and phosphoprotein of Hendra virus. PLoS Pathog 9:e1003631
Habchi J, Blangy S, Mamelli L, Ringkjobing Jensen M, Blackledge M, Darbon H, Oglesbee M, Shu Y, Longhi S (2011) Characterization of the interactions between the nucleoprotein and the phosphoprotein of Henipaviruses. J Biol Chem 286:13583–13602
Kingston RL, Gay LS, Baase WS, Matthews BW (2008) Structure of the nucleocapsid-binding domain from the mumps virus polymerase; an example of protein folding induced by crystallization. J Mol Biol 379:719–731
Bloyet L, Brunel J, Dosnon M, Hamon V, Erales J, Gruet A, Lazert C, Bignon C, Roche P, Longhi S, Gerlier D (2016) Modulation of re-initiation of measles virus transcription at intergenic regions by PXD to NTAIL binding strength. PLoS Pathog 12:e1006058
Krumm SA, Takeda M, Plemper RK (2013) The measles virus nucleocapsid protein tail domain is dispensable for viral polymerase recruitment and activity. J Biol Chem 288:29943–29953
Llorente MT, Barreno-Garcia B, Calero M, Camafeita E, Lopez JA, Longhi S, Ferron F, Varela PF, Melero JA (2006) Structural analysis of the human respiratory syncitial virus phosphoprotein: characterization of an a-helical domain involved in oligomerization. J Gen Virol 87:159–169
Tran TL, Castagne N, Bhella D, Varela PF, Bernard J, Chilmonczyk S, Berkenkamp S, Benhamo V, Grznarova K, Grosclaude J, Nespoulos C, Rey FA, Eleouet JF (2007) The nine C-terminal amino acids of the respiratory syncytial virus protein P are necessary and sufficient for binding to ribonucleoprotein complexes in which six ribonucleotides are contacted per N protein protomer. J Gen Virol 88:196–206
Yegambaram K, Bulloch EM, Kingston RL (2013) Protein domain definition should allow for conditional disorder. Protein Sci 22:1502–1518
D’Urzo A, Konijnenberg A, Rossetti G, Habchi J, Li J, Carloni P, Sobott F, Longhi S, Grandori R (2015) Molecular basis for structural heterogeneity of an intrinsically disordered protein bound to a partner by combined ESI-IM-MS and modeling. J Am Soc Mass Spectrom 26:472–481
Bonetti D, Camilloni C, Visconti L, Longhi S, Brunori M, Vendruscolo M, Gianni S (2016) Identification and structural characterization of an intermediate in the folding of the measles virus X domain. J Biol Chem 291:10886–10892
Rahaman A, Srinivasan N, Shamala N, Shaila MS (2004) Phosphoprotein of the rinderpest virus forms a tetramer through a coiled coil region important for biological function. A structural insight. J Biol Chem 279:23606–23614
Blocquel D, Habchi J, Durand E, Sevajol M, Ferron F, Erales J, Papageorgiou N, Longhi S (2014) Coiled-coil deformations in crystal structures: the measles virus phosphoprotein multimerization domain as an illustrative example. Acta Crystallogr D 70:1589–1603
Cox R, Green TJ, Purushotham S, Deivanayagam C, Bedwell GJ, Prevelige PE, Luo M (2013) Structural and functional characterization of the mumps virus phosphoprotein. J Virol 87:7558–7568
Bruhn-Johannsen JF, Barnett K, Bibby J, Thomas J, Keegan R, Rigden D, Bornholdt ZA, Saphire EO (2014) Crystal structure of the Nipah virus phosphoprotein tetramerization domain. J Virol 88:758–762
Leyrat C, Renner M, Harlos K, Grimes JM (2013) Solution and crystallographic structures of the central region of the phosphoprotein from human metapneumovirus. PLoS One 8:e80371
Castagne N, Barbier A, Bernard J, Rezaei H, Huet JC, Henry C, Da Costa B, Eleouet JF (2004) Biochemical characterization of the respiratory syncytial virus P–P and P–N protein complexes and localization of the P protein oligomerization domain. J Gen Virol 85:1643–1653
Kolakofsky D, Le Mercier P, Iseni F, Garcin D (2004) Viral DNA polymerase scanning and the gymnastics of Sendai virus RNA synthesis. Virology 318:463–473
Llorente MT, Taylor IA, Lopez-Vinas E, Gomez-Puertas P, Calder LJ, Garcia-Barreno B, Melero JA (2008) Structural properties of the human respiratory syncytial virus P protein: evidence for an elongated homotetrameric molecule that is the smallest orthologue within the family of paramyxovirus polymerase cofactors. Proteins 72:946–958
Luthra P, Jordan DS, Leung DW, Amarasinghe GK, Basler CF (2015) Ebola virus VP35 interaction with dynein LC8 regulates viral RNA synthesis. J Virol 89:5148–5153
Beltrandi M, Blocquel D, Erales J, Barbier P, Cavalli A, Longhi S (2015) Insights into the coiled-coil organization of the Hendra virus phosphoprotein from combined biochemical and SAXS studies. Virology 477:42–55
Blocquel D, Beltrandi M, Erales J, Barbier P, Longhi S (2013) Biochemical and structural studies of the oligomerization domain of the Nipah virus phosphoprotein: evidence for an elongated coiled-coil homotrimer. Virology 446:162–172
Salvamani S, Goh Z, Ho K, Tey B, Tan W (2013) Oligomerization state of the multimerization domain of Nipah virus phosphoprotein. Process Biochem 48:1476–1480
Dutta K, Alexandrov A, Huang H, Pascal SM (2001) pH-induced folding of an apoptotic coiled coil. Protein Sci 10:2531–2540
Lupas AN, Gruber M (2005) The structure of alpha-helical coiled coils. Adv Protein Chem 70:37–78
Oshaben KM, Salari R, McCaslin DR, Chong LT, Horne WS (2012) The native GCN4 leucine-zipper domain does not uniquely specify a dimeric oligomerization state. Biochemistry 51:9581–9591
Curran J (1998) A role for the Sendai virus P protein trimer in RNA synthesis. J Virol 72:4274–4280
Curran J, Boeck R, Lin-Marq N, Lupas A, Kolakofsky D (1995) Paramyxovirus phosphoproteins form homotrimers as determined by an epitope dilution assay, via predicted coiled coils. Virology 214:139–149
Bruhn JF, Kirchdoerfer RN, Urata SM, Li S, Tickle IJ, Bricogne G, Saphire EO (2017) Crystal structure of the Marburg virus VP35 oligomerization domain. J Virol 91. doi:10.1128/JVI.01085-16
Asenjo A, Mendieta J, Gomez-Puertas P, Villanueva N (2008) Residues in human respiratory syncytial virus P protein that are essential for its activity on RNA viral synthesis. Virus Res 132:160–173
Chen M, Ogino T, Banerjee AK (2006) Mapping and functional role of the self-association domain of vesicular stomatitis virus phosphoprotein. J Virol 80:9511–9518
Choudhary SK, Malur AG, Huo Y, De BP, Banerjee AK (2002) Characterization of the oligomerization domain of the phosphoprotein of human parainfluenza virus type 3. Virology 302:373–382
Jacob Y, Real E, Tordo N (2001) Functional interaction map of lyssavirus phosphoprotein: identification of the minimal transcription domains. J Virol 75:9613–9622
Warnes A, Fooks AR, Dowsett AB, Wilkinson GW, Stephenson JR (1995) Expression of the measles virus nucleoprotein gene in Escherichia coli and assembly of nucleocapsid-like structures. Gene 160:173–178
Bhella D, Ralph A, Yeo RP (2004) Conformational flexibility in recombinant measles virus nucleocapsids visualised by cryo-negative stain electron microscopy and real-space helical reconstruction. J Mol Biol 340:319–331
Milles S, Jensen MR, Communie G, Maurin D, Schoehn G, Ruigrok RW, Blackledge M (2016) Self-assembly of measles virus nucleocapsid-like particles: kinetics and RNA sequence dependence. Angew Chem Int Ed Engl 55:9356–9360
Jensen MR, Bernado P, Houben K, Blanchard L, Marion D, Ruigrok RW, Blackledge M (2010) Structural disorder within Sendai virus nucleoprotein and phosphoprotein: insight into the structural basis of molecular recognition. Protein Pept Lett 17:952–960
Erales J, Blocquel D, Habchi J, Beltrandi M, Gruet A, Dosnon M, Bignon C, Longhi S (2015) Order and disorder in the replicative complex of Paramyxoviruses. Adv Exp Med Biol 870:351–381
Heggeness MH, Scheid A, Choppin PW (1980) Conformation of the helical nucleocapsids of paramyxoviruses and vesicular stomatitis virus: reversible coiling and uncoiling induced by changes in salt concentration. Proc Natl Acad Sci USA 77:2631–2635
Heggeness MH, Scheid A, Choppin PW (1981) The relationship of conformational changes in the Sendai virus nucleocapsid to proteolytic cleavage of the NP polypeptide. Virology 114:555–562
Ringkjøbing Jensen M, Communie G, Ribeiro ED Jr, Martinez N, Desfosses A, Salmon L, Mollica L, Gabel F, Jamin M, Longhi S, Ruigrok RW, Blackledge M (2011) Intrinsic disorder in measles virus nucleocapsids. Proc Natl Acad Sci USA 108:9839–9844
Bankamp B, Horikami SM, Thompson PD, Huber M, Billeter M, Moyer SA (1996) Domains of the measles virus N protein required for binding to P protein and self-assembly. Virology 216:272–277
Liston P, Batal R, DiFlumeri C, Briedis DJ (1997) Protein interaction domains of the measles virus nucleocapsid protein (NP). Adv Virol 142:305–321
Alayyoubi M, Leser GP, Kors CA, Lamb RA (2015) Structure of the paramyxovirus parainfluenza virus 5 nucleoprotein–RNA complex. Proc Natl Acad Sci USA 112:E1792–E1799
Tawar RG, Duquerroy S, Vonrhein C, Varela PF, Damier-Piolle L, Castagné N, MacLellan K, Bedouelle H, Bricogne G, Bhella D, Eleouet JF, Rey FA (2009) 3D structure of a nucleocapsid-like nucleoprotein–RNA complex of respiratory syncytial virus. Science 326:1279–1283
Severin C, Terrell JR, Zengel JR, Cox R, Plemper RK, He B, Luo M (2016) Releasing the genomic RNA sequestered in the mumps virus nucleocapsid. J Virol. doi:10.1128/JVI.01422-16
Barbet-Massin E, Felletti M, Schneider R, Jehle S, Communie G, Martinez N, Jensen MR, Ruigrok RW, Emsley L, Lesage A, Blackledge M, Pintacuda G (2014) Insights into the structure and dynamics of measles virus nucleocapsids by 1H-detected solid-state NMR. Biophys J 107:941–946
Noton SL, Fearns R (2015) Initiation and regulation of paramyxovirus transcription and replication. Virology 479–480:545–554
Cox R, Pickar A, Qiu S, Tsao J, Rodenburg C, Dokland T, Elson A, He B, Luo M (2014) Structural studies on the authentic mumps virus nucleocapsid showing uncoiling by the phosphoprotein. Proc Natl Acad Sci USA 111:15208–15213
Wang Y, Chu X, Longhi S, Roche P, Han W, Wang E, Wang J (2013) Multiscaled exploration of coupled folding and binding of an intrinsically disordered molecular recognition element in measles virus nucleoprotein. Proc Natl Acad Sci USA 110:E3743–E3752
Morin B, Bourhis JM, Belle V, Woudstra M, Carrière F, Guigliarelli B, Fournel A, Longhi S (2006) Assessing induced folding of an intrinsically disordered protein by site-directed spin-labeling EPR spectroscopy. J Phys Chem B 110:20596–20608
Belle V, Rouger S, Costanzo S, Liquiere E, Strancar J, Guigliarelli B, Fournel A, Longhi S (2008) Mapping alpha-helical induced folding within the intrinsically disordered C-terminal domain of the measles virus nucleoprotein by site-directed spin-labeling EPR spectroscopy. Proteins Struct Funct Bioinform 73:973–988
Jensen MR, Houben K, Lescop E, Blanchard L, Ruigrok RW, Blackledge M (2008) Quantitative conformational analysis of partially folded proteins from residual dipolar couplings: application to the molecular recognition element of Sendai virus nucleoprotein. J Am Chem Soc 130:8055–8061
Serrano L, Fersht AR (1989) Capping and alpha-helix stability. Nature 342:296–299
Blocquel D, Habchi J, Gruet A, Blangy S, Longhi S (2012) Compaction and binding properties of the intrinsically disordered C-terminal domain of Henipavirus nucleoprotein as unveiled by deletion studies. Mol BioSyst 8:392–410
Hazy E, Tompa P (2009) Limitations of induced folding in molecular recognition by intrinsically disordered proteins. ChemPhysChem 10:1415–1419
Martinho M, Habchi J, El Habre Z, Nesme L, Guigliarelli B, Belle V, Longhi S (2013) Assessing induced folding within the intrinsically disordered C-terminal domain of the Henipavirus nucleoproteins by site directed spin labeling EPR spectroscopy. J Biomol Struct Dyn 31:453–471
Baronti L, Erales J, Habchi J, Felli IC, Pierattelli R, Longhi S (2015) Dynamics of the intrinsically disordered C-terminal domain of the Nipah virus nucleoprotein and interaction with the X domain of the phosphoprotein as unveiled by NMR spectroscopy. ChemBioChem 16:268–276
Dosnon M, Bonetti D, Morrone A, Erales J, di Silvio E, Longhi S, Gianni S (2015) Demonstration of a folding after binding mechanism in the recognition between the measles virus NTAIL and X domains. ACS Chem Biol 10:795–802
Bourhis JM, Receveur-Bréchot V, Oglesbee M, Zhang X, Buccellato M, Darbon H, Canard B, Finet S, Longhi S (2005) The intrinsically disordered C-terminal domain of the measles virus nucleoprotein interacts with the C-terminal domain of the phosphoprotein via two distinct sites and remains predominantly unfolded. Protein Sci 14:1975–1992
Belle V, Rouger S, Costanzo S, Longhi S, Fournel A (2010) Site-directed spin labeling EPR spectroscopy. In: Uversky VN, Longhi S (eds) Instrumental analysis of intrinsically disordered proteins: assessing structure and conformation. Wiley, Hoboken
Erales J, Beltrandi M, Roche J, Maté M, Longhi S (2015) Insights into the Hendra virus NTAIL-XD complex: evidence for a parallel organization of the helical MoRE at the XD surface stabilized by a combination of hydrophobic and polar interactions. Biochim Biopys Acta 1854:1038–1053
Schreiber G, Haran G, Zhou HX (2009) Fundamental aspects of protein–protein association kinetics. Chem Rev 109:839–860
Xue Y, Yuwen T, Zhu F, Skrynnikov NR (2014) Role of electrostatic interactions in binding of peptides and intrinsically disordered proteins to their folded targets. 1. NMR and MD characterization of the complex between the c-Crk N-SH3 domain and the peptide Sos. Biochemistry 53:6473–6495
Meszaros B, Tompa P, Simon I, Dosztanyi Z (2007) Molecular principles of the interactions of disordered proteins. J Mol Biol 372:549–561
Tsai CD, Ma B, Kumar S, Wolfson H, Nussinov R (2001) Protein folding: binding of conformationally fluctuating building blocks via population selection. Crit Rev Biochem Mol Biol 36:399–433
Tsai CJ, Ma B, Sham YY, Kumar S, Nussinov R (2001) Structured disorder and conformational selection. Proteins Struct Funct Bioinform 44:418–427
Shoemaker BA, Portman JJ, Wolynes PG (2000) Speeding molecular recognition by using the folding funnel: the fly-casting mechanism. Proc Natl Acad Sci USA 97:8868–8873
Gianni S, Dogan J, Jemth P (2014) Distinguishing induced fit from conformational selection. Biophys Chem 189:33–39
Schneider R, Maurin D, Communie G, Kragelj J, Hansen DF, Ruigrok RW, Jensen MR, Blackledge M (2015) Visualizing the molecular recognition trajectory of an intrinsically disordered protein using multinuclear relaxation dispersion NMR. J Am Chem Soc 137:1220–1229
Fuxreiter M, Tompa P (2009) Fuzzy interactome: the limitations of models in molecular biology. Trends Biochem Sci 34:3
Fuxreiter M (2012) Fuzziness: linking regulation to protein dynamics. Mol BioSyst 8:168–177
Longhi S (2012) The measles virus N(TAIL)-XD complex: an illustrative example of fuzziness. Adv Exp Med Biol 725:126–141
Kavalenka A, Urbancic I, Belle V, Rouger S, Costanzo S, Kure S, Fournel A, Longhi S, Guigliarelli B, Strancar J (2010) Conformational analysis of the partially disordered measles virus NTAIL-XD complex by SDSL EPR spectroscopy. Biophys J 98:1055–1064
Zhang X, Bourhis JM, Longhi S, Carsillo T, Buccellato M, Morin B, Canard B, Oglesbee M (2005) Hsp72 recognizes a P binding motif in the measles virus N protein C-terminus. Virology 337:162–174
Couturier M, Buccellato M, Costanzo S, Bourhis JM, Shu Y, Nicaise M, Desmadril M, Flaudrops C, Longhi S, Oglesbee M (2010) High affinity binding between Hsp70 and the C-terminal domain of the measles virus nucleoprotein requires an Hsp40 co-chaperone. J Mol Recognit 23:301–315
Zhang X, Glendening C, Linke H, Parks CL, Brooks C, Udem SA, Oglesbee M (2002) Identification and characterization of a regulatory domain on the carboxyl terminus of the measles virus nucleocapsid protein. J Virol 76:8737–8746
Carsillo T, Zhang X, Vasconcelos D, Niewiesk S, Oglesbee M (2006) A single codon in the nucleocapsid protein C terminus contributes to in vitro and in vivo fitness of Edmonston measles virus. J Virol 80:2904–2912
Oglesbee M (2007) Nucleocapsid protein interactions with the major inducible heat shock protein. In: Longhi S (ed) Measles virus nucleoprotein. Nova Publishers Inc., Hauppage, pp 53–98
Carsillo T, Traylor Z, Choi C, Niewiesk S, Oglesbee M (2006) hsp72, a host determinant of measles virus neurovirulence. J Virol 80:11031–11039
Hagiwara K, Sato H, Inoue Y, Watanabe A, Yoneda M, Ikeda F, Fujita K, Fukuda H, Takamura C, Kozuka-Hata H, Oyama M, Sugano S, Ohmi S, Kai C (2008) Phosphorylation of measles virus nucleoprotein upregulates the transcriptional activity of minigenomic RNA. Proteomics 8:1871–1879
Sugai A, Sato H, Yoneda M, Kai C (2013) Phosphorylation of measles virus nucleoprotein affects viral growth by changing gene expression and genomic RNA stability. J Virol 87:11684–11692
Huang M, Sato H, Hagiwara K, Watanabe A, Sugai A, Ikeda F, Kozuka-Hata H, Oyama M, Yoneda M, Kai C (2011) Determination of a phosphorylation site in Nipah virus nucleoprotein and its involvement in virus transcription. J Gen Virol 92:2133–2141
Gruet A, Dosnon M, Vassena A, Lombard V, Gerlier D, Bignon C, Longhi S (2013) Dissecting partner recognition by an intrinsically disordered protein using descriptive random mutagenesis. J Mol Biol 425:3495–3509
Gruet A, Dosnon M, Blocquel D, Brunel J, Gerlier D, Das RK, Bonetti D, Gianni S, Fuxreiter M, Longhi S, Bignon C (2016) Fuzzy regions in an intrinsically disordered protein impair protein–protein interactions. FEBS J 283:576–594
Green TJ, Luo M (2009) Structure of the vesicular stomatitis virus nucleocapsid in complex with the nucleocapsid-binding domain of the small polymerase cofactor, P. Proc Natl Acad Sci USA 106:11713–11718
Oglesbee M, Ringler S, Krakowka S (1990) Interaction of canine distemper virus nucleocapsid variants with 70K heat-shock proteins. J Gen Virol 71:1585–1590
Oglesbee M, Tatalick L, Rice J, Krakowka S (1989) Isolation and characterization of canine distemper virus nucleocapsid variants. J Gen Virol 70(Pt 9):2409–2419
Shu Y, Habchi J, Costanzo S, Padilla A, Brunel J, Gerlier D, Oglesbee M, Longhi S (2012) Plasticity in structural and functional interactions between the phosphoprotein and nucleoprotein of measles virus. J Biol Chem 287:11951–11967
Brunel J, Chopy D, Dosnon M, Bloyet LM, Devaux P, Urzua E, Cattaneo R, Longhi S, Gerlier D (2014) Sequence of events in measles virus replication: role of phosphoprotein–nucleocapsid interactions. J Virol 88:10851–10863
Huang H, Sarai A (2012) Analysis of the relationships between evolvability, thermodynamics, and the functions of intrinsically disordered proteins/regions. Comput Biol Chem 41:51–57
Kirchdoerfer RN, Moyer CL, Abelson DM, Saphire EO (2016) The Ebola virus VP30-NP interaction is a regulator of viral RNA synthesis. PLoS Pathog 12:e1005937
Sebolt-Leopold JS, English JM (2006) Mechanisms of drug inhibition of signalling molecules. Nature 441:457–462
Lagerstrom MC, Schioth HB (2008) Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat Rev Drug Discov 7:339–357
Betzi S, Restouin A, Opi S, Arold ST, Parrot I, Guerlesquin F, Morelli X, Collette Y (2007) Protein protein interaction inhibition (2P2I) combining high throughput and virtual screening: application to the HIV-1 Nef protein. Proc Natl Acad Sci USA 104:19256–19261
Cox R, Plemper RK (2015) The paramyxovirus polymerase complex as a target for next-generation anti-paramyxovirus therapeutics. Front Microbiol 6:459
Cheng Y, Legall T, Oldfield CJ, Mueller JP, Van YY, Romero P, Cortese MS, Uversky VN, Dunker AK (2006) Rational drug design via intrinsically disordered protein. Trends Biotechnol 24:435–442
Uversky VN (2010) Targeting intrinsically disordered proteins in neurodegenerative and protein dysfunction diseases: another illustration of the D(2) concept. Expert Rev Proteomics 7:543–564
Dunker AK, Uversky VN (2010) Drugs for ‘protein clouds’: targeting intrinsically disordered transcription factors. Curr Opin Pharmacol 10:782–788
Uversky VN (2012) Intrinsic disorder-based protein interactions and their modulators. Curr Pharm, Des
Uversky VN (2012) Intrinsically disordered proteins and novel strategies for drug discovery. Expert Opin Drug Discov 7:475–488
Marasco D, Scognamiglio PL (2015) Identification of inhibitors of biological interactions involving intrinsically disordered proteins. Int J Mol Sci 16:7394–7412
Joshi P, Vendruscolo M (2015) Druggability of intrinsically disordered proteins. Adv Exp Med Biol 870:383–400
Lo Conte L, Chothia C, Janin J (1999) The atomic structure of protein–protein recognition sites. J Mol Biol 285:2177–2198
Gunasekaran K, Tsai CJ, Nussinov R (2004) Analysis of ordered and disordered protein complexes reveals structural features discriminating between stable and unstable monomers. J Mol Biol 341:1327–1341
Klein C, Vassilev LT (2004) Targeting the p53-MDM2 interaction to treat cancer. Br J Cancer 91:1415–1419
Vassilev LT (2004) Small-molecule antagonists of p53-MDM2 binding: research tools and potential therapeutics. Cell Cycle 3:419–421
Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, Fotouhi N, Liu EA (2004) In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303:844–848
Krishnan N, Koveal D, Miller DH, Xue B, Akshinthala SD, Kragelj J, Jensen MR, Gauss CM, Page R, Blackledge M, Muthuswamy SK, Peti W, Tonks NK (2014) Targeting the disordered C terminus of PTP1B with an allosteric inhibitor. Nat Chem Biol 10:558–566
Monaghan AE, McEwan IJ (2016) A sting in the tail: the N-terminal domain of the androgen receptor as a drug target. Asian J Androl 18:687–694
Dey S, Pal A, Chakrabarti P, Janin J (2010) The subunit interfaces of weakly associated homodimeric proteins. J Mol Biol 398:146–160
Bourgeas R, Basse MJ, Morelli X, Roche P (2010) Atomic analysis of protein–protein interfaces with known inhibitors: the 2P2I database. PLoS One 5:e9598
Dunker AK, Cortese MS, Romero P, Iakoucheva LM, Uversky VN (2005) Flexible nets. FEBS J 272:5129–5148
Uversky VN, Oldfield CJ, Dunker AK (2005) Showing your ID: intrinsic disorder as an ID for recognition, regulation and cell signaling. J Mol Recognit 18:343–384
Haynes C, Oldfield CJ, Ji F, Klitgord N, Cusick ME, Radivojac P, Uversky VN, Vidal M, Iakoucheva LM (2006) Intrinsic disorder is a common feature of hub proteins from four eukaryotic interactomes. PLoS Comput Biol 2:e100
Sato H, Masuda M, Miura R, Yoneda M, Kai C (2006) Morbillivirus nucleoprotein possesses a novel nuclear localization signal and a CRM1-independent nuclear export signal. Virology 352:121–130
Iwasaki M, Takeda M, Shirogane Y, Nakatsu Y, Nakamura T, Yanagi Y (2009) The matrix protein of measles virus regulates viral RNA synthesis and assembly by interacting with the nucleocapsid protein. J Virol 83:10374–10383
Watanabe A, Yoneda M, Ikeda F, Sugai A, Sato H, Kai C (2011) Peroxiredoxin 1 is required for efficient transcription and replication of measles virus. J Virol 85:2247–2253
De BP, Banerjee AK (1999) Involvement of actin microfilaments in the transcription/replication of human parainfluenza virus type 3: possible role of actin in other viruses. Microsc Res Tech 47:114–123
Moyer SA, Baker SC, Horikami SM (1990) Host cell proteins required for measles virus reproduction. J Gen Virol 71:775–783
tenOever BR, Servant MJ, Grandvaux N, Lin R, Hiscott J (2002) Recognition of the measles virus nucleocapsid as a mechanism of IRF-3 activation. J Virol 76:3659–3669
Colombo M, Bourhis JM, Chamontin C, Soriano C, Villet S, Costanzo S, Couturier M, Belle V, Fournel A, Darbon H, Gerlier D, Longhi S (2009) The interaction between the measles virus nucleoprotein and the Interferon Regulator Factor 3 relies on a specific cellular environment. Virol J 6:59
Laine D, Bourhis J, Longhi S, Flacher M, Cassard L, Canard B, Sautès-Fridman C, Rabourdin-Combe C, Valentin H (2005) Measles virus nucleoprotein induces cell proliferation arrest and apoptosis through NTAIL/NR and NCORE/FcgRIIB1 interactions, respectively. J Gen Virol 86:1771–1784
Laine D, Trescol-Biémont M, Longhi S, Libeau G, Marie J, Vidalain P, Azocar O, Diallo A, Canard B, Rabourdin-Combe C, Valentin H (2003) Measles virus nucleoprotein binds to a novel cell surface receptor distinct from FcgRII via its C-terminal domain: role in MV-induced immunosuppression. J Virol 77:11332–11346
Chen M, Cortay JC, Gerlier D (2003) Measles virus protein interactions in yeast: new findings and caveats. Virus Res 98:123–129
Devaux P, Priniski L, Cattaneo R (2013) The measles virus phosphoprotein interacts with the linker domain of STAT1. Virology 444:250–256
Curran J, Marq JB, Kolakofsky D (1995) An N-terminal domain of the Sendai paramyxovirus P protein acts as a chaperone for the NP protein during the nascent chain assembly step of genome replication. J Virol 69:849–855
Tapparel C, Maurice D, Roux L (1998) The activity of Sendai virus genomic and antigenomic promoters requires a second element past the leader template regions: a motif (GNNNNN)3 is essential for replication. J Virol 72:3117–3128
Dunker AK, Garner E, Guilliot S, Romero P, Albrecht K, Hart J, Obradovic Z, Kissinger C, Villafranca JE (1998) Protein disorder and the evolution of molecular recognition: theory, predictions and observations. Pac Symp Biocomput 3:473–484
Wright PE, Dyson HJ (1999) Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J Mol Biol 293:321–331
Dunker AK, Obradovic Z (2001) The protein trinity–linking function and disorder. Nat Biotechnol 19:805–806
Dunker AK, Brown CJ, Obradovic Z (2002) Identification and functions of usefully disordered proteins. Adv Protein Chem 62:25–49
Uversky VN, Li J, Souillac P, Jakes R, Goedert M, Fink AL (2002) Biophysical properties of the synucleins and their propensities to fibrillate: inhibition of alpha-synuclein assembly by beta- and gamma- synucleins. J Biol Chem 25:25
Gunasekaran K, Tsai CJ, Kumar S, Zanuy D, Nussinov R (2003) Extended disordered proteins: targeting function with less scaffold. Trends Biochem Sci 28:81–85
Fink AL (2005) Natively unfolded proteins. Curr Opin Struct Biol 15:35–41
Dyson HJ, Wright PE (2005) Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol 6:197–208
Pancsa R, Fuxreiter M (2012) Interactions via intrinsically disordered regions: what kind of motifs? IUBMB Life 64:513–520
Jordan IK, Sutter BA, McClure MA (2000) Molecular evolution of the Paramyxoviridae and Rhabdoviridae multiple-protein-encoding P gene. Mol Biol Evol 17:75–86
Narechania A, Terai M, Burk RD (2005) Overlapping reading frames in closely related human papillomaviruses result in modular rates of selection within E2. J Gen Virol 86:1307–1313
Rancurel C, Khosravi M, Dunker KA, Romero PR, Karlin D (2009) Overlapping genes produce proteins with unusual sequence properties and offer insight into de novo protein creation. J Virol 83:10719–10736
Kovacs E, Tompa P, Liliom K, Kalmar L (2010) Dual coding in alternative reading frames correlates with intrinsic protein disorder. Proc Natl Acad Sci USA 107:5429–5434
Bourhis JM, Canard B, Longhi S (2006) Structural disorder within the replicative complex of measles virus: functional implications. Virology 344:94–110
Xue B, Blocquel D, Habchi J, Uversky AV, Kurgan L, Uversky VN, Longhi S (2014) Structural disorder in viral proteins. Chem Rev 114:6880–6911
Tokuriki N, Oldfield CJ, Uversky VN, Berezovsky IN, Tawfik DS (2009) Do viral proteins possess unique biophysical features? Trends Biochem Sci 34:53–59
Xue B, Williams RW, Oldfield CJ, Goh GK, Dunker AK, Uversky VN (2010) Viral disorder or disordered viruses: do viral proteins possess unique features? Protein Pept Lett 17:932–951
Afonso CL, Amarasinghe GK, Banyai K, Bao Y, Basler CF, Bavari S, Bejerman N, Blasdell KR, Briand FX, Briese T, Bukreyev A, Calisher CH, Chandran K, Cheng J, Clawson AN, Collins PL, Dietzgen RG, Dolnik O, Domier LL, Durrwald R, Dye JM, Easton AJ, Ebihara H, Farkas SL, Freitas-Astua J, Formenty P, Fouchier RA, Fu Y, Ghedin E, Goodin MM, Hewson R, Horie M, Hyndman TH, Jiang D, Kitajima EW, Kobinger GP, Kondo H, Kurath G, Lamb RA, Lenardon S, Leroy EM, Li CX, Lin XD, Liu L, Longdon B, Marton S, Maisner A, Muhlberger E, Netesov SV, Nowotny N, Patterson JL, Payne SL, Paweska JT, Randall RE, Rima BK, Rota P, Rubbenstroth D, Schwemmle M, Shi M, Smither SJ, Stenglein MD, Stone DM, Takada A, Terregino C, Tesh RB, Tian JH, Tomonaga K, Tordo N, Towner JS, Vasilakis N, Verbeek M, Volchkov VE, Wahl-Jensen V, Walsh JA, Walker PJ, Wang D, Wang LF, Wetzel T, Whitfield AE, Xie JT, Yuen KY, Zhang YZ, Kuhn JH (2016) Taxonomy of the order Mononegavirales: update 2016. Adv Virol 161:2351–2360
DeLano WL (2002) The PyMOL molecular graphics system. Proteins Struct Funct Bioinform 30:442–454
Acknowledgements
S.L. wishes to thank all the members of her lab and her co-workers for their critical contribution to the studies herein summarized. Within her group, she thanks David Karlin, François Ferron, Jean-Marie Bourhis, Kenth Johansson, Antoine Gruet, Johnny Habchi, David Blocquel, Jenny Erales, Lorenzo Baronti, Marion Dosnon, Jennifer Roche and Matilde Beltrandi (previous members), and Christophe Bignon (present members). The authors also wish to thank David Blocquel who is the author of Fig. 9c. Among her numerous past and present co-workers, she thanks Bruno Canard (AFMB, Marseille, France), Maria Maté (AFMB, Marseille, France), Michael Oglesbee (Ohio State University, Columbus, USA), Hélène Valentin (CIRI, Lyon, France), Valerie Belle and Bruno Guigliarelli (BIP, Marseille, France), Janez Strancar (Jozef Stefan Institute, Ljubljana, Slovenia), Gary Daughdrill (University of South Florida, USA), Martin Blackledge, Malene Ringkjobin-Jensen and Guillaume Communie (Institut de Biologie Structurale, Grenoble, France), Jin Wang (Changchun Institute of Applied Chemistry, Chinese Academy of Sciences, Changchun, China), Roberta Pierattelli and Isabella Felli (CERM, Florence, Italy), Rita Grandori (Universita’ degli Studi Milano-Bicocca, Milan, Italy), Andrea Cavalli (IRB, Bellinzona, Switzerland), Pascale Barbier (CRO2 UMR_S911, Marseille, France), Paolo Carloni (Institute for Advanced Simulation IAS-5 and Institute of Neuroscience and Medicine INM-9, Jülich, Germany), Joanna Brunel (CIRI, Lyon, France), Daniela Bonetti (Sapienza, Universita’ of Rome, Italy), and Carlo Camilloni and Michele Vendruscolo (Department of Chemistry, University of Cambridge, UK). She is particularly grateful to Vladimir Uversky (University of South Florida, USA) for the numerous stimulating discussions and for his useful advice on various issues. The studies herein reviewed were carried out with the financial support of the Agence Nationale de la Recherche, specific programs “Physico-Chimie du Vivant”, ANR-08-PCVI-0020-01, and “ASTRID”, ANR-11-ASTR-003-01 to S.L and D.G. They also benefited from support from the CNRS, the Direction Générale de l’Armement (DGA) and the Fondation pour la Recherche Médicale (FRM). Work partly supported by grants from the Italian Ministero dell’Istruzione dell’Università e della Ricerca (Progetto di Interesse ‘Invecchiamento’ to S.G.) and Sapienza University of Rome (C26A155S48 to S.G). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Longhi, S., Bloyet, LM., Gianni, S. et al. How order and disorder within paramyxoviral nucleoproteins and phosphoproteins orchestrate the molecular interplay of transcription and replication. Cell. Mol. Life Sci. 74, 3091–3118 (2017). https://doi.org/10.1007/s00018-017-2556-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-017-2556-3