Abstract
The IL-10 family of cytokines is comprised of IL-10, IL-19, IL-20, IL-22, IL-24, IL-26, and IFN-λs (IL-28A, IL-28B, and IL-29). The IL-10 family members bind to shared class II cytokine receptor chains that associate in various combinations in heterodimeric complexes. Upon interleukin/receptor complex formation, these proteins switch on the Jak/STAT pathway and elicit pleiotropic biological responses whose variety sharply contrasts with their structural similarities. IL-10 family members are involved in several human diseases and health conditions and hence their structural analyses may provide valuable information to design specific therapeutic strategies. In this review, we describe the human interleukin-10 family of cytokines, focusing on their structures and functions, with particular attention given to IL-22 and IL-10. We report on the recently published structures of IL-10 cytokine family members and their complexes with cognate transmembrane and soluble receptors as well as on interleukin physiology and physiopathology.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Immunomodulation is a primary physiological function of interleukins such as interleukin-10 (IL-10), which was originally described as a factor that inhibits IFN-γ production by T lymphocytes [1]. Ten years after the original identification of IL-10, various genetic approaches led to the cloning of a series genes coding for IL-10-related factors that are now collectively described as the IL-10 family, which comprises IL-10, IL-19, IL-20, IL-22, IL-24, IL-26, and interferon-λs: IL-28A (IFN-λ2), IL-28B (IFN-λ3), and IL-29 (IFN-λ1). These cytokines were grouped together based on sequence alignment, structural homology, and binding to shared class II cytokine receptor chains that activate the JAK/STAT pathway [2–5]. IL-10 family members are helical proteins formed by six or seven helices and connecting loops [6–8]. Each IL-10 member can associate with the extracellular portion of two transmembrane proteins from the class II cytokine receptor, forming a heterodimeric complex. There are six different receptor chains for nine ligands, four with a long cytoplasmic tail (IL-10R1, IL-20R1, IL-22R1, and IL-28R) and two with a short cytoplasmic domain (IL-10R2 and IL-20R2), and each of the members of the IL-10 family bind to either IL-10R2 or IL-20R2 associated with a long receptor chain [9–13]. Besides these transmembrane proteins, a soluble receptor has also been described and designated IL-22 binding protein (IL-22BP) because it binds and neutralizes IL-22 in vitro. The type I IFN-s and IFN-γ also share some structural characteristics with this family [14], but they do not share any of these receptor chains, and will not be described in this review.
IL-10 family proteins are involved in a broad range of biological processes that include antiviral activity, secretion of antibacterial proteins, cell-growth stimulation, acute phase response, wound healing, antitumoral activity, and apoptosis induction. These important immunological activities are implicated in several human diseases such as psoriasis [15–17], rheumatoid arthritis [18–20], and inflammatory lung and bowel diseases [21, 22]. In this context, the study of IL-10 members, their interaction with receptors, and elicited cell signaling cascade, are important to understand the basis of both immunoregulation and associated diseases, establishing strategies to modulate or to control these health conditions and illnesses.
In this review, we discuss the human IL-10 family of cytokines starting from a structural point of view. First, we describe the relationships among the IL-10 members, their amino acid sequence alignment, as well as the similarities and clustering of these cytokines. Subsequently, we devote one section to every member of the family, in which a brief biological introduction precedes the structural considerations.
We also present the mechanism by which the IL-10 members signal throughout the target cell surfaces by interacting with their cognate receptors. The IL-10 receptors are introduced and the possible ternary complexes formed by the interleukins and the different receptors are discussed. Three-dimensional structures of the recently published IL-10 and IL-22 complexes with their receptors (IL-10:IL-10R1; IL-22:IL-22R1 and IL-22:IL-22BP) are described in detail, and their interfaces and amino acid residues involved in protein–protein interactions are analyzed. We also compare different IL-10 member binary complexes, which shed light onto the molecular basis of receptor selectivity and/or promiscuity.
Finally, we review important publications which use molecular modeling tools and site direct mutagenesis analysis to pinpoint the second receptor interfaces on the surface of IL-10 and IL-22 binary complexes. Similar analysis performed for the IL-10 members with unknown binary complex structures is also presented.
The IL-10 members
Primary structures
The sequence alignment of IL-10 members (Fig. 1) shows that these cytokines share 15–39% amino acid sequence identity, except for the interferon-λ cluster, which displays less than 10% identity with the other IL-10 members and shows a much higher amino acid conservation within the cluster (>70%). IL-20 and IL-24 are very similar to IL-19, with 39 and 30% of amino acid identity with IL-19, respectively, thereby forming another cluster. IL-10 and IL-26 share relatively high identity (24%) and are also proposed to form a separate cluster within the IL-10 family, whereas IL-22 seems to be relatively more closely related to the IFN-λs than the other IL-10 family members [23].
Alignment of the IL-10 family member amino acid sequences. a Sequence alignment colored according to the ClustalW/Jalview color code [169]. The potential cysteines involved in disulfides and the non-interferon IL-10 member sequence signature are contoured by solid and dashed lines, respectively. The secondary structures features are shown as found in the IL-22 crystal structure (PDB ID 1M4R [8]). The potential IL-22 [25, 56] and IL-19 [24] glycosylation sites are contoured by double lines. b A phylogram tree extract from this alignment. Alignment was performed using ClustalW [170]
A common hydrophobic signal peptide, about 20 amino acids in length, is found at the N-terminal region of all IL-10 members. Another common feature of these cytokines is a sequence signature K A/S X G/S E X D XX (where X is a hydrophobic residue) identified in all non-interferon IL-10 members (reviewed in [8]; Fig. 1a).
The cysteine residues are partially conserved and have a somewhat complex distribution pattern (Table 1). All members of the family possess at least one Cys residue at the N-terminal portion and a highly conserved cysteine is observed in the DE loop region (IL-24 being the only exception). IL-10, IL-19, IL-20, and IL-26 all display an additional Cys residue at the DE loop; however IL-19 and IL-20 also contain one more cysteine (a total of three Cys residues) located at the DE loop. The non-interferon members also contain another highly conserved Cys residue at the helix C. Notably, IL-22 is the unique non-interferon IL-10 member that contains a Cys residue at the helix F.
The IFN-λ group displays quite a different distribution pattern of cysteine residues. All three IFN-λs have highly conserved cysteines at the AB loop, helix F, C-termini, and N-termini regions. IFN-λ2 and IFN-λ3 also display an additional cysteine at the C-termini of their helices F2, promoting the formation of an additional S–S bond in the C-terminal part of the protein (Fig. 1a; Table 1).
Three-dimensional structure considerations
The crystal structures of IL-10 [6, 7], IL-19 [24], IL-22 [8, 25], and, more recently, IFN-λ3 [23] have been determined. Based on the information extracted from these 3D structures, as well as sequence alignments and secondary structure prediction, one can draw several conclusions about IL-10 family members’ secondary, tertiary, and quaternary organization.
IL-10 family members are helical cytokines formed by six or seven helices and connecting loops. A four-helix bundle, generally formed by helices A, C, D, and F, comprises the protein core, which is a characteristic feature of all these helical cytokines [13, 23, 24, 26]. Despite the partially conserved cysteine positions among the members of the IL-10 family, the organizations of the disulfide networks are quite different, as revealed by the crystal structures of IL-10, IL-19, IL-22, and IFN-λ3. A characteristic helix F kink is observed in all known 3D structures of IL-10 family members. This kink divides helix F into two parts, generally designated helix F1 and F2. As it will be discussed below, the kink exposes important residues that contribute to the interaction sites with the respective receptors [27, 28].
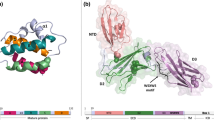
From the quaternary structure point of view, IL-10 family members can be found either as monomers [8, 23, 24] or intimately intertwined dimers [6, 13]. The preference for dimer or monomer formation is most probably a consequence of S–S interactions with the DE loop and/or the length of this loop. In particular, disulfide interactions in the beginning of helix E may influence the flexibility of this helix. As a consequence of these interactions, helices E and F could not fold back onto the same monomer [26] and instead form a separated EF domain that is donated to the homodimerization partner in the process of dimer formation. IL-10, for example, forms an intertwined dimer in which one compact domain is composed of helices A–D and helices E′ and F′ from its intimate dimeric counterpart [13] (Fig. 2a). A similar dimeric conformation has been described for IFN-γ [6, 13]. Alternatively, depending on the length and flexibility of the DE loop [26] and/or the orientation promoted by additional disulfide bridges [23], all the six to seven helices can fold up as an individual monomer, as observed in IL-19, IL-22, and IFN-λ3 crystal structures (Fig. 2b).
IL-10 family members with known crystal structures. a The intertwined IL-10 dimer (PDB ID 2ILK [7]). b The independent monomers of IL-22 (PDB ID 1M4R [8]), IL-19 (PDB ID 2N1F [24]), and IL-28B (IFN-λ3; PDB ID 3HHC [23]). Helices are named and the cysteine residues involved in disulfide linkages are shown as sticks and numbered according Table 1. Figures were generated with the PyMOL software (DeLano Scientific) [171]
Below, we will describe in detail each IL-10 family member. We divided this section into two parts based on their quaternary structures: the monomeric cytokines are designated as ‘the IL-19 subfamily’ (IL-19, IL-20, IL-22, IL-24, and INF-λs) and ‘the IL-10 family members that form intertwined dimers’ which includes IL-10 and IL-26 descriptions. A brief introduction presenting the biological aspects of each family member is given, followed by a detailed structural description of the cytokines with the known 3D structure: IL-10, IL-19, IL-22, and IFN-λ3. We will also briefly discuss the possible structural architectures of the IL-10 members whose 3D structures are unknown, as well as sequence alignments and secondary structure predictions.
The IL-19 subfamily
The non-interferon IL-10 family members are IL-19, IL-20, IL-22, and IL-24. From a structural point of view, these cytokines compose the IL-19 sub-family, whose member’s main feature is their quaternary assembly as individual monomers [24]. Recently, the IFN-λ group of cytokines was also included in the IL-19 sub-family, after IFN-λ3 crystal structure determination [23] and based on the structural comparison (reviewed in [29]). All these proteins are found as individual monomers, and do not form intertwined dimers characteristic for IL-10 and IL-26 [8, 13, 29].
IL-22
IL-22 was first identified as a gene induced by interleukin-9 (IL-9) in murine T lymphoma cells [30]. Because it encodes a glycosylated protein that bears 22% identity to interleukin-10 (IL-10) its original designation was IL-10-related T-cell-derived inducible factor (IL-TIF). The human IL-22 gene is located on chromosome 12q15, in the vicinity of interferon-γ (IFN-γ) and interleukin-26 (IL-26) genes, forming an interleukin gene cluster [31]. IL-22 genomic structure is composed of five introns and six exons that are similar in size and organization when compared to IL-10.
IL-22 expression was found to be induced by IL-9 in thymic lymphomas, T helper cells, and mast cells, and also upon ConA activation of freshly isolated splenocytes. Similar induction was observed after lipopolysaccharide injection in mice, and that was the first indication of IL-22 involvement in the inflammatory response in vivo [32]. Its first biological activity described was the induction of acute phase reactants such as serum amyloid A, α1-antichymotrypsin, and haptoglobin in hepatocytes [32] and in pancreatic cell lines [33].
More recently, it was shown that IL-22 and IL-17 can be coexpressed by Th17 cells [34, 35]. These cells correspond to a separate lineage of effector CD4+ T cells, defined on the basis of their high IL-17 production [36]. The expression of both cytokines in these cells enhances cooperative secretion of antimicrobial peptides involved in skin innate immunity [34, 35], and initially pointed to a proinflammatory role of the interleukin.
The dual biological role of IL-22
As soon as it was found that IL-22 mediates its activity on hepatocytes through binding to the IL-22R1/IL-10R2 receptor complex, screening for IL-22R1 receptor expression was conducted to identify the potential biological targets of this cytokine. IL-10R2 is expressed constitutively by almost every cell type. IL-22R1 expression turned out to be more restricted and was mostly found in cells from the skin, digestive, and respiratory tracts. Interestingly, IL-22 stimulates the production of β-defensin 2 and 3 in keratinocytes [37]. These small cationic proteins kill bacteria and fungi, and are normally expressed in skin and mucosal epithelia, acting as an innate immunity barrier against microbial infections [38, 39]. Besides inducing the expression of antimicrobial genes, IL-22 also regulates cellular differentiation and motility of keratinocytes, thereby exerting a spectrum of activities that are beneficial during wound healing and infectious processes, but that contribute to skin lesions of patients with psoriasis disease [16, 35, 37, 40–42].
Whether these biological activities should be considered as proinflammatory remains an open question. On the one hand, in the context of psoriasis lesions, IL-22 definitely plays a proinflammatory role. On the other hand, IL-22 exerts a protective effect in inflammatory and infectious processes affecting the intestinal and respiratory mucosa, both by inducing the production of anti-microbial proteins and by promoting homeostasis of epithelial cells [43–47]. IL-22 also plays a protective role in an hepatitis model by promoting hepatocyte survival [48, 49].
Altogether, these findings indicate that upon bacterial infection in physiologically outer barriers, where IL-22R1 receptor is mostly expressed such as the skin and mucosal epithelia, IL-22 secretion promotes a response aiming at wound healing and pathogen killing [50]. In this context, a TH17 lymphocyte response contributes to tissue homeostasis, but other cell lineages might be involved in IL-22 production as well. Recently, three independent studies described mucosal NKp46+ cells as a subpopulation of natural killer cells that secrete IL-22 in the intestinal mucosa [51–53]. Moreover, NK-22 is another subset of natural killer cells that when stimulated with IL-23 secretes IL-22, IL-26, and leukemia inhibitory factor in mucosa-associated lymphoid tissues such as tonsils and Peyer’s patches (located in the lowest portion of the small intestine ileum) [54]. For both TH17, NK, or NK-like cells, IL-23 appears to be a major inducer of IL-22 production, suggesting that IL-22 is a key mediator in IL-23-dependent immune reactions in skin and mucosal epithelia by stimulating innate antimicrobial responses as well as promoting tissue repair.
IL-22 structures
The crystal structures of human IL-22 were solved independently for IL-22 proteins expressed in E. coli (ECIL-22, PDB ID 1M4R [8]) and D. melanogaster (DMIL-22 PDB ID 1YKB [25]). The ECIL-22 and DMIL-22 show minimal structural divergences [25, 26].
The IL-22 monomer is composed of six helices named A–F and a small N-terminal helix termed helix preA. IL-22 helices A–F are folded together forming a compact six-helix bundle, of which helices A, C, D, and F form the characteristic class II cytokines four-helix bundle. The N-terminal portion of IL-22 (residues 44–47) acquires a 310-helix secondary structure (helix preA) [8, 25]. The preA structure is oriented perpendicular to the IL-22 helix A, and is stabilized by hydrogen bonds between residues Asp43–Ser45, Asp43–Asn46, and Ser45–Gln49 [25]. Despite the preA kink, the helix A is straight and continuous, as well as all the other IL-22 helices, with the exception of helix F. IL-22 displays the characteristic helix F bend found in all class II alpha-helical cytokines [26, 55]. As a consequence, helix F can be divided into two parts, helix F1 and helix F2. In IL-22 the helix F bend occurs at the position of the residue Glu166, exposing to solvent this and the neighboring residues charged side chains (Figs. 1a, 2b).
The DMIL-22 carries several carbohydrates attached to some of the predicted IL-22 glycosylation sites, which provides additional information on the carbohydrate positions, compositions, and their importance to IL-22 structure and putative interactions with IL-22 receptors [25]. Three potential glycosylation sites (Asn-X-Ser/Thr, X = any amino acid), which correspond to Asn54, Asn68, and Asn97, map to IL-22 helix A, AB loop and helix C, respectively (Fig. 1a). Mass spectroscopy analysis revealed that hexasaccharides (two N-acetyl glucosamines, three mannoses, and one fucose) are attached to the IL-22 protein expressed in D. melanogaster [56]. However, only glycosylation at Asn54 and Asn97 position was observed in the DMIL-22 crystal structure [25]. By analysis of the six different IL-22 monomers packed into the DMIL-22 crystal asymmetric unit, as well as their comparison to the ECIL-22 crystal structure, Xu et al. [25] pinpointed IL-22 regions susceptible to conformational changes. These include the helix preA, helix A, the AB loop, and helix D. In particular, Gln49 and Pro50, located at the beginning of helix A, are considered as a ‘flexible pivot point’, which allows the preA and helix A to undergo rigid body conformational motions. Notably, Pro50 is conserved in all IL-22 sequences from different organisms known to date [25, 26]. The rigid body conformational changes allow, for example, reorientation of the Tyr51 side chain (helix A) as well as the Tyr114–Glu117 main chains (helix D). These IL-22 structural adjustments are most probably important for the binary and ternary complex formation, as will be discussed further in this review.
In the ECIL-22 and DMIL-22 crystal structures the six helices form a single IL-22 monomer. Asymmetric unit cells of the ECIL-22 and DMIL-22 crystals contain two and six IL-22 monomers, respectively. The absence of intertwined dimers, as observed in IL-10, and the monomer contacts organization in IL-22 crystal structures supported the idea that IL-22 is a monomer in solution [8, 25]. The IL-22 monomer has two disulfide bonds (Fig. 2b; Table 1). The first S–S bridge (Cys40–Cys132) connects the IL-22 N-terminal portion to the DE loop. The second one (Cys89–Cys178) buckles helix C to helix F, and is proposed to influence the F1 and F2 position, as well as the helix F bend [26]. The presence of only one disulfide bond in helix D region, instead of two as observed in IL-10, gives more flexibility to helix E and F to fold back onto the IL-22 A–D helices bundle, thus completing IL-22 monomeric structure formation [26].
Surface plasmon resonance studies [57] also confirmed the IL-22 assembly as monomers. However, recent SAXS data [58] revealed that at high concentrations, IL-22 is capable of forming dimers or even tetramers in solution. Nevertheless, the IL-22 dimers are formed by interactions between individual monomers, which is very different from the intertwined dimers observed for IL-10. An interesting feature of the IL-22 SAXS envelopes is that the shape of the IL-22 dimer is remarkably similar to the shape of the IL-10, including the 90° opening angle formed between the two monomer sub-units of the dimer. This might indicate that the functionally relevant form of IL-22 that is recognized by its cognate receptors at the cell surface may indeed be a dimeric form [58]. However, the physiological relevance of these findings still remains to be elucidated.
A possible connection between IL-22 dimerization and human physiology [16] can be examined using via, for example, peptide interference, perturbing the putative interface of IL-22 dimerization with a small homolog peptide that would inhibit homodimer formation [59]. Alternatively, one might force receptor dimerization using a similar approach to what has been described for the glycoprotein gp130 [60]. In this study, gp130 was truncated 15 amino acids above its transmembrane domain and the extracellular portion replaced by the leucine zipper region of the human c-jun gene, and stabilized by the addition of a disulfide bridge [60]. Similar work could be done with IL-22R1, and its interactions with IL-22 tested by JAK-STAT transcriptional activation in cells expressing IL-10R2.
IL-19
IL-19 is expressed in monocytes upon induction with bacterial lipopolysaccharide (LPS), but not by T lymphocytes nor by NK cells [2, 9]. IL-19 expression showed transcriptional variants [2, 61] that are detected 2–3 h after induction, indicating a feedback role in inflammation, as observed for IL-10 [2]. The secreted protein has 21% amino acid identity with IL-10 [2] and shows different levels of glycosylation [61]. IL-19 signals through IL-20R1/IL-20R2 complex [10], activating STAT3 transcription factor [62]. At the mRNA level, the genes encoding the IL-20R1/IL-20R2 complex appear to be expressed in heart, lung, testis, adrenal gland, and skin, suggesting that these tissues may be potential targets for IL-19 [62]. However, many activities ascribed to IL-19 based on in vitro observation do not fit at all with the tissue distribution of its receptor chains, raising questions that need to be solved in order to draw conclusions about the actual biological significance and mode of action of this cytokine.
For instance, monocytes do not seem to express IL-20R1 [9, 62], and yet IL-19 has been reported to stimulate IL-6 and TNF release from monocytes, resulting in TNF-α-mediated cell apoptosis [61], or to regulate its own production and production of IL-10 [63]. Similarly, IL-19 was reported to promote the maturation of T cells towards Th2 polarization in mouse and human models [64, 65]. IL-19 levels were reported to be two-fold higher in patients with asthma as compared to healthy controls and this correlated with the expression of IL-4 and IL-13, involved in mucus secretion and airway hypersensitivity [65].
IL-19 is also expressed in human injured arteries and in vascular smooth muscle cells (VSMC) by inflammatory cytokines, and IL-19 was reported to have antiproliferative activity in VSMC [66]. In addition, through induction of SOCS5 and inhibition of signal transduction, IL-19 expression in VSMCs was suggested to mediate protective, autocrine response of VSMCs to inflammatory stimuli and vascular injury [66].
A role of IL-19 in skin inflammation is more in line with the expression pattern of its receptor complex, which is expressed by keratinocytes [62–64]. In human psoriatic lesions, IL-19 is expressed in epidermis above the dermal papillae, as shown in the first report to demonstrate the presence of this cytokine in keratinocytes [67]. Moreover, the same work demonstrated that the cyclosporin A, and calcipotriol, a derivate of vitamin D, both used in treatment of psoriasis, abolished IL-19 mRNA expression. The precise involvement of IL-19 in psoriasis is still to be determined, but this cytokine might represent a therapeutical possibility for treatment of this disease.
IL-19 structural description
The crystal structure of IL-19 was solved by Chang and co-workers in 2003 and deposited in the Protein Data Bank under code 1N1F [24]. Although Chang et al. named the pre helix A as helix A in their paper, here we will adopt the same nomenclature used in the IL-22 structure description. Interleukin-19 is structurally organized by a unique six-helix bundle, formed by helices A–F and the N-terminal helix preA (Fig. 2b). Helices, A, C, D, and F form the characteristic cytokine four-helix bundle and helix F displays the typical helix F bend found in all other helical cytokines. As also observed in the IL-10 3D structure, IL-19 also has a kink of helix A [24]. Superposition of the IL-19 crystal structure with IL-22 monomer or one of the domains of IL-10 leads to the conclusion that they are relatively similar, with r.m.s.d. of 1.7 Ǻ [24]. However, because of the orientation of IL-19 helix A and N-terminal region, IL-19 is more compact than IL-10. IL-19 helix D also acquires a different position in comparison to its counterpart in IL-10.
Three disulfide bonds are present in IL-19 and two of those are structurally conserved in IL-10 (Fig. 2; Table 1). IL-19 helix A, covering the molecule, is stabilized by a disulfide bridge with the C-terminal portion of helix D (Cys10–Cys103). The other two disulfide bonds (Cys57–Cys109 and Cys58–Cys111) link the N-terminal portion of helix C with the DE loop [13, 24]. The additional disulfide bridge observed in the IL-19 DE loop (S–S interaction 3; Fig. 2b, Table 1) is probably the main structural feature that contributes to helices EF refolding onto helices A–D, enabling the IL-19 fold as a compact monomer. Two putative glycosylation sites are predicted in the IL-19 amino acid sequence, Asn38–Val39–Thr40, and Asn117–Ala118–Tyr119 (Fig. 1a); although only glycosylation at Asn38 side chain was found in the IL-19 crystal structure [24].
An ionic interaction between Lys27 and Asp143, connecting helices A and F, is observed at the surface of IL-19 monomer. This interaction may contribute to IL-19 helix A bend. It is important to note that the aspartate residue at the helix F bend is conserved in all non-interferon IL-10 family members, although in IL-22 and IL-10 crystal structures the helix A Lys side chain is turned away from the helix F (data not shown). Nevertheless, the conservation of charged residues at the equivalent structural position on the surface of all non-interferon members of IL-10 family may be important for the recognition of their cognate receptors, as will be discussed in the following chapters.
IL-20
IL-20 is a cytokine that bears 28% sequence identity to IL-10 and is located in a chromosomal region (1q32) that defines a cytokine cluster, comprised of IL-10, IL-19, IL-20, and IL-24 [68]. In contrast to IL-22, which is mainly produced by TH17, NK, and NK-like cells, IL-20 is mainly a monocyte and dendritic cell product, like IL-19 [9, 69]. Bacterial LPS also induces IL-20 expression in glial cells in the brain [70]. By tissue microarray analysis performed on human tissues, anti-IL-20 antibodies showed a positive signal in several non-neoplastic tissue types that include skin, small intestine, colon, liver, lung, spleen, kidney, prostate, uterus, and brain [71].
IL-20 binds to two different receptor complexes designated type 1 and type 2 IL-20 receptor complexes and consisting of the IL-20R1/IL20R2 or IL-22R1/IL-20R2, respectively [68]. These receptor complexes are expressed by keratinocytes in normal skin and at lower levels in other tissues like ovary and placenta. The skin is considered so far as the major target for IL-20, which induces hyperproliferation of keratinocytes [68]. Its overexpression in transgenic mice resulted in neonatal lethality with aberrant epidermal differentiation, resulting in lesions that mimic psoriasis lesions in human patients [68]. Interestingly, both IL-20 and IL-22 transgenic mice showed a similar psoriasis-like phenotype, have very similar effects on human keratinocytes, and are coexpressed in human psoriasis lesions [17], suggesting that their common receptor chain, IL-22R1, might be a key target for psoriasis.
IL-20 structural considerations
IL-20 shares high primary structure identity (44.1%) and similarity (52.4%) with IL-19. The position of IL-20 cysteine residues is equivalent to those in IL-19 and therefore it is likely that the three S–S interactions found in IL-19 also exist in IL-20 [24]. Furthermore, IL-20 and IL-19 can both signal through type 1 IL-20 receptor complexes composed of IL-20R1 and IL-20R2 [10]. Both IL-19 and IL-20 are observed as monomers in solution, even at high protein concentration [72]. Collectively, these observations suggest that IL-20 is structurally very similar to IL-19.
IL-24
IL-24 is a unique gene among those of the IL-10 family. Originally cloned by a strategy of differentiation induction subtraction hybridization (DISH), and named melanoma differentiation-associated gene-7 (mda7) [73] because its expression correlated with differentiation of melanoma cells upon addition of IFN-β and mezerein, an antitumor compound isolated from the Daphne species of plants [74]. This induction leads to growth suppression, melanin synthesis, and widespread alterations in gene expression in melanocytes [75]. Interestingly, the mda-7 gene was originally not recognized to encode a secreted protein and was considered as a cancer-specific apoptosis-inducing gene in which antitumoral activity has been intensely studied [76–79].
In normal human tissue, IL-24 transcripts are detected in spleen, thymus, and peripheral blood leukocytes, an indication of expression restricted to the immune system [80]. The mouse IL-24 orthogue, originally called FISP for (IL-4-induced secreted protein), was described as a TH2 cytokine [81]. The rat orthologue of mda-7/IL-24, originally called c49a, is induced during wound repair [82], suggesting that it may play the same kind of activity as IL-22 or IL-20. In line with this hypothesis, IL-24 has been described to bind to the same receptor complexes as IL-20 [10, 83], and IL-24 transgenic mice recapitulate most of the features of IL-20- and IL-22-transgenic mice, namely neonatal lethality, epidermal hyperplasia, and abnormality in keratinocyte differentiation, supporting a largely redundant role in epidermal functions for IL-20, IL-22, and IL-24, which seem to be IL-22R1-dependent [84].
The anti-tumor activity of IL-24 is more controversial [85]. This effect does not correlate with the expression of receptor complexes binding the secreted form of this protein, but an appealing hypothesis proposes that the mda-7 gene encodes both the secreted IL-24 form and a cytoplasmic isoform that is able to induce apoptosis in cancer cells [86] but not in normal cells. This effect is observed when the protein is expressed without its secretion signal peptide. The mechanism by which IL-24 specifically induces apoptosis of tumor cells but not in normal cells remains to be described. Both the secreted and the signal peptide-deficient IL-24 isoforms might accumulate in the ER and Golgi compartments, thereby generating a signal that induces “ER-stress apoptosis”. Mitochondrial function might be an important player in this signaling, either by generating reactive oxygen species (ROS), release of cytochrome c, or by loss of mitochondrial transmembrane potential [87, 88].
IL-24 has been evaluated in several clinical trials. The first clinical phase I study used direct intratumoral injection of adenoviruses containing IL-24 in patients with advanced carcinoma. The injection was well tolerated and IL-24 mRNA was detected in 100% of injected lesions, and this expression induced apoptosis in a high percentage of the cells [89, 90]. The desirable clinical response was observed only with repetition of the injection. Similar to what is found in culture cells, the results of these gene therapies showed induction of tumor cell-specific apoptosis, antitumor bystander effect, and also modulation of the immune response. The clinical trials confirmed possibilities of medical applications of IL-24 in cancer therapy. Further investigations of the mechanisms by which IL-24 controls cell apoptosis intracellularly might further improve the clinical strategies of such treatments.
Besides its direct antitumoral activity, IL-24 is able to prevent angiogenesis, the growth of new blood vessels that sustain tumor development and metastasis. This could be achieved by two mechanisms, one through interaction with IL-20R2/IL-22R1 receptor complexes, which affects endothelial cells [91], and the other by inhibiting vascular endothelial growth factor (VEGF) production by cancer cells [92]. Finally, like IL-19, several reports have described activities of the secreted form of IL-24 upon cell types that are not supposed to express its functional receptor complexes. For instance, IL-24 reportedly promoted the secretion of TNF-α, IFN-γ, and IL-6 from peripheral blood mononuclear cells (PBMC) [93], and inhibits the plasma cell differentiation program in human germinal center B cells [94]. Further studies are definitely required to better understand the molecular mechanisms underlying these atypical effects of IL-24, and their potential pathophysiological relevance.
IL-24 structural considerations
Similar to IL-22 and IL-19, the secreted form of human IL-24 is also glycosylated and shows an apparent molecular mass of 25.30 kDa [73]. IL-24 has only two cysteines: Cys59, at the N-terminal region, and Cys106, at helix C. The amino acid sequence alignment of IL-24 with IL-19 and comparison with the IL-19 crystal structure suggest that IL-24 most probably monomerize in solution, particularly if Cys59 and Cys106 form a disulfide bond (reviewed in [13]). However, if Cys106 from one IL-24 polypeptide chain interacts with Cys106 from the other IL-24 polypeptide chain, IL-24 may also form an intertwined dimer exemplified by IL-10 structure. Such helix C/helix C′ disulfide bond has already been observed in the viral IL-10-related protein encoded by the cytomegalovirus genome [95]. However, in this case, Cys59 could not be involved in S–S bond formation, which is unusual for helical cytokines [24].
The interferon lambda subgroup
IFN-λs, also called type III interferons, display a modest but significant sequence identity with IFN-α2, a type I interferon, mainly in what concerns the cysteine residues positions. Although IFN-λs sequence similarity to IL-10 family members is even lower (~20%) than to IFN-α2 or IFN-β (~30%), these cytokines are generally considered to belong to the IL-10 family, mainly because they bind to and signal via the class II cytokine receptor IL-10R2 [4, 5, 96]. In this review, we will follow this somewhat broader definition of the IL-10 family, which appeared to be supported by the 3D structure of human IFN-λ3 [23].
At the biological level, IFN-λ1, IFN-λ2, and IFN-λ3 cytokines are more related to type I IFN, sharing the establishment of antiviral protection [97], antiproliferative [96], and antitumor [98–100] activities (reviewed in [101]). These cytokines activate the same JAK-STAT pathway, leading to the activation of STAT1/STAT2/IRF9 transcription factor complexes [4, 5, 96]. Therefore, both type I and type III IFNs are involved in the induction of a common set of genes, but differ by the duration of signaling and the tissue specificity of the expression of their respective receptors [102, 103]. Likewise, analysis of gene expression using DNA microarrays indicated similar induction in hepatocyte and B cell lines stimulated with either IFN-λ1 or IFN-α cytokines, and this involves phosphorylation of STAT proteins STAT1, -2, -3, -4, and STAT5 [104].
On the cell surface, IFN-λs bind to the IFN-λR1 receptor (also called IL-28R1) associated with the common receptor IL-10R2 for signaling, which is also used by IL-10, IL-22, and IL-26 [105]. The tissue and cell specificities of the responses to systemic IFN-α/β and IFN-λ have been recently compared by the group of Michiels [106]. The response to IFN-λ correlated with expression of IFN-λR1. The IFN-λ response was prominent in the stomach, intestine, and lungs, and was mainly restricted to epithelial cells. In contrast, the response to IFN-α/β was more ubiquitous and was observed in various cell types [106]. These results raised the hypothesis that the IFN-λ system probably evolved to specifically protect epithelia. Thus, IFN-λs might represent key contributors to the prevention of viral invasion through skin and mucosal surfaces, whereas IL-22 might be a key mediator of anti-bacterial and anti-fungi responses at the very same surfaces of the body, as discussed above.
The antiviral protection conferred by IFN-λs has been confirmed in various cell lines, including HeLa, HuH7 hepatoma, and HT-29 colon epithelial cells and with different viruses, including Sindbis virus, dengue virus, vesicular stomatitis virus, and human cytomegalovirus [97, 107]. Mouse deficient in the gene encoding IFN-λR1, recently allowed to demonstrate the respective roles of type I and type II IFN in an influenza infection model. Mice lacking functional IFN-λR1 were only slightly more susceptible to influenza virus than wild-type mice. However, mice lacking functional receptors for both IFN-α/β and IFN-λ were hypersensitive and even failed to restrict usually non-pathogenic influenza virus mutants lacking the IFN-antagonistic factor NS1 [108].
When using interferons as therapeutic agents, the most important limiting factor is specificity. Indeed, IFN-α has been used for therapy of carcinomas but this strategy has a large range of side effects, which include myelosuppression [109]. Because of the restricted tissue expression of the IFN-λR1 receptor, IFN-λs might be used as therapeutic agents without this type of toxicity. Hepatitis C virus could be an important target of IFN-λs, as suggested by genetics studies showing that genetic variation in the gene encoding IFN-λ3 predicts hepatitis C treatment-induced viral clearance [110–113]. Although mouse liver cells might not respond to IFN-λs, it has been shown that IFN-λ1 in combination with either IFN-α or IFN-γ is able to block hepatitis C virus replication in human hepatocytes, indicating that IFN-λs may be used for treatment of this disease [114].
IFN-λ3 structural studies
Very recently, Gad et al. [23] solved and published the human IFN-λ3 crystal structure (PDB ID 3HHC). Analysis of IFN-λ3 X-ray structure reveals that this type III interferon is more structurally related to IL-10 family members than to other interferons. Furthermore, IFN-λ3 is very similar to IL-22, which displays parallel functions in skin as discussed above [23]. IFN-λ3 contains the six α-helical domain (helices A to F), characteristic of all class II cytokines. The four-helix bundle, typical for the IL-10 family, is found in the IFN-λ3 core. This comprises helices A, C, D, and F. The later helix, helix F, also has the characteristic bend found in other IL-10 family members.
Seven cysteine residues are found in IFN-λ3 structure, and six of these form three disulfide bonds (Fig. 2b; Table 1). Cys4 is bound to Cys103, connecting the N-terminal region to helix D. It is important to note that this S–S bond is conserved among the IL-10 family members. However, the other two S–S bridges have no equivalents in other IL-10 family proteins. The Cys38–Cys136 bond, which connects the AB loop with the beginning of helix F1, is restricted to IFN I and IFN III groups; whereas Cys155–Cys162 bridge, found in the end of helix F2, is only observed in the IFN-λs group (with the exception of the human IFN-λ1) [23].
IFN-λ1 and IFN-λ2: structural considerations
Since IFN-λ2 shares 94% of amino acid sequence identity with IFN-λ3, it is to be expected that their 3D structures are almost identical, including the disulfide bonds pattern [23]. At the same time, IFN-λ1 has very high amino acid sequence identity with IFN-λ3 (69%), therefore, it is highly probable that IFN-λ1 is also structurally very similar to IFN-λ3 [23]. The structural organization of IFN-λ3 [23] and previous experimental observations [13] suggest that IFN-λs form monomers in solution.
The IL-10 family members that form intertwined dimers: IL-10 and IL-26
IL-10
Originally termed cytokine synthesis inhibitor factor [115], interleukin-10 (IL-10) is a notably important cytokine with pleiotropic immunoregulatory effects. It was first identified as a product of T-helper type 2 (Th2) cells, but since its discovery, a large variety of cells were identified as the source of IL-10, which include macrophages, monocytes, B cells, dendritic cells (DC), CD8+ T cells, regulatory T cells (Tregs), Th1 cells, and Th17 cells [116]. After secretion, on the surface of its target cells, dimeric IL-10 molecules bind to the high-affinity IL-10R1 receptor and this complex recruits the low-affinity receptor IL-10R2 [117]. This sequential mechanism allows the intracellular activation of Janus kinase 1 (Jak1) and also tyrosine kinase 2 (Tyk2). Once activated, these kinases phosphorylate signal transducer and activator of transcription proteins (STAT1 and STAT3), which are the main transcription factors involved in IL-10-mediated immune responses [1]. In turn, the transcription factors activate the expression of SOCS-3 (suppressor of cytokine signaling-3) and also SOCS-1 genes, which results in the inhibition of JAK-STAT-dependent signaling [118].
The immunomodulatory effects of IL-10 are complex, given that this cytokine presents both antiinflammatory and immunostimulatory activities. Its anti-inflammatory activity can be illustrated by a long list of genes that are downregulated by IL-10 including IFN-γ and IL-2 by Th1 lymphocytes, Th2-cell derived cytokines (IL-4 and IL-5), IFN-γ and TNF-α by NK cells, and proinflammatory cytokines (IL-1β, TNF-α, and IL-6) by mononuclear phagocytes [1], while IL-10 promotes the production of cytokine antagonists such as IL-1RA [119] and soluble TNF p75 receptors [120]. It was also described that IL-10 molecules are capable of inhibiting the production of chemokines such as MIP-1α (monocyte inflammatory protein-1α), IL-8, and RANTES (regulated on activation, normal T expressed and secreted) [121, 122]. Moreover, IL-10 inhibits the expression of the major histocompatibility complex (MHC) class II molecules, intercellular adhesion molecule 1 and costimulatory molecules CD80 and CD86 in antigen-presenting cells (APC) [123].
Immunostimulatory effects elicited by IL-10 include promotion of the survival of human B cells, which occurs simultaneously with increasing levels of anti-apoptotic bcl-2 protein [124] and an increase of NK cell activity [125]. Increased IL-10 production has been observed in systemic lupus erythematosus (SLE) patients and serum IL-10 values reflect SLE disease activity, suggesting that overexpression of IL-10 might play a pathogenic role in severe lupus disease [126]. In vitro, interleukin-10 blockade corrects impaired cellular immune responses of SLE patients [127], suggesting that anti-IL-10 antibodies might represent a meaningful therapeutic approach in this disease. Indeed, clinical trials already tested murine anti-IL-10 for treatment of lupus erythematosus during 3 weeks [128]. This treatment resulted in rapid and long-lasting amelioration of the disease.
Altogether, IL-10 effects predominantly limit inflammatory responses in order to minimize tissue damage initiated by microorganism challenges. Such an anti-inflammatory activity represents an attractive perspective for inflammatory diseases such as rheumatoid arthritis. In a murine model of collagen-induced arthritis, injection of human adenovirus type 5 expressing IL-10 had a protective effect in the knee joint. This overexpression altered TNF-α and IL-1α production in inflamed synovium and probably has an effect in collagen arthritis in the knee [129]. These results suggest that IL-10 cytokine is likely to be suitable for treatment of rheumatoid arthritis.
IL-10 structural description
The crystal structure of IL-10 was the first reported three-dimensional structure of an IL-10 family member [6, 7], which revealed an unexpected similarity with the IFN-γ 3D fold [7]. Differently from what is observed for the IL-19 subfamily members, IL-10 forms intimate intertwined homodimers. Each IL-10 subunit has two disulfide bonds connecting the DE loop to helix C (Cys62–Cys114) and to the N-terminal region (Cys12–Cys108). These S–S interactions, mainly the one between Cys62 and Cys114, promote a certain degree of rigidity of helix E. As a consequence, helices E and F fold as an independent IL-10 domain that makes a right angle with the other domain composed of helices A–D [6, 7, 130, 131]. This promotes two IL-10 polypeptide chains to join together to form an intertwined V-shaped dimer. Each of the two domains of the IL-10 dimer contains four helices from one polypeptide chain (helices A–D) and two other helices from the other polypeptide chain (helices E′ and F′, Fig. 2a). In IFN-γ, a short DE loop is the main reason for a similar structural architecture; however, in IFN-γ, the angle formed between the two monomers is ~60° [131–134]. In IL-10, the 90° inter-domain angle allows helix C to be exposed, creating a potential interaction site with its receptors [6].
The quaternary structure arrangement of IL-10 in the form of intimate intertwined dimer is one of the main structural differences that distinguishes it from the IL-19 sub-family members (reviewed in [13]). However, similar to all other class II cytokines, IL-10 forms a four-helix bundle (helices A, C, D, and F′), which is characteristic feature of this class of proteins. These four helices form an extensive hydrophobic core that accounts for 85% of all the hydrophobic residues of IL-10 [130].
In IL-10, helices A, C, and F display significant bends, which were inferred to improve the IL-10 fold [6]. Helix A is kinked at the residues Leu23 and Phe30, which disrupts the hydrogen bond pattern of helix A, forming a new network with the residues of helix D. Helix C contains the well-conserved Pro78 residue, found in all known IL-10 amino acid sequences. Pro78 is responsible for a 23° bend of helix C, which is centered at Val-76 and leads to the exposure of Glu75 side chain [6]. The conserved kink at helix F is found in all other IL-10 family members with known 3D structures. In IL-10, helix F is turned ~129°, exposing the side chain of Glu142 and braking helix F in helix F1 and helix F2, in exactly the same way as observed for IL-22.
The IL-10 helix F presents a canonical IL-10 family sequence signature K A/S X G/S E X D X X (where X is a hydrophobic residue), also found in IL-22. However, there are several amino acid substitutions in IL-10 primary structure in comparison to the same region of IL-22 amino acid sequence (Fig. 1a).
IL-26
IL-26 was found in a screen searching for phenotypic alterations of T-cells upon simian rhadinovirus herpesvirus saimiri (HVS) infection [135, 136]. This interleukin has 47% similarity to IL-10 and it is secreted as intimate homodimers in solution as also found for IL-10 [137]. IL-26 secretion is not observed in B cells [138] but it is detected by RT-PCR in T and NK cells [9] and also in various T-cell lines transformed with HVS, but not in Jurkat, HeLa, SupT1, MT2, HuT-102, C91PL, B/JAB, and Tera2 cells [138].
The target cells of IL-26 are colon carcinoma cells and keratinocytes [139]. A typical feature of IL-26 is its high affinity for heparin that suggests similar affinity for glycosaminoglycans moieties on proteoglycans that could enrich the cytokine at the cell surface and work as IL-26 co-receptors. Enriched cytokines would bind to IL-20R1 and IL-10R2 receptors combination [139, 140] and activate STAT1 and STAT3 transcription factors. This particular receptor combination is what gives IL-26 its specificity since the other IL-10 family members are not able to signal via this complex. Moreover, this specificity is determined by IL-20R1 expression in specific tissue and not by IL-10R2, which is broadly expressed.
The participation of IL-26 in human diseases remains unclear, and the absence of any IL-26 orthologue in the mouse genome does not facilitate the evaluation of its biological activities. IL-26 maps to human chromosomal region 12q15 in a close vicinity to IL-22 and IFN- γ [138]. Polymorphisms in this region are related to sex bias in rheumatoid arthritis [141]. A more compelling result indicates that IL-26 protein expression is higher in patients with Crohn’s disease [142]. IL-26 stimulates the expression of proinflammatory mediators (TNF-α e IL-8) in IECs (intestinal epithelial cells) [142]. Moreover, overexpression of IL-26 in colonic lesions of patients with Crohn’s disease is correlated to IL-22 and IL-8 production, which are also involved in inflammation. Whether IL-26 plays an actual role in the pathogenesis or just represents a useful marker for inflammatory bowel disease remains to be established.
IL-26 structural considerations
IL-26 has 24.7% amino acid sequence identity and 47% similarity to IL-10. Based on the highly conserved cysteine positions found between IL-26 and IL-10, as well as in vitro analysis of recombinant IL-26 [137, 138], it was proposed that IL-26 forms IL-10-like intercalated homodimers [138].
Cell signaling by IL-10 family members
The signaling of IL-10 family members is initiated by the recognition of a soluble interleukin by specific transmembrane receptors. IL-10 family members bind to the extracellular domain of two transmembrane receptors, forming a ternary complex constituted of the interleukin, one receptor chain with a long cytoplasmic domain, and one receptor chain with a short cytoplasmic domain.
Both receptor chains possess cognate Janus-family tyrosine kinases (JAK1, JAK2, or Tyk2) associated with their intracellular domain. Interleukin binding to the two specific receptors activates JAK proteins, either by allowing ideal protein/protein contacts, or by inducing conformational changes (Fig. 3). It is important to note that a single JAK molecule cannot phosphorylate any specific residues on its own chain, due to intra-chain mobility problems (reviewed in [143]). However, upon the ternary IL/IL-R1/IL-R2 complex formation, the two JAKs molecules associated with IL-R1 and IL-R2 can phosphorylate each other, which increases their phosphorylation activity. The activated JAKs then phosphorylate several residues in the receptor chain, signaling to signal transducers and activator of transcription (STATs), which results in phosphorylation of specific STAT residues.
Schematic representation of IL-10 family members signaling pathway, based on the IL-22 set of receptors. 1 Inactivated long- and short-chain class II cytokine receptors are bound to Janus-family tyrosine kinases (JAK1 and Tyk2). Inactivated STAT homodimers are found in the cytoplasm. 2 Interleukin binding on the extracellular portion of R1 and R2 activates intracellular JAK/Tyk phosphorylation. Tyrosine residues on either Jak/Tyk and on the receptors are phosphorylated. However, a soluble decoy receptor (IL-22BP) can interact with the IL-22 lacking transmembrane receptor activation. 3 STATs are recruited and phosphorylated, forming activated STAT dimers. 4 STAT dimers migrate to the nucleus activating cytokine specific induced genes
The phosphorylated STATs form homo- or heterodimers then migrate to the nucleus where they specifically activate transcription. Phosphorylated STAT dimers specifically bind to GAS (IFN-γ activation site), a palindromic enhancer with a consensus sequence (N5-GAS) found in many promoters of cytokine-responsive genes [143, 144] (Fig. 3). In the case of signaling by IFN-λs, a complex consisting of STAT1, STAT2, and p48 (IRF9), termed ISGF3, binds to interferon-stimulated response element (ISRE) promoting interferon-specific gene transcription [145, 146]. All other members of the IL-10 family activate mainly STAT3.
Receptors involved in IL-10 family members signaling
As alluded to above, a ternary complex formation involving the interleukin and its cognate type 1 and type 2 receptors is a crucial step in activation of the effective interleukin signaling. All IL-10 family members signal through the combination of a long chain (IL-10R1, IL-22R1, IL-20R1, or IFN-λR) and a short-chain (IL-10R2 or IL20-R2) class II cytokine receptor (reviewed in [143, 145], Table 2). Additionally, IL-22 can interact with high affinity with the IL-22 binding protein (IL-22BP), a soluble type 1 receptor that can antagonizes IL-22 signaling by impairing its interaction with the transmembrane receptor IL-22R1 [147]; Fig. 3.
The class II cytokine receptor family (CRF2) comprises transmembrane glycoproteins with a single membrane spanning region of 20–25 amino acid residues. The CRF2s can be separated into three domains, the extracellular (~200 amino acids long), the intracellular (~60 to ~300 amino acids long, for the short- and long-chain receptors, respectively) and the transmembrane domain. Although low residue conservation is observed in the transmembrane and intracellular domains of the CRF2s, several important features are preserved in the extracellular domain.
The extracellular portion of CRF2s contains two highly conserved disulfide bridges and several conserved proline and tryptophan residues. This domain is generally organized into two fibronectin type III (FNIII) sub-domains in tandem, which have an immunoglobulin-like fold with seven β-strands organized in two β-sheets (reviewed in [143, 145]). The extracellular domain of class II cytokine receptors is responsible for its ligand binding and specificity. There is a partial overlap of CRF2s combination in IL-10 family signaling, but specificity is still attained by a specific receptor and/or interleukin expression by different cell types [145, 146].
The cytoplasmic domain of cytokine receptors generally contains short α-helices organized in boxes named box I and box II close to the transmembrane regions. These ‘boxes’ are responsible for stable interactions with JAKs [148] (Table 3). Several tyrosine residues are found in the cytokine receptor cytoplasmic portion, and these are phosphorylated by the JAKs associated kinases after receptor stimulation (reviewed in [143]). JAK phosphorylation leads to STAT activation, mostly STAT1 and STAT3, but STAT5 phosphorylation can also be induced by IL-10, IL-22 and the IFN-λ group that also signals through STAT2 (reviewed in [143]). A unique characteristic of the IL-22R1 chain is that the C terminus of this receptor chain recruits in a tyrosine-independent manner the coiled-coil domain of STAT3. Mutation of all IL-22R cytoplasmic tyrosines did not abolish activation of STAT3 in response to IL-22. By contrast, deletion of the C-terminal part of IL-22R1 dramatically decreased its ability to activate STAT3 and to mediate IL-22 activity in cell lines, demonstrating that pre-association of STAT3 with this cytokine receptor, independent from the interaction between the Src homology 2 domain and phosphotyrosines, is required for its full activity [149].
In terms of crystal structures, limited information is currently available about IL-10 family members’ binary complexes, and no crystallographic data exists for the ternary complexes to date. Overall, the crystal structures of IL-10/IL-10R1 [117] and IL-22/IL-22R1 [27, 28] binary complexes have already been solved. Very recently, the 3D structure of IL-22 bound to its decoy soluble receptor IL-22BP has also been determined [150].
Based on the available crystal structures of IL-10s binary complexes, here we will discuss the interleukin/receptor interfaces, conformational changes caused by the type 1 receptor binding and the potential type 2 receptor binding sites formed upon the binary complex formation. Since the ternary complex’s structures are not yet available, we will refer to theoretical models and site-directed mutagenesis analysis to discuss the second receptor-binding interfaces and the importance of cytokine conformational changes for the ternary complex formation. We will also review the published data that focused on the interleukin/receptor hotspots in the other IL-10 family members.
Binary complexes with known three-dimensional structure
IL-10 and IL-22 binary and ternary complexes have been extensively studied over the last decade [27, 28, 57, 150–153]. However, due to the intrinsic difficulties to work with the entire transmembrane receptor, these studies were mostly conducted with the receptor extracellular domain. It has been shown that IL-10 and IL-22 display high affinity for their long-chain cognate receptors (IL-10R1 and IL-22R1, respectively); while the interaction of the free interleukin with the second receptor chain, IL-10R2, is weak [57, 151].
It was also demonstrated that the first binary complex formation facilitates interactions with IL-10R2. Experimental data obtained using surface plasmon resonance (SPR) demonstrated that the dissociation constant of the soluble sIL-10R2 and IL-22 complex is 120 μM, as compared to 14.4 μM for the IL-22/IL-22R1 binary complex, [151]. In a similar way, the dissociation constant for IL-10R2 and IL-10 is in the millimolar range whereas that of IL-10/IL-10R1 binary complex is in the micromolar range [57]. These findings suggest that conformational changes of the cytokines, caused by the first receptor binding, and/or the formation of new interaction sites on the binary complex, may contribute to the second receptor binding, in a sequential cooperative process involving the binary and ternary complexes formation.
The IL-22/IL-22R1 structure and interfaces
Two crystal structures of IL-22 complexes with the soluble extracellular domain of IL-22R1 (sIL-22R1) independently determined by two different groups have been reported recently (PDB ID: 3DGC [27] and 3DLQ [28]). Both structures revealed that the sIL-22R1 is composed of two fibronectin type III (FBN-III) domains, termed D1 (N-terminal) and D2 (C-terminal) [27, 28]. The D1 and D2 domains make an angle of approximately 120° to each other [28]. Each IL-22R1 domain presents seven β-strands (A, B, C, C′, E, F and G) forming a sandwich of two antiparallel β-sheets connected by small helix and loops. In addition, IL-22R1 three-dimensional structure contains a small N-terminal helix (Fig. 4a).
The IL-22/IL-22R1, IL-22/IL-22BP, and IL-10/IL-10R1 binary complex interfaces. a IL-22/IL-22R1 (PDB entry 3DLQ [28]). b IL-22/IL-22BP (PDB entry 3G9V [150]). c IL-10/IL-10R1 (PDB entry 17JV [117]). The cysteine residues involved in disulfide bonds are shown as light grey sticks and labeled. Sites 1a and 1b are highlighted and amplified. The potential polar contacts are displayed as dashed lines, with the hydrogen bonds in yellow and the ionic interactions in red. The selected water molecules are shown as red spheres. Oxygen atoms are shown in red, nitrogen in blue, and sulfur in yellow. Figures were generated with the PyMOL software (DeLano Scientific)
Residues from IL-22 helix A, AB loop, and helix F make contacts with IL-22R1 loops L2 to L6, which are located at the interface of D1 and D2 domains. A total of 25 amino acid residues are involved in IL-22 and IL-22R1 interactions [28], and these are mainly mediated by polar contacts. Two sub-sites can be recognized in the IL-22/IL-22R1 interface, namely site 1a and 1b [27]. Site 1a is characterized by hydrogen bond and ionic interactions between IL-22 and the D1 N-terminal domain from IL-22R1. An extensive water-mediated hydrogen bond network is present between residues from IL-22R1 L2–L4 loops and residues from IL-22 AB loop and helix F. The IL-22R1 Tyr60 (loop L2) and the IL-22 Arg73 (AB loop) residues are found at the center of two discernible clusters of this network, located, respectively, at the upper and lower portion of the site 1a. Residue Tyr60 inserts itself into a cleft formed by IL-22 helices A and F and the AB loop, contacting IL-22 residues from helix F1 (Lys162 and Glu166) and AB loop (Asp67, Asp71, and Arg73). It is important to note that Lys162 and Glu166 make part of the IL-10 signature (KAIGELDLL). IL-22R1 residues Glu62, Ser64, and Gly61 from loop L2 and Tyr93 and Arg112 from loop L4 also participate in this network. On the other portion of site 1a, Arg73 side chain is stabilized by a water molecule that also interacts with the IL-22R1 Tyr60 main chain oxygen atom. Arg73 projects itself into a cluster of water molecules and polar side chains. Arg73, Asp168 (helix F2), and Gly165 (helix F bent) from IL-22 participate in the hydrogen bond network by interacting with residues from IL-22R1 loop L3 (Glu90, Thr89 and Lys88) and loop L4 (Ser114). Additionally, three ionic contacts are found in IL-22/IL-22R1 site 1a, involving residues Asp71, Asp67, and Arg112 from IL-22 and residues Lys58, Arg112, and Glu90 from IL-22R1 (connected by the red lines in Fig. 4a).
On the other hand, the IL-22/IL-22R1 interactions at site 1b are less extensive, but nevertheless appear very important for IL-22 conformational changes induced by IL-22R1 binding. A hydrogen bond network is formed, which involves IL-22 residues from helices A and F and IL-22R1 residues from loops L5 and L6 in its D2 C-terminal domain. Phe47 and Thr53 from IL-22 helix A interact with Thr207 from IL-22R1 loop L6 through a water molecule, whereas the IL-22R1 Thr207 side chain interacts directly with the Arg175 side chain located on IL-22 helix F. Phe47, Gln49, and Thr53 from IL-22 helix A interact with the backbone oxygen from Pro206 on IL-22R1 loop L5. A hydrophobic cluster, involving IL-22 residues positioned at helix F (Met172 and Arg175) and IL-22R1 residues from loop L6 (Thr207 and Trp208), also contributes to IL-22/IL-22R1 binding and recognition.
Mutagenesis analysis revealed that IL-22R1 residue Tyr60 is essential for IL-22/IL-22R1 complex formation [28]. Mutation of Tyr60 to either Arg or Ala impaired STAT3-dependent responses in HEK-293 cells expressing the IL-22R1 point mutants [28]. On the other hand, cells expressing the IL-22R1 Lys58Ala point mutant still responded to IL-22 stimulation, however with a 100-fold decrease in specific activity [28]. This observation suggests that lost of ionic interaction between IL-22R1 Lys58 and IL-22 Asp71 is important, but somewhat less fundamental for IL-22/IL-22R1 complex formation.
The IL-22/IL-22BP structure and interfaces
In 2001, a gene encoding a protein of 231 amino acid length and sharing 33% amino acid identity with the extracellular domain of IL-22R1 has been identified [154, 155]. The protein, lacking the transmembrane and cytoplasmic domains of typical class II cytokine receptors, displays structural homology with the extracellular portion of IL-22R1 [155]. The purified protein migrated as a 35 to 45-kDa band on SDS-PAGE gels, suggesting existence of glycosylation. Indeed, bioinformatics analysis predicts five N-glycosylation sites (Asn-X-Thr/Ser) based on the protein amino acid sequence. The recombinant protein binds to and inhibits IL-22 activity in hepatocytes and intestinal epithelial cells [155]. Furthermore, cross-linking experiments revealed that the protein binds IL-22 and prevents the formation of IL-22/IL-22R1/IL-10R2 ternary complex [154]. This protein was termed IL-22 binding protein (IL-22BP) [154, 155].
IL-22BP has higher affinity to IL-22 (~1 pM) than IL-22R1 (~1 nM) and does not bind any other family member, such as IL-10, IL-19, IL-20, or IL-24. Thus, it seems that the IL-22BP expression is one of the mechanisms to specifically regulate IL-22 activity and probably modulates local inflammation [27, 57].
The IL-22BP human gene is organized in six exons and located on chromosome 6, 100 kb apart from IL-20R1 and 24 kb from the gene encoding IFN-γR1 [147, 154, 155]. Tissue distribution of IL-22BP mRNA showed the highest expression levels in breast, placenta, and skin; but high expression was also found in spleen, gastrointestinal tract, and lungs, and lower levels were detected in brain, heart, thymus, pancreas, testis, and prostate [147, 155]. Nothing is currently known about the distribution of the protein, its concentration in various body fluids, and its correlation with pathophysiological processes.
The structure of IL-22BP bound to IL-22 has been solved very recently ([150], PDB entry 3G9V). Not surprisingly, the overall IL-22BP structure organization is very similar to the extracellular part of IL-22R1 (r.m.s.d. = 1.68 Å). IL-22BP molecule folds in an L-shaped structure consisting of two FBN-III domains (a N-terminal D1 domain and a C-terminal D2 domain), connected by a small 310 helix and making a 125° opening angle with respect to each other. Each FBN-III domain forms a β-sandwich composed by two anti-parallel sheets formed from seven β-strands, named A, B, E and C, C′, F, and G (Fig. 4b).
IL-22BP D1 domain is more similar to IL-22R1 D1 (r.m.s.d. = 0.81 Å) than are their D2 domains (r.m.s.d. = 3.57 Å). This is in line with the lower amino acid sequence identity between IL-22R1 and IL-22BP at the D2 domain regions [150]. There are also structural divergences in the disulfide bond pattern between these two receptors within the D2 domain region. In IL-22BP, Cys206 (β-strand F) is probably connected to Cys227 (β-strand G), although Cys227 has not been observed in the electron density of the IL-22/IL-22BP crystal complex. However, in IL-22R1, Cys128 (β-strand A) is linked to Cys217 (β-strand G). This is a particularity of the IL-22R1, since the disulfide bond pattern found in IL-22BP D2 domain is conserved in all other class II cytokine receptors. Another disulfide bond found in IL-22BP structure occurs in the D1 domain: Cys78 interacts with Cys86, connecting strands C′ and E. Equivalent S–S bridges are found in the IL-22R1 and IL-10R2 D1 domains.
Comparison of IL-22/IL-22BP and IL-22/IL-22R1 crystal structures show that both receptors display overlapping IL-22 binding interfaces, which is consistent with the inhibitory role played by IL-22BP. As observed for the IL-22/IL-22R1 and IL-10/IL-10R1 complexes, IL-22BP contacts IL-22 using loops L2 to L6 forming two distinct binding interfaces: site 1a and site 1b.
Residues from IL-22BP loops L2–L4 contact IL-22 residues from loop AB (Asp67, Thr70, Asp71, Val72, Arg73, Glu77) and helix F1 (Lys162, Gly165, Glu166), forming the IL-22/IL-22BP site 1a. Tyr67 from IL-22BP loop L2, which is equivalent to IL-22R1 Tyr60, inserts itself into a cleft formed by residues from IL-22 helices A and F1 and the AB loop, including Lys162 and Glu166 that are part of the IL-10 family signature. IL-22BP Tyr67 makes direct contacts with Asp67, Asp166, and Lys162 side chains and Val72, Arg73, and Thr70 main chains. Furthermore, water mediates the interaction between IL-22BP Tyr67 main chain oxygen atom and IL-22 Glu77 side chain. Lys65, Glu97, and Arg119 from IL-22BP loops L2, L3, and L4, respectively, form ionic interactions with IL-22 residues Asp71, Arg73 and Asp67 located on the AB loop (Fig. 4b).
On the other hand, there are fewer polar contacts at the IL-22/IL-22BP site 1b. IL-22BP residues Met215 and Leu216 from the loop L6 interact with IL-22 residues from the helices pre-A (Phe47) and A (Thr53 and Asn54). Interestingly, IL-22BP residue Glu168 and IL-22 helix F2 residue Arg175 form an ionic interaction that is not observed in the IL-22/IL-22R1 site 1b interface, due to an alternative side chain conformation of Arg175 in the IL-22/IL-22R1 binary complex structure [28]. Additionally, IL-22BP Trp123, Trp124, Met215, and Leu216 are closely positioned to the hydrophobic IL-22 residues Phe57, and Met172, thus forming a hydrophobic cluster that further contributes to IL-22/IL-22BP stabilization at the site 1b (Fig. 4b).
Structural analysis of the IL-22/IL-22BP interface and its comparison to IL-22/IL-22R1 binary complex revealed a number of structural divergences concerning their binding interfaces. Overall, the IL-22BP polar contacts with IL-22 are higher in number and smaller in distances than the observed to IL-22R1 and these are mainly direct instead of water mediated contacts. Furthermore, IL-22/IL-22BP complex contains a larger hydrophobic cluster centered at site 1b as compared to IL-22/IL-22R1. These structural differences might explain dissimilarities in affinity of IL-22 binding to IL-22BP and IL-22R1.
The critical residues identified from the structural analysis of the IL-22/IL-22BP complex allowed the design of site-directed mutagenesis for functional studies [89]. Two IL-22BP residues (Tyr67 and Arg119) were chosen due to their participation in important IL-22/IL-22BP interactions. Both IL-22BP Tyr67 and Arg119 point mutants completely abolished IL-22 binding. Mutations in equivalent residues of IL-22R1 led to similar results. These findings indicate that Tyr67 (Tyr60 in IL-22R1) and Arg119 (Arg112 in IL-22R1) are crucial for IL-22BP and IL-22R1 binding to IL-22 [28, 150].
The IL-10/IL-10R1 structure and interfaces
The three-dimensional structure of IL-10 in complex with its cognate receptor sIL-10R1 was solved by Josephson and co-workers in 2001 (PDB ID 1J7V [117]). Despite the low amino acid sequence identity between IL-22R1 and IL-10R1, they are structurally similar. The main differences observed between these two structures are related to the orientation of the IL-10R1 D2 domain with respect to its D1 domain. The IL10R1 D1 domain is rotated about 30° with respect to the D2 domain in comparison to the orthogonal mutual orientation of D1/D2 domains of the IL-22R1 structure [27, 28]. Furthermore, IL-10R1 contains a WSXWS-like motif, consisting of residues His87, Ser88, Gln89, Trp90, and Thr91, in its D1 domain (Fig. 4c). The WSXWS motif is found in class 1 cytokine receptors [156]. In IL-10R1, the HSQWT sequence divides the G strand of D1 domain in two equivalent sub-parts, named G1 and G2 [117]. Differently from the classic class 1 receptors containing the WSXWS sequence, the hydrogen bond pattern of the IL-10R1 positions the G1 and G2 segments parallel to each other, orienting the L2 and L4 loops position. It was proposed that the residues in or close to the WSXWS-like motif may influence domain orientation, as well interleukin/receptor interactions [117]. It is important to note that, in IL-22R1, there is no equivalent sequence and the G strand is continuous (Fig. 4a). Furthermore, the disulfide bonds found in IL-10R1 are not structurally equivalent to those observed in IL-22R1.
Despite the low sequence identity and several secondary structure mismatches, the overall IL-10/IL-10R1 interactions are very similar to those found in the IL-22/IL-22R1 complex. The same two interfaces described to IL-22/IL-22R1, sites 1a and 1b, occur in the IL-10/IL-10R1 complex, involving structurally equivalent regions. Site 1a includes the IL-10R1 loops L2-L4 and IL-10 AB loop and helix F; whereas site 1b is composed by the IL-10R1 loops L5, L6, and the N-terminal portion of helix A and the C-terminal region of IL-10 (Fig. 4c).
The main interactions observed in IL-22/IL-22R1 site 1a are conserved in the equivalent site of the IL-10/IL-10R1 crystal complex. IL-10 Tyr43 play the role of IL-22 Tyr60 being positioned into the cleft formed by the interleukin helix A, the AB loop and helix F, interacting with the IL-10 residues Glu142 and Lys138, from the IL-10 sequence signature region, through hydrogen bonds [117]. Thus, the main epitope observed in IL-22/IL-22R1 binary complex is conserved in IL-10/IL-10R1.
However, when comparing the IL-10/IL-10R1 to the IL-22/IL-22R1 site 1a interface, several differences can be identified. The second cluster of polar residues found in the IL-22/IL-22R1 1a interface is absent in the IL-10 binary complex. The charged Arg73 found in IL-22 is replaced by an apolar residue, Leu46, in the IL-10 AB loop. This substitution impairs IL-10 interactions with the IL-10R1 loop L3 (Fig. 4).
The IL-22R1 residues Tyr93 and Lys58 have their structural equivalent counterparts in IL-10R1, Arg76, and Leu41. Arg76 is found in two alternative conformations in the IL-10/IL-10R1 crystal complex. In one of its conformations, Arg76 plays the role of Tyr93 at the IL-22/IL-22R1 interface, being hydrogen bonded to the main chain of IL-10 Gln38 and to a water molecule. On the other hand, the second Arg76 side chain conformation substitutes for the IL-22R1 Lys58 side chain, making an ionic interaction with Asp44 at IL-10 AB loop and a hydrogen bond interaction with Gln42 side chain. In IL-10R1, a smaller and uncharged residue (Leu41) is at IL-22R1 Lys58 equivalent position. Thus, the L2 loop residue Arg76, instead of the strand C residue Lys58 (in IL-22R1), forms the ionic interaction at the upper portion of the IL-10/IL-10R1 complex.
Another interesting feature found in the IL-10/IL-10R1 crystal complex is related to IL-10R1 L4 loop position. If one superposes the equivalent loop region of IL-22R1 with IL-10R1, it can be observed that the IL-10R1 loop L4 is in a more extended conformation than the same loop in the IL-22R1 structure. The above-mentioned WSXWS-like motif present in IL-10R1 G strand projects its L4 loop in the direction of IL-10. As a result, the IL-10R1 Arg96 side chain is positioned close to the negative-charged side chains of Asp144 and Glu142 placed on IL-10 helix F, contributing to the binding of the interleukin to its receptor. Furthermore, Arg96 also interacts with the IL-10 Gln38 side chain and Ser141 main chain. However, in IL-22R1, the Arg112 side chain—that is equivalent to IL-10R1 Arg96—is positioned at the upper portion of the IL-22/IL-22R1 site 1a due to the shorter L4 loop extension, and, as a consequence, the interactions with IL-22 helix F are mainly mediated by water molecules.
In the site 1b, the IL-10/IL-10R1 contacts are more extensive than IL-22/IL22R1 crystal complex interactions. Comparison of IL-10 and IL-22 free structures (Fig. 2) reveals that these two interleukins display different orientations of their N-terminal regions and helix A. These features are important for receptor specificity and binding to site 1b. In IL-10/IL-10R1 complex, the IL-10 N-terminal portion of helix A is continuous and there are two arginine side chains projected in the direction of the receptor (Arg24 and Arg27). These Arg side chains form a hydrogen bond network with the main chains of Arg191 and Ser190 of the IL-10R1 L6 loop. Furthermore, the Arg27 side chain from IL-10 forms an ionic interaction with Glu151 from IL-10, stabilizing Glu151 side chain on IL-10 helix F bent. Therefore, Glu151 is at the ideal distance to interact with the receptor Ser190 and Arg191 side chains (Fig. 4c) at site 1b. However, in IL-22R1 Trp208, found in the equivalent position of IL-10R1 Arg191, makes hydrophobic contacts with Met172 on IL-22 helix F. Furthermore, in IL-22, the N-terminal portion of helix A is rotated, forming the preA helix. The preA helix is extended away from the receptor, and most of the IL-22/IL-22R1 interactions in this region are mediated by water molecules. Similar to the IL-22/IL-22R1 crystal complex, IL-10/IL10R1 binary complex also contains a small hydrophobic cluster. The side chain of Phe143, placed on IL-10R1 L5 loop, inserts itself into a cleft formed by IL-10 Pro20 (helix A) and Ile158 (helix F), making additional contacts in the IL-10/IL-10R1 site 1b (Fig. 4c).
Although the binding constants of IL-10 to IL-10R1 are in the same order of magnitude to the IL-22/IL-22R1 constants (K d ~1 nM), the IL-10/IL-10R1 complex half-life (T 1/2) is higher (T 1/2 = 44 min) than the observed for the IL-22/IL-22R1 (T 1/2 = 7.4 min) [57, 157]. This is proposed to be a consequence of the weaker contacts found in the IL-22/IL-22R1 site 1b.
The IL-10 family members binary complexes: structural basis of receptor specificity and promiscuity
Comparison of the IL-10/IL-10R1 and the IL-22/IL-22R1 binary complexes show conservation of the main features of recognition between the ligand and its receptor. As discussed above, different amino acid residues and loop positions are observed at the two interleukin/receptor interfaces. The maintenance of similar epitopes, conserving the overall type and number of interactions, is possible since both the interleukin and their receptors suffered continuous sequence adaptations during evolution. This guarantees receptor, and consequently, cell response specificity to each interleukin. Besides the discrepancies found in site 1b, the contacts found in site 1a, mainly mediated by Tyr60 in IL-22R1 and Tyr43 in IL-10R1, are also proposed to be a crucial determinant in interleukin/receptor binding specificity [27, 28, 117]. The geometry of the hydrogen bond network found in site 1a is not reproducible in different interleukin/receptor pairs without changing of their mutual orientation. Therefore, ideal contacts at site 1a become incompatible with the simultaneous match at 1b interface [27] in non-cognate interleukin/receptor pairs that in turn confers receptor specificity.
Aiming to verify these suggestions, Jones et al. [27] compared the AB loop sequences found in other IL-10 family members. They found that the interleukins identified as IL-22R1 binders (IL-20 and IL-24) display structural conserved residues with IL-22. IL-20 contains equivalent residues at IL-22 Arg73 and Asp71 position, whereas IL-24 conserves the IL-22 Arg73 and Thr70. Accordingly, in IL-10, IL-19 and IL-26, which do not bind IL-22R1, the equivalent residues are absent on loop AB.
Unfortunately, the 3D structures of the IL-10s ternary complexes are not yet available. However, site-direct mutagenesis followed by cell-based assays and binding studies [151, 153, 158] have generated experimental data to propose structural models for the interleukin ternary complex. The 3D structures of the IL-22/IL-22R1 [27, 28] and IL-10/IL-10R1 [117] binary complexes confirm previous suggestions of the overall location of the first receptor binding sites on IL-10 and IL-22 and also contribute to a better understanding of the second receptor interface, and hence, the overall ternary complex structure [27, 28, 117, 153]. In the following and last section of this review, we discuss recently published data on IL-10 members ternary complexes analysis, describing the main interactions and binding interfaces pinpointed from these studies.
Binary and ternary complexes models
Putative IL-10R2 binding site at the IL-22:IL-22R1 binary complex
Several lines of evidence suggest that the IL-10R2 binding site is located at helices A and D of IL-22 [27, 28, 151, 158]. Initially, site-directed mutagenesis and surface-plasmon resonance analysis revealed that IL-22 residues (placed on helices A and D) Tyr51, Asn54, Arg55, Tyr114, and Glu117 are important for IL-10R2 engagement with IL-22/IL-22R1 binary complex [151]. Later, an extensive mutagenesis study screening of 146 different IL-22 residues also included IL-22 Ile14, Thr56, Lys61, Ala66, Val83, Arg88, Pro113, Phe121, Leu122, and Met172 as important residues for IL-10R2 binding [158].
Very recently, based on the IL-22 binary complex crystal structures, two independent models of IL-10R2 interaction with IL-22/IL-22R1 were proposed [27, 28]. Jones and collaborators used mutagenesis scanning analysis [27] as a starting point to construct their model, whereas Bleicher and co-workers used the recent crystal structures of IL-4 and IL-13 ternary complexes (IL-4/γc/IL-4Rα, IL-4/IL-13Rα1/IL-4Rα and IL-13/IL-13Rα1/IL-4Rα [159]) as an initial scaffold to model IL-10R2 interactions with the IL-22/IL-22R1 binary complex [28]. These two independent tertiary complex models appear to be very similar. In both models, IL-10R2 is proposed to interact with IL-22 through two binding sites, site 2a and site 2b, that are adjacent to the IL-22R1 sites 1a and 1b (Fig. 5a). These IL-10R2 binding sites are formed by IL-22 residues placed on helices A and D, in line with alanine scanning mutagenesis analysis [151, 158]. Furthermore, it was proposed that IL-10R2 also contacts IL-22R1 via their respective D2 domains and the three IL-10R2 binding interfaces were named sites 2a, 2b, and 2c [27].
IL-22 ternary complex interfaces. a IL-22R1 and putative IL-10R2 interfaces mapped on the IL-22 surface (PDB ID 2M4R [8]). Site 1a is colored orange, site 1b red, site 2a cyan, and site 2b blue. b Detailed view of the potential IL-10R2 binding interfaces at IL-10 (PDB ID 2YLK [130]; colored green) and IL-22 (PDB ID 2M4R [8]; colored light grey) crystal structures. Oxygen atoms are shown in red and nitrogen in blue. Figures were generated with PyMOL software (DeLano Scientific)
Site 2a consists of putative IL-22/IL-10R2 contacts at IL-22 C-terminal portion of helix A (Tyr51, Asn54, Arg55, Glu63) and N-terminal portion of helix D (Tyr114). It is important to note that Arg55 probably plays a central role in the IL-22 binding with IL-10R2 by forming a stable ionic interaction with IL-10R2 Asp84 on site 2a. Site 2b is located on the helix A bent, at the terminal portion of helix preA. In this region, a proline (Pro50) and a glutamine (Gln48) residue are proposed to mediate interactions with, respectively, Trp152 and Asn150 from IL-10R2 [27, 28] (Fig. 5b). Additional residues placed nearby IL-22 helix A bent, such as Met172, were also identified in mutagenesis screening analysis [158] and are therefore likely to contribute to IL-10R2 stabilization on site 2b. On the other hand, site 2c is formed by residues located in the IL-22R1 and IL-10R2 D2 domains. These extra contacts promoted by receptor/receptor interactions further contribute to the ternary complex formation. IL-22R1 residues Glu168, Gln176, Asn172, Thr174 and IL-10R2 Lys146, Leu132, Arg130, Gln119, Glu121 are presumably involved in these interactions [27, 28]. Importantly, the IL-22R1 residue Asn174, which is presumably involved in interaction at site 2c, can be glycosylated, which in turn can also modulate IL-22R1 contacts with IL-10R2 [27].
Small overall structural adjustments are observed when superposing the crystal structures of free and bound IL-22 structures (IL-22/IL-22R1) (r.m.s.d. < 0.7 Å). However, much more significant conformational changes are observed at the IL-22 N-terminal region and helix A (r.m.s.d. ~ 4 Å) [28]. These structural adaptations might result in better accommodation of IL-10R2 at the IL-22 surface (sites 2a and 2b). These conformational changes are in concert with the notion that IL-10R2 interactions with the IL-22R1 D2 domain contribute to the higher affinity to the IL-22/IL-22R1 binary complex (K d = 14 μM) than to IL-22 alone (K d = 120 μM) [27, 151].
Putative IL-10R2 binding site at the IL-10:IL-10R1 binary complex
Overall, the IL-10R2 interactions with the IL-10/IL-10R1 binary complex are likely to be similar to those proposed for the IL-22/IL-22R1/IL-10R2 ternary complex. However, structural differences on the molecular level must explain the higher affinity displayed by IL-10R2 when binding to IL-22 (K d = 120 μM (free IL-22) and K d = 14 μM (IL-22/IL-22R1 [151]) in comparison to IL-10 (K d ~ 1 mM (free IL-10) and K d = 234 μM (IL-10/IL-10R1) [153]).
According to the proposed IL-10 and IL-22 ternary complexes, it has been argued that the flatter IL-10 helix A is the main cause for weakening the IL-10R2 contacts with IL-10 on site 2a and 2b [27]. In IL-22, Tyr51 stabilizes and orients Arg55 that forms an ionic interaction with Asp84 from IL-10R2 on site 2a. In contrast, in IL-10, the Arg32 residue (which plays the role of Arg55 in IL-22) is not stabilized by any additional interactions. Furthermore, IL-10 Arg32 is one turn of a helix away from IL-22 Arg55 (Fig. 5b), and its side chain must be reoriented to interact with IL-10R2 Asp84. Therefore, it is proposed that this salt bridge is weakened in IL-10/IL-10R2 site 2a, and might be one of the main reasons for the low free IL-10 affinity to IL-10R2 [27].
Conformational changes at the IL-10 N-terminal portion of helix A are observed when comparing free and IL-10R1 bound IL-10 crystal structures. Residues 18–22 suffer a rigid body movement (>5 Å) produced by rotating (26°) Leu23 phi\psi angles. As a result, in the receptor-bound IL-10, helix A is longer and straighter than in free IL-10. These conformational changes are proposed to be essential for IL-10R2 binding to IL-10 [153] and corroborate with the experimental IL-10R2 binding affinities to free and bound IL-10.
The IL-10R2 binding interface was investigated by different methodologies [27, 117, 151, 153]. Based on IL-10 binary complex crystal structure [117] and on previous IL-22 mutagenesis analysis [151], Yoon and co-workers (2006) [153] designed IL-10 alanine mutants to search for the potential IL-10R2 binding site on this interleukin. They found that, similar to IL-22, IL-10 residues placed on the C-terminal portion of helix A (Ser31 and Arg32) and residues from helix D (His90 and Ser93) are involved in site 2a contacts with IL-10R2, whereas, the N-terminal portion of helix A (Asn21, Met22, and Arg24) is involved in contacts related to site 2b. Additionally, structural modeling of IL-10R2 on IL-10/IL-10R1 binary complex [27] revealed that, as observed to IL-22, IL-10R2 can also contact IL-10R1 through interactions with their D2 domains, thus forming site 2c, which further contributes to IL-10 ternary complex stabilization.
Although the overall IL-10R2 binding interfaces at the IL-22 and IL-10 ternary complexes are similar, the molecular details are quite different [160]. It appears that site 2b is the most important IL-10R2 interface in IL-10 ternary complex; whereas site 2a is central for IL-22 ternary complex formation [153]. IL-10 Met22 seems to play a primary role in IL-10R2 binding, by helping the rigidification of IL-10 N-terminal region and by interacting directly with IL-10R2 through hydrophobic interactions. Furthermore, Arg24, which also participates in IL-10/IL-10R1 site 1b interactions, is probably involved in polar contacts with IL-10R2 L5 loop on site 2b [153].
Putative IL-19, IL-20, and IL-24 binary and ternary complexes: structural models
Unfortunately, the crystal structures of IL-19 or IL-20 complexes with receptors are currently not available, and neither are IL-20R1 nor IL-20R2 structures. However, due to IL-19 structural similarities with IL-10, Chang et al. [24] generated a model describing the potential hot-spots in IL-19, based on the IL-10/IL-10R1 crystal structure [117]. One year later, in 2004, Preimeland and Sticht [161, 162] also proposed a model for IL-20R1 interaction with IL-19, using the IFN-γ binary complex crystal structure as a starting model [163]. In both hypothetical 3D structures, IL-19 AB loop interacts with IL-20R1 loops, in a similar mode to the one observed for IL-10, with contributions from IL-19 helices A and F [24, 161]. Chang and co-workers found that superposition of IL-19 with IL-10 in the IL-10/IL-10R1 binary complex shows that the majority of the interactions found in IL-10/IL-10R1 site 1a and 1b are allowed [24]. However, a clash between IL-10R1 Glu46 and the glycosylated IL-19 Asn38 is observed. Comparisons of this model with IL-20 and IL-24 amino acid sequences (which also bind to IL-20R1) indicate that the IL-20R1 interaction sites are probably conserved in these two interleukins [161]. Along the same lines, the predicted IL-20R2 binding sites to IL-19 are similar to IL-10R2 binding site to IL-10. Thus, the possible IL-19 receptor binding sites are probably located at the IL-19 helix A, loop AB, helices D and F, and at its C-terminal portion [24].
Differently from IL-10 and IL-22, experimental evidence suggests that IL-19 and IL-20 display high affinity to IL-20R2, and that IL-20R1 binding occurs after the binary complex formation with the short-chain receptor (IL-20R2) [68, 72]. However, the molecular basis of these events is currently unknown.
Putative IFN-λ3 residues involved in binary and ternary complex formation: site-direct mutagenesis analysis
Based on the known IL-10/IL-10R1 and IL-22/IL-22R1 binary complexes 3D structures, on the putative IL-10R2 interfaces and on the recent IFN-λ3 crystal structure, Gad and co-workers designed IFN-λ3 Ala mutants to pinpoint binding hotspots [23]. They found that the interface related to site 1b of IL-10 and IL-22 plays the major role in IFN-λ3/IFN-λR1 interactions. In IFN-λ3, the polar residues positioned at IFN-λ3 helix A (Arg33, Arg34, Lys36, and Asp37) and hydrophobic residues of the helix F (Phe155 and Phe158) are the most sensitive for Ala mutations. Furthermore, IFN-λ3 mutants were constructed to shed light on the putative IFN-λ3/IFN-λR1/IL-10R2 interface. Residues at IFN-λ3 helix D (at the vicinity of Gln100) were shown to be essential for IL-10R2 binding. Moreover, the IFN-λR1 and IL-10R2 interfaces are probably placed at IFN-λ3 helix A [23].
Considerations about IL-26 binary and ternary complexes
Experimental evidence suggests that IL-26, differently from IL-19, IL-20 and IL-24, uses IL-20R1 and IL-10R2 for signaling. It is interesting to note that IL-26 is the only IL-10 family member that employs this pair of receptors in its function [13, 138–140]. However, little experimental data is currently available concerning crystal structures of IL-26 and its complexes with the cognate receptors. Therefore, at this point, it would be far too speculative to argue about binary and ternary complexes of IL-26 and atomic details of its interactions with the cognate IL-20R1 and IL-10R2 receptors.
References
Moore KW, de Waal Malefyt R, Coffman RL, O’Garra A (2001) Interleukin-10 and the interleukin-10 receptor. Annu Rev Immunol 19:683–765
Gallagher G, Dickensheets H, Eskdale J, Izotova LS, Mirochnitchenko OV, Peat JD, Vazquez N, Pestka S, Donnelly RP, Kotenko SV (2000) Cloning, expression and initial characterization of interleukin-19 (IL-19), a novel homologue of human interleukin-10 (IL-10). Genes Immun 1:442–450
Dumoutier L, Renauld JC (2002) Viral and cellular interleukin-10 (IL-10)-related cytokines: from structures to functions. Eur Cytokine Netw 13:5–15
Sheppard P, Kindsvogel W, Xu W, Henderson K, Schlutsmeyer S, Whitmore TE, Kuestner R, Garrigues U, Birks C, Roraback J, Ostrander C, Dong D, Shin J, Presnell S, Fox B, Haldeman B, Cooper E, Taft D, Gilbert T, Grant FJ, Tackett M, Krivan W, McKnight G, Clegg C, Foster D, Klucher KM (2003) IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat Immunol 4:63–68
Kotenko SV (2002) The family of IL-10-related cytokines and their receptors: related, but to what extent? Cytokine Growth Factor Rev 13:223–240
Walter MR, Nagabhushan TL (1995) Crystal structure of interleukin-10 reveals an interferon gamma-like fold. Biochemistry 34:12118–12125
Zdanov A, Schalk-Hihi C, Gustchina A, Tsang M, Weatherbee J, Wlodawer A (1995) Crystal structure of interleukin-10 reveals the functional dimer with an unexpected topological similarity to interferon gamma. Structure 3:591–601
Nagem RA, Colau D, Dumoutier L, Renauld JC, Ogata C, Polikarpov I (2002) Crystal structure of recombinant human interleukin-22. Structure 10:1051–1062
Wolk K, Kunz S, Asadullah K, Sabat R (2002) Cutting edge: immune cells as sources and targets of the IL-10 family members? J Immunol 168:5397–5402
Dumoutier L, Leemans C, Lejeune D, Kotenko SV, Renauld JC (2001) Cutting edge: STAT activation by IL-19, IL-20 and mda-7 through IL-20 receptor complexes of two types. J Immunol 167:3545–3549
Liu Y, Wei SH, Ho AS, de Waal Malefyt R, Moore KW (1994) Expression cloning and characterization of a human IL-10 receptor. J Immunol 152:1821–1829
Kotenko SV, Krause CD, Izotova LS, Pollack BP, Wu W, Pestka S (1997) Identification and functional characterization of a second chain of the interleukin-10 receptor complex. EMBO J 16:5894–5903
Zdanov A (2006) Structural studies of the interleukin-19 subfamily of cytokines. Vitam Horm 74:61–76
Renauld JC (2003) Class II cytokine receptors and their ligands: key antiviral and inflammatory modulators. Nat Rev Immunol 3:667–676
Griffiths CE, Barker JN (2007) Pathogenesis and clinical features of psoriasis. Lancet 370:263–271
Wolk K, Witte E, Wallace E, Docke WD, Kunz S, Asadullah K, Volk HD, Sterry W, Sabat R (2006) IL-22 regulates the expression of genes responsible for antimicrobial defense, cellular differentiation, and mobility in keratinocytes: a potential role in psoriasis. Eur J Immunol 36:1309–1323
Wolk K, Haugen HS, Xu W, Witte E, Waggie K, Anderson M, Vom Baur E, Witte K, Warszawska K, Philipp S, Johnson-Leger C, Volk HD, Sterry W, Sabat R (2009) IL-22 and IL-20 are key mediators of the epidermal alterations in psoriasis while IL-17 and IFN-gamma are not. J Mol Med 87:523–536
Choy EH, Panayi GS (2001) Cytokine pathways and joint inflammation in rheumatoid arthritis. N Engl J Med 344:907–916
Ikeuchi H, Kuroiwa T, Hiramatsu N, Kaneko Y, Hiromura K, Ueki K, Nojima Y (2005) Expression of interleukin-22 in rheumatoid arthritis: potential role as a proinflammatory cytokine. Arthritis Rheum 52:1037–1046
Geboes L, Dumoutier L, Kelchtermans H, Schurgers E, Mitera T, Renauld JC, Matthys P (2009) Proinflammatory role of the Th17 cytokine interleukin-22 in collagen-induced arthritis in C57BL/6 mice. Arthritis Rheum 60:390–395
Martinez JA, King TE Jr, Brown K, Jennings CA, Borish L, Mortenson RL, Khan TZ, Bost TW, Riches DW (1997) Increased expression of the interleukin-10 gene by alveolar macrophages in interstitial lung disease. Am J Physiol 273:L676–L683
Whittington HA, Armstrong L, Uppington KM, Millar AB (2004) Interleukin-22: a potential immunomodulatory molecule in the lung. Am J Respir Cell Mol Biol 31:220–226
Gad HH, Dellgren C, Hamming OJ, Vends S, Paludan SR, Hartmann R (2009) Interferon-λ is functionally an interferon but structurally related to the interleukin-10 family. J Biol Chem 284:20869–20875
Chang C, Magracheva E, Kozlov S, Fong S, Tobin G, Kotenko S, Wlodawer A, Zdanov A (2003) Crystal structure of interleukin-19 defines a new subfamily of helical cytokines. J Biol Chem 278:3308–3313
Xu T, Logsdon NJ, Walter MR (2005) Structure of insect-cell-derived IL-22. Acta Crystallogr D Biol Crystallogr 61:942–950
Nagem RA, Ferreira Junior JR, Dumoutier L, Renauld JC, Polikarpov I (2006) Interleukin-22 and its crystal structure. Vitam Horm 74:77–103
Jones BC, Logsdon NJ, Walter MR (2008) Structure of IL-22 bound to its high-affinity IL-22R1 chain. Structure 16:1333–1344
Bleicher L, de Moura PR, Watanabe L, Colau D, Dumoutier L, Renauld JC, Polikarpov I (2008) Crystal structure of the IL-22/IL-22R1 complex and its implications for the IL-22 signaling mechanism. FEBS Lett 582:2985–2992
Zdanov A (2004) Structural features of the interleukin-10 family of cytokines. Curr Pharm Des 10:3873–3884
Dumoutier L, Louahed J, Renauld JC (2000) Cloning and characterization of IL-10-related T cell-derived inducible factor (IL-TIF), a novel cytokine structurally related to IL-10 and inducible by IL-9. J Immunol 164:1814–1819
Dumoutier L, Van Roost E, Ameye G, Michaux L, Renauld JC (2000) IL-TIF/IL-22: genomic organization and mapping of the human and mouse genes. Genes Immun 1:488–494
Dumoutier L, Van Roost E, Colau D, Renauld JC (2000) Human interleukin-10-related T cell-derived inducible factor: molecular cloning and functional characterization as an hepatocyte-stimulating factor. Proc Natl Acad Sci USA 97:10144–10149
Aggarwal S, Xie MH, Maruoka M, Foster J, Gurney AL (2001) Acinar cells of the pancreas are a target of interleukin-22. J Interferon Cytokine Res 21:1047–1053
Liang SC, Tan XY, Luxenberg DP, Karim R, Dunussi-Joannopoulos K, Collins M, Fouser LA (2006) Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J Exp Med 203:2271–2279
Boniface K, Bernard FX, Garcia M, Gurney AL, Lecron JC, Morel F (2005) IL-22 inhibits epidermal differentiation and induces proinflammatory gene expression and migration of human keratinocytes. J Immunol 174:3695–3702
Harrington LE, Hatton RD, Mangan PR, Turner H, Murphy TL, Murphy KM, Weaver CT (2005) Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat Immunol 6:1123–1132
Wolk K, Kunz S, Witte E, Friedrich M, Asadullah K, Sabat R (2004) IL-22 increases the innate immunity of tissues. Immunity 21:241–254
Lehrer RI, Ganz T (2002) Defensins of vertebrate animals. Curr Opin Immunol 14:96–102
Zasloff M (2002) Antimicrobial peptides of multicellular organisms. Nature 415:389–395
Boniface K, Guignouard E, Pedretti N, Garcia M, Delwail A, Bernard FX, Nau F, Guillet G, Dagregorio G, Yssel H, Lecron JC, Morel F (2007) A role for T cell-derived interleukin 22 in psoriatic skin inflammation. Clin Exp Immunol 150:407–415
Ma HL, Liang S, Li J, Napierata L, Brown T, Benoit S, Senices M, Gill D, Dunussi-Joannopoulos K, Collins M, Nickerson-Nutter C, Fouser LA, Young DA (2008) IL-22 is required for Th17 cell-mediated pathology in a mouse model of psoriasis-like skin inflammation. J Clin Invest 118:597–607
Zheng Y, Danilenko DM, Valdez P, Kasman I, Eastham-Anderson J, Wu J, Ouyang W (2007) Interleukin-22, a T(H)17 cytokine, mediates IL-23-induced dermal inflammation and acanthosis. Nature 445:648–651
Zheng B, Switzer K, Marinova E, Zhang J, Han S (2008) Exacerbation of autoimmune arthritis by copolymer-I through promoting type 1 immune response and autoantibody production. Autoimmunity 41:363–371
Sugimoto K, Ogawa A, Mizoguchi E, Shimomura Y, Andoh A, Bhan AK, Blumberg RS, Xavier RJ, Mizoguchi A (2008) IL-22 ameliorates intestinal inflammation in a mouse model of ulcerative colitis. J Clin Invest 118:534–544
Aujla SJ, Chan YR, Zheng M, Fei M, Askew DJ, Pociask DA, Reinhart TA, McAllister F, Edeal J, Gaus K, Husain S, Kreindler JL, Dubin PJ, Pilewski JM, Myerburg MM, Mason CA, Iwakura Y, Kolls JK (2008) IL-22 mediates mucosal host defense against Gram-negative bacterial pneumonia. Nat Med 14:275–281
Pickert G, Neufert C, Leppkes M, Zheng Y, Wittkopf N, Warntjen M, Lehr HA, Hirth S, Weigmann B, Wirtz S, Ouyang W, Neurath MF, Becker C (2009) STAT3 links IL-22 signaling in intestinal epithelial cells to mucosal wound healing. J Exp Med 206:1465–1472
Zenewicz LA, Yancopoulos GD, Valenzuela DM, Murphy AJ, Stevens S, Flavell RA (2008) Innate and adaptive interleukin-22 protects mice from inflammatory bowel disease. Immunity 29:947–957
Pan H, Hong F, Radaeva S, Gao B (2004) Hydrodynamic gene delivery of interleukin-22 protects the mouse liver from concanavalin A-, carbon tetrachloride-, and Fas ligand-induced injury via activation of STAT3. Cell Mol Immunol 1:43–49
Radaeva S, Sun R, Pan HN, Hong F, Gao B (2004) Interleukin 22 (IL-22) plays a protective role in T cell-mediated murine hepatitis: IL-22 is a survival factor for hepatocytes via STAT3 activation. Hepatology 39:1332–1342
Wolk K, Sabat R (2006) Interleukin-22: a novel T- and NK-cell derived cytokine that regulates the biology of tissue cells. Cytokine Growth Factor Rev 17:367–380
Luci C, Reynders A, Ivanov II, Cognet C, Chiche L, Chasson L, Hardwigsen J, Anguiano E, Banchereau J, Chaussabel D, Dalod M, Littman DR, Vivier E, Tomasello E (2009) Influence of the transcription factor RORgammat on the development of NKp46+ cell populations in gut and skin. Nat Immunol 10:75–82
Sanos SL, Bui VL, Mortha A, Oberle K, Heners C, Johner C, Diefenbach A (2009) RORgammat and commensal microflora are required for the differentiation of mucosal interleukin 22-producing NKp46+ cells. Nat Immunol 10:83–91
Satoh-Takayama N, Vosshenrich CA, Lesjean-Pottier S, Sawa S, Lochner M, Rattis F, Mention JJ, Thiam K, Cerf-Bensussan N, Mandelboim O, Eberl G, Di Santo JP (2008) Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity 29:958–970
Cella M, Fuchs A, Vermi W, Facchetti F, Otero K, Lennerz JK, Doherty JM, Mills JC, Colonna M (2009) A human natural killer cell subset provides an innate source of IL-22 for mucosal immunity. Nature 457:722–725
Pestka S, Krause CD, Sarkar D, Walter MR, Shi Y, Fisher PB (2004) Interleukin-10 and related cytokines and receptors. Annu Rev Immunol 22:929–979
Xu T, Logsdon NJ, Walter MR (2004) Crystallization and X-ray diffraction analysis of insect-cell-derived IL-22. Acta Crystallogr D Biol Crystallogr 60:1295–1298
Logsdon NJ, Jones BC, Josephson K, Cook J, Walter MR (2002) Comparison of interleukin-22 and interleukin-10 soluble receptor complexes. J Interferon Cytokine Res 22:1099–1112
de Oliveira Neto M, Ferreira JR Jr, Colau D, Fischer H, Nascimento AS, Craievich AF, Dumoutier L, Renauld JC, Polikarpov I (2008) Interleukin-22 forms dimers that are recognized by two interleukin-22R1 receptor chains. Biophys J 94:1754–1765
Loiarro M, Sette C, Gallo G, Ciacci A, Fanto N, Mastroianni D, Carminati P, Ruggiero V (2005) Peptide-mediated interference of TIR domain dimerization in MyD88 inhibits interleukin-1-dependent activation of NF-kappa B. J Biol Chem 280:15809–15814
Stuhlmann-Laeisz C, Lang S, Chalaris A, Krzysztof P, Enge S, Eichler J, Klingmuller U, Samuel M, Ernst M, Rose-John S, Scheller J (2006) Forced dimerization of gp130 leads to constitutive STAT3 activation, cytokine-independent growth, and blockade of differentiation of embryonic stem cells. Mol Biol Cell 17:2986–2995
Liao YC, Liang WG, Chen FW, Hsu JH, Yang JJ, Chang MS (2002) IL-19 induces production of IL-6 and TNF-alpha and results in cell apoptosis through TNF-alpha. J Immunol 169:4288–4297
Parrish-Novak J, Xu W, Brender T, Yao L, Jones C, West J, Brandt C, Jelinek L, Madden K, McKernan PA, Foster DC, Jaspers S, Chandrasekher YA (2002) Interleukins 19, 20, and 24 signal through two distinct receptor complexes. Differences in receptor-ligand interactions mediate unique biological functions. J Biol Chem 277:47517–47523
Jordan WJ, Eskdale J, Boniotto M, Lennon GP, Peat J, Campbell JD, Gallagher G (2005) Human IL-19 regulates immunity through auto-induction of IL-19 and production of IL-10. Eur J Immunol 35:1576–1582
Gallagher G, Eskdale J, Jordan W, Peat J, Campbell J, Boniotto M, Lennon GP, Dickensheets H, Donnelly RP (2004) Human interleukin-19 and its receptor: a potential role in the induction of Th2 responses. Int Immunopharmacol 4:615–626
Liao SC, Cheng YC, Wang YC, Wang CW, Yang SM, Yu CK, Shieh CC, Cheng KC, Lee MF, Chiang SR, Shieh JM, Chang MS (2004) IL-19 induced Th2 cytokines and was up-regulated in asthma patients. J Immunol 173:6712–6718
Tian Y, Sommerville LJ, Cuneo A, Kelemen SE, Autieri MV (2008) Expression and suppressive effects of interleukin-19 on vascular smooth muscle cell pathophysiology and development of intimal hyperplasia. Am J Pathol 173:901–909
Romer J, Hasselager E, Norby PL, Steiniche T, Thorn Clausen J, Kragballe K (2003) Epidermal overexpression of interleukin-19 and -20 mRNA in psoriatic skin disappears after short-term treatment with cyclosporine a or calcipotriol. J Invest Dermatol 121:1306–1311
Blumberg H, Conklin D, Xu WF, Grossmann A, Brender T, Carollo S, Eagan M, Foster D, Haldeman BA, Hammond A, Haugen H, Jelinek L, Kelly JD, Madden K, Maurer MF, Parrish-Novak J, Prunkard D, Sexson S, Sprecher C, Waggie K, West J, Whitmore TE, Yao L, Kuechle MK, Dale BA, Chandrasekher YA (2001) Interleukin 20: discovery, receptor identification, and role in epidermal function. Cell 104:9–19
Wolk K, Witte K, Witte E, Proesch S, Schulze-Tanzil G, Nasilowska K, Thilo J, Asadullah K, Sterry W, Volk HD, Sabat R (2008) Maturing dendritic cells are an important source of IL-29 and IL-20 that may cooperatively increase the innate immunity of keratinocytes. J Leukoc Biol 83:1181–1193
Hosoi T, Wada S, Suzuki S, Okuma Y, Akira S, Matsuda T, Nomura Y (2004) Bacterial endotoxin induces IL-20 expression in the glial cells. Brain Res Mol Brain Res 130:23–29
Hsing CH, Ho CL, Chang LY, Lee YL, Chuang SS, Chang MS (2006) Tissue microarray analysis of interleukin-20 expression. Cytokine 35:44–52
Pletnev S, Magracheva E, Kozlov S, Tobin G, Kotenko SV, Wlodawer A, Zdanov A (2003) Characterization of the recombinant extracellular domains of human interleukin-20 receptors and their complexes with interleukin-19 and interleukin-20. Biochemistry 42:12617–12624
Jiang H, Lin JJ, Su ZZ, Goldstein NI, Fisher PB (1995) Subtraction hybridization identifies a novel melanoma differentiation associated gene, mda-7, modulated during human melanoma differentiation, growth and progression. Oncogene 11:2477–2486
Barton K, Randall G, Sagone AL Jr (1989) The effects of the anti-tumor agent mezerein on the cytotoxic capacity and oxidative metabolism of human blood cells. Invest New Drugs 7:179–188
Fisher PB, Prignoli DR, Hermo H Jr, Weinstein IB, Pestka S (1985) Effects of combined treatment with interferon and mezerein on melanogenesis and growth in human melanoma cells. J Interferon Res 5:11–22
Fisher PB (2005) Is mda-7/IL-24 a “magic bullet” for cancer? Cancer Res 65:10128–10138
Fisher PB, Gopalkrishnan RV, Chada S, Ramesh R, Grimm EA, Rosenfeld MR, Curiel DT, Dent P (2003) mda-7/IL-24, a novel cancer selective apoptosis inducing cytokine gene: from the laboratory into the clinic. Cancer Biol Ther 2:S23–S37
Lebedeva IV, Sauane M, Gopalkrishnan RV, Sarkar D, Su ZZ, Gupta P, Nemunaitis J, Cunningham C, Yacoub A, Dent P, Fisher PB (2005) mda-7/IL-24: exploiting cancer’s Achilles’ heel. Mol Ther 11:4–18
Sauane M, Gopalkrishnan RV, Sarkar D, Su ZZ, Lebedeva IV, Dent P, Pestka S, Fisher PB (2003) MDA-7/IL-24: novel cancer growth suppressing and apoptosis inducing cytokine. Cytokine Growth Factor Rev 14:35–51
Huang EY, Madireddi MT, Gopalkrishnan RV, Leszczyniecka M, Su Z, Lebedeva IV, Kang D, Jiang H, Lin JJ, Alexandre D, Chen Y, Vozhilla N, Mei MX, Christiansen KA, Sivo F, Goldstein NI, Mhashilkar AB, Chada S, Huberman E, Pestka S, Fisher PB (2001) Genomic structure, chromosomal localization and expression profile of a novel melanoma differentiation associated (mda-7) gene with cancer specific growth suppressing and apoptosis inducing properties. Oncogene 20:7051–7063
Schaefer G, Venkataraman C, Schindler U (2001) Cutting edge: FISP (IL-4-induced secreted protein), a novel cytokine-like molecule secreted by Th2 cells. J Immunol 166:5859–5863
Soo C, Shaw WW, Freymiller E, Longaker MT, Bertolami CN, Chiu R, Tieu A, Ting K (1999) Cutaneous rat wounds express c49a, a novel gene with homology to the human melanoma differentiation associated gene, mda-7. J Cell Biochem 74:1–10
Wang M, Tan Z, Zhang R, Kotenko SV, Liang P (2002) Interleukin 24 (MDA-7/MOB-5) signals through two heterodimeric receptors, IL-22R1/IL-20R2 and IL-20R1/IL-20R2. J Biol Chem 277:7341–7347
He M, Liang P (2010) IL-24 transgenic mice: in vivo evidence of overlapping functions for IL-20, IL-22, and IL-24 in the epidermis. J Immunol 184:1793–1798
Kreis S, Philippidou D, Margue C, Behrmann I (2008) IL-24: a classic cytokine and/or a potential cure for cancer? J Cell Mol Med 12:2505–2510
Sauane M, Lebedeva IV, Su ZZ, Choo HT, Randolph A, Valerie K, Dent P, Gopalkrishnan RV, Fisher PB (2004) Melanoma differentiation associated gene-7/interleukin-24 promotes tumor cell-specific apoptosis through both secretory and nonsecretory pathways. Cancer Res 64:2988–2993
Breckenridge DG, Germain M, Mathai JP, Nguyen M, Shore GC (2003) Regulation of apoptosis by endoplasmic reticulum pathways. Oncogene 22:8608–8618
Pahl HL (1999) Signal transduction from the endoplasmic reticulum to the cell nucleus. Physiol Rev 79:683–701
Cunningham CC, Chada S, Merritt JA, Tong A, Senzer N, Zhang Y, Mhashilkar A, Parker K, Vukelja S, Richards D, Hood J, Coffee K, Nemunaitis J (2005) Clinical and local biological effects of an intratumoral injection of mda-7 (IL24; INGN 241) in patients with advanced carcinoma: a phase I study. Mol Ther 11:149–159
Tong AW, Nemunaitis J, Su D, Zhang Y, Cunningham C, Senzer N, Netto G, Rich D, Mhashilkar A, Parker K, Coffee K, Ramesh R, Ekmekcioglu S, Grimm EA, van Wart Hood J, Merritt J, Chada S (2005) Intratumoral injection of INGN 241, a nonreplicating adenovector expressing the melanoma-differentiation associated gene-7 (mda-7/IL24): biologic outcome in advanced cancer patients. Mol Ther 11:160–172
Ramesh R, Mhashilkar AM, Tanaka F, Saito Y, Branch CD, Sieger K, Mumm JB, Stewart AL, Boquoi A, Dumoutier L, Grimm EA, Renauld JC, Kotenko S, Chada S (2003) Melanoma differentiation-associated gene 7/interleukin (IL)-24 is a novel ligand that regulates angiogenesis via the IL-22 receptor. Cancer Res 63:5105–5113
Inoue S, Branch CD, Gallick GE, Chada S, Ramesh R (2005) Inhibition of Src kinase activity by Ad-mda7 suppresses vascular endothelial growth factor expression in prostate carcinoma cells. Mol Ther 12:707–715
Caudell EG, Mumm JB, Poindexter N, Ekmekcioglu S, Mhashilkar AM, Yang XH, Retter MW, Hill P, Chada S, Grimm EA (2002) The protein product of the tumor suppressor gene, melanoma differentiation-associated gene 7, exhibits immunostimulatory activity and is designated IL-24. J Immunol 168:6041–6046
Maarof G, Bouchet-Delbos L, Gary-Gouy H, Durand-Gasselin I, Krzysiek R, Dalloul A (2010) Interleukin-24 inhibits the plasma cell differentiation program in human germinal center B cells. Blood 115:1718–1726
Jones BC, Logsdon NJ, Josephson K, Cook J, Barry PA, Walter MR (2002) Crystal structure of human cytomegalovirus IL-10 bound to soluble human IL-10R1. Proc Natl Acad Sci USA 99:9404–9409
Dumoutier L, Tounsi A, Michiels T, Sommereyns C, Kotenko SV, Renauld JC (2004) Role of the interleukin (IL)-28 receptor tyrosine residues for antiviral and antiproliferative activity of IL-29/interferon-lambda 1: similarities with type I interferon signaling. J Biol Chem 279:32269–32274
Kotenko SV, Gallagher G, Baurin VV, Lewis-Antes A, Shen M, Shah NK, Langer JA, Sheikh F, Dickensheets H, Donnelly RP (2003) IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol 4:69–77
Zitzmann K, Brand S, Baehs S, Goke B, Meinecke J, Spottl G, Meyer H, Auernhammer CJ (2006) Novel interferon-lambdas induce antiproliferative effects in neuroendocrine tumor cells. Biochem Biophys Res Commun 344:1334–1341
Numasaki M, Tagawa M, Iwata F, Suzuki T, Nakamura A, Okada M, Iwakura Y, Aiba S, Yamaya M (2007) IL-28 elicits antitumor responses against murine fibrosarcoma. J Immunol 178:5086–5098
Sato A, Ohtsuki M, Hata M, Kobayashi E, Murakami T (2006) Antitumor activity of IFN-lambda in murine tumor models. J Immunol 176:7686–7694
Li M, Liu X, Zhou Y, Su SB (2009) Interferon-lambdas: the modulators of antivirus, antitumor, and immune responses. J Leukoc Biol 86:23–32
Maher SG, Sheikh F, Scarzello AJ, Romero-Weaver AL, Baker DP, Donnelly RP, Gamero AM (2008) IFNalpha and IFNlambda differ in their antiproliferative effects and duration of JAK/STAT signaling activity. Cancer Biol Ther 7:1109–1115
Meager A, Visvalingam K, Dilger P, Bryan D, Wadhwa M (2005) Biological activity of interleukins-28 and -29: comparison with type I interferons. Cytokine 31:109–118
Doyle SE, Schreckhise H, Khuu-Duong K, Henderson K, Rosler R, Storey H, Yao L, Liu H, Barahmand-pour F, Sivakumar P, Chan C, Birks C, Foster D, Clegg CH, Wietzke-Braun P, Mihm S, Klucher KM (2006) Interleukin-29 uses a type 1 interferon-like program to promote antiviral responses in human hepatocytes. Hepatology 44:896–906
Donnelly RP, Sheikh F, Kotenko SV, Dickensheets H (2004) The expanded family of class II cytokines that share the IL-10 receptor-2 (IL-10R2) chain. J Leukoc Biol 76:314–321
Sommereyns C, Paul S, Staeheli P, Michiels T (2008) IFN-lambda (IFN-lambda) is expressed in a tissue-dependent fashion and primarily acts on epithelial cells in vivo. PLoS Pathog 4:e1000017
Brand S, Beigel F, Olszak T, Zitzmann K, Eichhorst ST, Otte JM, Diebold J, Diepolder H, Adler B, Auernhammer CJ, Goke B, Dambacher J (2005) IL-28A and IL-29 mediate antiproliferative and antiviral signals in intestinal epithelial cells and murine CMV infection increases colonic IL-28A expression. Am J Physiol Gastrointest Liver Physiol 289:G960–G968
Mordstein M, Kochs G, Dumoutier L, Renauld JC, Paludan SR, Klucher K, Staeheli P (2008) Interferon-lambda contributes to innate immunity of mice against influenza A virus but not against hepatotropic viruses. PLoS Pathog 4:e1000151
Plockinger U, Rindi G, Arnold R, Eriksson B, Krenning EP, de Herder WW, Goede A, Caplin M, Oberg K, Reubi JC, Nilsson O, Delle Fave G, Ruszniewski P, Ahlman H, Wiedenmann B (2004) Guidelines for the diagnosis and treatment of neuroendocrine gastrointestinal tumours. A consensus statement on behalf of the European Neuroendocrine Tumour Society (ENETS). Neuroendocrinology 80:394–424
Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, Heinzen EL, Qiu P, Bertelsen AH, Muir AJ, Sulkowski M, McHutchison JG, Goldstein DB (2009) Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature 461:399–401
Thomas DL, Thio CL, Martin MP, Qi Y, Ge D, O’Huigin C, Kidd J, Kidd K, Khakoo SI, Alexander G, Goedert JJ, Kirk GD, Donfield SM, Rosen HR, Tobler LH, Busch MP, McHutchison JG, Goldstein DB, Carrington M (2009) Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature 461:798–801
Suppiah V, Moldovan M, Ahlenstiel G, Berg T, Weltman M, Abate ML, Bassendine M, Spengler U, Dore GJ, Powell E, Riordan S, Sheridan D, Smedile A, Fragomeli V, Muller T, Bahlo M, Stewart GJ, Booth DR, George J (2009) IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet 41:1100–1104
Tanaka Y, Nishida N, Sugiyama M, Kurosaki M, Matsuura K, Sakamoto N, Nakagawa M, Korenaga M, Hino K, Hige S, Ito Y, Mita E, Tanaka E, Mochida S, Murawaki Y, Honda M, Sakai A, Hiasa Y, Nishiguchi S, Koike A, Sakaida I, Imamura M, Ito K, Yano K, Masaki N, Sugauchi F, Izumi N, Tokunaga K, Mizokami M (2009) Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet 41:1105–1109
Pagliaccetti NE, Eduardo R, Kleinstein SH, Mu XJ, Bandi P, Robek MD (2008) Interleukin-29 functions cooperatively with interferon to induce antiviral gene expression and inhibit hepatitis C virus replication. J Biol Chem 283:30079–30089
Fiorentino DF, Bond MW, Mosmann TR (1989) Two types of mouse T helper cell. IV. Th2 clones secrete a factor that inhibits cytokine production by Th1 clones. J Exp Med 170:2081–2095
O’Garra A, Vieira P (2007) T(H)1 cells control themselves by producing interleukin-10. Nat Rev Immunol 7:425–428
Josephson K, Logsdon NJ, Walter MR (2001) Crystal structure of the IL-10/IL-10R1 complex reveals a shared receptor binding site. Immunity 15:35–46
Riley JK, Takeda K, Akira S, Schreiber RD (1999) Interleukin-10 receptor signaling through the JAK-STAT pathway. Requirement for two distinct receptor-derived signals for anti-inflammatory action. J Biol Chem 274:16513–16521
Jenkins JK, Malyak M, Arend WP (1994) The effects of interleukin-10 on interleukin-1 receptor antagonist and interleukin-1 beta production in human monocytes and neutrophils. Lymphokine Cytokine Res 13:47–54
Dickensheets HL, Freeman SL, Smith MF, Donnelly RP (1997) Interleukin-10 upregulates tumor necrosis factor receptor type-II (p75) gene expression in endotoxin-stimulated human monocytes. Blood 90:4162–4171
Marfaing-Koka A, Maravic M, Humbert M, Galanaud P, Emilie D (1996) Contrasting effects of IL-4, IL-10 and corticosteroids on RANTES production by human monocytes. Int Immunol 8:1587–1594
Berkman N, John M, Roesems G, Jose PJ, Barnes PJ, Chung KF (1995) Inhibition of macrophage inflammatory protein-1 alpha expression by IL-10. Differential sensitivities in human blood monocytes and alveolar macrophages. J Immunol 155:4412–4418
Asadullah K, Sterry W, Volk HD (2003) Interleukin-10 therapy–review of a new approach. Pharmacol Rev 55:241–269
Levy Y, Brouet JC (1994) Interleukin-10 prevents spontaneous death of germinal center B cells by induction of the bcl-2 protein. J Clin Invest 93:424–428
Cai G, Kastelein RA, Hunter CA (1999) IL-10 enhances NK cell proliferation, cytotoxicity and production of IFN-gamma when combined with IL-18. Eur J Immunol 29:2658–2665
Houssiau FA, Lefebvre C, Vanden Berghe M, Lambert M, Devogelaer JP, Renauld JC (1995) Serum interleukin-10 titers in systemic lupus erythematosus reflect disease activity. Lupus 4:393–395
Lauwerys BR, Garot N, Renauld JC, Houssiau FA (2000) Interleukin-10 blockade corrects impaired in vitro cellular immune responses of systemic lupus erythematosus patients. Arthritis Rheum 43:1976–1981
Llorente L, Richaud-Patin Y, Garcia-Padilla C, Claret E, Jakez-Ocampo J, Cardiel MH, Alcocer-Varela J, Grangeot-Keros L, Alarcon-Segovia D, Wijdenes J, Galanaud P, Emilie D (2000) Clinical and biologic effects of anti-interleukin-10 monoclonal antibody administration in systemic lupus erythematosus. Arthritis Rheum 43:1790–1800
Lubberts E, Joosten LA, Van Den Bersselaar L, Helsen MM, Bakker AC, Xing Z, Richards CD, Van Den Berg WB (2000) Intra-articular IL-10 gene transfer regulates the expression of collagen-induced arthritis (CIA) in the knee and ipsilateral paw. Clin Exp Immunol 120:375–383
Zdanov A, Schalk-Hihi C, Wlodawer A (1996) Crystal structure of human interleukin-10 at 1.6 Å resolution and a model of a complex with its soluble receptor. Protein Sci 5:1955–1962
Ealick SE, Cook WJ, Vijay-Kumar S, Carson M, Nagabhushan TL, Trotta PP, Bugg CE (1991) Three-dimensional structure of recombinant human interferon-gamma. Science 252:698–702
Randal M, Kossiakoff AA (2000) The 2.0 Å structure of bovine interferon-gamma; assessment of the structural differences between species. Acta Crystallogr D Biol Crystallogr 56:14–24
Samudzi CT, Rubin JR (1993) Structure of recombinant bovine interferon-gamma at 3.0 Å resolution. Acta Crystallogr D Biol Crystallogr 49:513–521
Samudzi CT, Burton LE, Rubin JR (1991) Crystal structure of recombinant rabbit interferon-gamma at 2.7-Å resolution. J Biol Chem 266:21791–21797
Fickenscher H, Biesinger B, Knappe A, Wittmann S, Fleckenstein B (1996) Regulation of the herpesvirus saimiri oncogene stpC, similar to that of T-cell activation genes, in growth-transformed human T lymphocytes. J Virol 70:6012–6019
Fickenscher H, Fleckenstein B (2001) Herpesvirus saimiri. Philos Trans R Soc Lond B Biol Sci 356:545–567
Knappe A, Hor S, Wittmann S, Fickenscher H (2000) Induction of a novel cellular homolog of interleukin-10, AK155, by transformation of T lymphocytes with herpesvirus saimiri. J Virol 74:3881–3887
Fickenscher H, Pirzer H (2004) Interleukin-26. Int Immunopharmacol 4:609–613
Hor S, Pirzer H, Dumoutier L, Bauer F, Wittmann S, Sticht H, Renauld JC, de Waal Malefyt R, Fickenscher H (2004) The T-cell lymphokine interleukin-26 targets epithelial cells through the interleukin-20 receptor 1 and interleukin-10 receptor 2 chains. J Biol Chem 279:33343–33351
Sheikh F, Baurin VV, Lewis-Antes A, Shah NK, Smirnov SV, Anantha S, Dickensheets H, Dumoutier L, Renauld JC, Zdanov A, Donnelly RP, Kotenko SV (2004) Cutting edge: IL-26 signals through a novel receptor complex composed of IL-20 receptor 1 and IL-10 receptor 2. J Immunol 172:2006–2010
Vandenbroeck K, Cunningham S, Goris A, Alloza I, Heggarty S, Graham C, Bell A, Rooney M (2003) Polymorphisms in the interferon-gamma/interleukin-26 gene region contribute to sex bias in susceptibility to rheumatoid arthritis. Arthritis Rheum 48:2773–2778
Dambacher J, Beigel F, Zitzmann K, De Toni EN, Goke B, Diepolder HM, Auernhammer CJ, Brand S (2009) The role of the novel Th17 cytokine IL-26 in intestinal inflammation. Gut 58:1207–1217
Haque SJ, Sharma P (2006) Interleukins and STAT signaling. Vitam Horm 74:165–206
Seidel HM, Milocco LH, Lamb P, Darnell JE Jr, Stein RB, Rosen J (1995) Spacing of palindromic half sites as a determinant of selective STAT (signal transducers and activators of transcription) DNA binding and transcriptional activity. Proc Natl Acad Sci USA 92:3041–3045
Kotenko SV, Langer JA (2004) Full house: 12 receptors for 27 cytokines. Int Immunopharmacol 4:593–608
Zhou Z, Hamming OJ, Ank N, Paludan SR, Nielsen AL, Hartmann R (2007) Type III interferon (IFN) induces a type I IFN-like response in a restricted subset of cells through signaling pathways involving both the Jak-STAT pathway and the mitogen-activated protein kinases. J Virol 81:7749–7758
Xu W, Presnell SR, Parrish-Novak J, Kindsvogel W, Jaspers S, Chen Z, Dillon SR, Gao Z, Gilbert T, Madden K, Schlutsmeyer S, Yao L, Whitmore TE, Chandrasekher Y, Grant FJ, Maurer M, Jelinek L, Storey H, Brender T, Hammond A, Topouzis S, Clegg CH, Foster DC (2001) A soluble class II cytokine receptor, IL-22RA2, is a naturally occurring IL-22 antagonist. Proc Natl Acad Sci USA 98:9511–9516
Bazan JF (1990) Structural design and molecular evolution of a cytokine receptor superfamily. Proc Natl Acad Sci USA 87:6934–6938
Dumoutier L, de Meester C, Tavernier J, Renauld JC (2009) New activation modus of STAT3: a tyrosine-less region of the interleukin-22 receptor recruits STAT3 by interacting with its coiled-coil domain. J Biol Chem 284:26377–26384
de Moura PR, Watanabe L, Bleicher L, Colau D, Dumoutier L, Lemaire MM, Renauld JC, Polikarpov I (2009) Crystal structure of a soluble decoy receptor IL-22BP bound to interleukin-22. FEBS Lett 583:1072–1077
Logsdon NJ, Jones BC, Allman JC, Izotova L, Schwartz B, Pestka S, Walter MR (2004) The IL-10R2 binding hot spot on IL-22 is located on the N-terminal helix and is dependent on N-linked glycosylation. J Mol Biol 342:503–514
Pletnev S, Magracheva E, Wlodawer A, Zdanov A (2005) A model of the ternary complex of interleukin-10 with its soluble receptors. BMC Struct Biol 5:10
Yoon SI, Logsdon NJ, Sheikh F, Donnelly RP, Walter MR (2006) Conformational changes mediate interleukin-10 receptor 2 (IL-10R2) binding to IL-10 and assembly of the signaling complex. J Biol Chem 281:35088–35096
Kotenko SV, Izotova LS, Mirochnitchenko OV, Esterova E, Dickensheets H, Donnelly RP, Pestka S (2001) Identification, cloning, and characterization of a novel soluble receptor that binds IL-22 and neutralizes its activity. J Immunol 166:7096–7103
Dumoutier L, Lejeune D, Colau D, Renauld JC (2001) Cloning and characterization of IL-22 binding protein, a natural antagonist of IL-10-related T cell-derived inducible factor/IL-22. J Immunol 166:7090–7095
Livnah O, Stura EA, Johnson DL, Middleton SA, Mulcahy LS, Wrighton NC, Dower WJ, Jolliffe LK, Wilson IA (1996) Functional mimicry of a protein hormone by a peptide agonist: the EPO receptor complex at 2.8 Å. Science 273:464–471
Kotenko SV, Izotova LS, Mirochnitchenko OV, Esterova E, Dickensheets H, Donnelly RP, Pestka S (2001) Identification of the functional interleukin-22 (IL-22) receptor complex: the IL-10R2 chain (IL-10Rbeta) is a common chain of both the IL-10 and IL-22 (IL-10-related T cell-derived inducible factor, IL-TIF) receptor complexes. J Biol Chem 276:2725–2732
Wu PW, Li J, Kodangattil SR, Luxenberg DP, Bennett F, Martino M, Collins M, Dunussi-Joannopoulos K, Gill DS, Wolfman NM, Fouser LA (2008) IL-22R, IL-10R2, and IL-22BP binding sites are topologically juxtaposed on adjacent and overlapping surfaces of IL-22. J Mol Biol 382:1168–1183
LaPorte SL, Juo ZS, Vaclavikova J, Colf LA, Qi X, Heller NM, Keegan AD, Garcia KC (2008) Molecular and structural basis of cytokine receptor pleiotropy in the interleukin-4/13 system. Cell 132:259–272
Wolk K, Witte E, Reineke U, Witte K, Friedrich M, Sterry W, Asadullah K, Volk HD, Sabat R (2005) Is there an interaction between interleukin-10 and interleukin-22? Genes Immun 6:8–18
Preimel D, Sticht H (2004) Molecular modeling of the interleukin-19 receptor complex. Novel aspects of receptor recognition in the interleukin-10 cytokine family. J Mol Model 10:290–296
Walter MR (2004) Structural analysis of IL-10 and Type I interferon family members and their complexes with receptor. Adv Protein Chem 68:171–223
Thiel DJ, le Du MH, Walter RL, D’Arcy A, Chene C, Fountoulakis M, Garotta G, Winkler FK, Ealick SE (2000) Observation of an unexpected third receptor molecule in the crystal structure of human interferon-gamma receptor complex. Structure 8:927–936
Parrish-Novak J, Foster DC, Holly RD, Clegg CH (2002) Interleukin-21 and the IL-21 receptor: novel effectors of NK and T cell responses. J Leukoc Biol 72:856–863
Xie MH, Aggarwal S, Ho WH, Foster J, Zhang Z, Stinson J, Wood WI, Goddard AD, Gurney AL (2000) Interleukin (IL)-22, a novel human cytokine that signals through the interferon receptor-related proteins CRF2–4 and IL-22R. J Biol Chem 275:31335–31339
Ozaki K, Spolski R, Feng CG, Qi CF, Cheng J, Sher A, Morse HC 3rd, Liu C, Schwartzberg PL, Leonard WJ (2002) A critical role for IL-21 in regulating immunoglobulin production. Science 298:1630–1634
Doyle WJ, Gentile DA, Cohen S (2006) Emotional style, nasal cytokines, and illness expression after experimental rhinovirus exposure. Brain Behav Immun 20:175–181
Marcello T, Grakoui A, Barba-Spaeth G, Machlin ES, Kotenko SV, MacDonald MR, Rice CM (2006) Interferons alpha and lambda inhibit hepatitis C virus replication with distinct signal transduction and gene regulation kinetics. Gastroenterology 131:1887–1898
Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ (2009) Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) ClustalW and ClustalX version 2.0. Bioinformatics 23:2947–2948
DeLano WL (2002) The PyMOL molecular graphics system. DeLano Scientific, San Carlos
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Trivella, D.B.B., Ferreira-Júnior, J.R., Dumoutier, L. et al. Structure and function of interleukin-22 and other members of the interleukin-10 family. Cell. Mol. Life Sci. 67, 2909–2935 (2010). https://doi.org/10.1007/s00018-010-0380-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-010-0380-0