Abstract
Soluble starch synthase (SSS) is a key enzyme in the biosynthesis of plant amylopectin. To reveal the starch synthesis pathway in lotus, we cloned a gene named NnSSSIII from Nelumbo nucifera in this study. A full-length cDNA of soluble starch synthase (SSS), designated as NnSSSIII, which was 4070 bp (GenBank accession NO. KR062069) containing a 3696 bp open reading frame and encoding a protein of 1231 amino acid. Twenty SNPs of NnSSSIII were found from three breeds of lotus, which resulted in 13 coding amino acid changes. The complete genomic structure of NnSSSIII comprises 17 exons and 16 introns and covers a successive region of 70.6 kb. Expression pattern and enzyme activity assay showed that NnSSSIII gene is mostly expressed in leaves and rhizomes at the time between 8th to 14th weeks after germination. Our results indicated that NnSSSIII may paly an important role in transient starch synthesis of photosynthetic tissue and storage starch synthase of storage tissue at the total developmental stages. In addition, characterization of a genetic polymorphism of NnSSSIII cDNA contributed more molecular markers about the SSSIII gene in lotus. This study elucidates more molecular information about the NnSSSIII gene and provided guidance for future improvement of starch quality and content in lotus.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
As a major storage carbohydrate molecule in plants, starch is an important decisive factor contributing to crop yield and quality which plays a vital role in food industry. In higher plant, at least six classes of enzymes are reported to be involved in starch biosynthesis: ADP glucose pyrophosphorylase (AGPase), granule-bound starch synthase (GBSS), soluble starch synthase (SSS), starch branching enzyme (SBE), starch debranching enzyme (DBE) and starch phosphorylase (SP) (Fujita et al. 2008). In the process of starch biosynthesis, catalyze substrate production, chain elongation and branching modification are executed by those enzymes, respectively. Among them, starch synthase catalyzes the transfer of a glucosyl moiety from ADP-glucose to the non-reducing end of elongating glucan chains.
Starch synthase catalyzing the transfer of the glucosyl unit of ADP-Glc to the nonreducing end of a glucan chain for chain elongation viaa-1,4-glycosidic linkages. There are two types of starch synthases in higher plants, GBSS and SSS (Hirose and Terao 2004). It has been assumed that GBSS functions specifically to elongate amylose, while SSS contributes to amylopectin synthesis. Nowadays, many studies focused on SSS, and it was isolated and characterized from many plants and algae now (Baba et al. 1993; Dian et al. 2005; Marshall et al. 1996). Multiple isoforms of SSS were found in plant, and different SSS isoforms play different roles in determination of amylopectin properties, such as length of branched chain. Currently, SSSI is reported to be responsible for the shortest glucan chains which are DP (DP: degree of polymerization) 6–12(Fujita et al. 2008). SSSII was responsible for elongating the longer chains (DP 13–26) while SSSIII was considered to produce the length of the medium amylopectin chains (DP > 30) mainly. The function of SSSIV mainly devoted to the synthesis of transient starch, which is related to controlling the number of starch grains in chloroplasts (Leterrier et al. 2008). Some further studies showed that the proportion of amylopectin with very long chains when loss of SSSIII expression reduce, and the amylopectin/amylase ratio would be affected (Zhang et al. 2005; Zhu et al. 2014).
Lotus (Nelumbo nucifera) is a perennial aquatic vegetable with important economic value. In China, lotus rhizome and seed could be directly eat or used to produce many food (Guo 2009). The edible rhizomes and seeds of lotus contain a large amount of starch. Lotus starch properties strongly influence the processing quality and consumption of lotus food. The taste quality of lotus rhizomes is very important for distinguishing various genotypes and cultivars of rhizome lotus, which is affected by the ratio of amylopectin and amylase (Li et al. 2006). Therefore, studies on lotus starch-related genes are important for lotus genetic research and breeding. Current researches about lotus starch are mainly concentrated on starch granules, physicochemical properties (Cai et al. 2014; Geng et al. 2007; Lin et al. 2006). At the genetic level, the Wx (encoded GBSSI) gene and AGPase gene were isolated and characterized (Cheng et al. 2014; Lu et al. 2012). Recently, transcriptomic analysis of regulation of rhizome formation in lotus found NnSSSIII expressed abundantly (Yang et al. 2015).
In this study, we isolated NnSSSIII from lotus by RACE technique, and then the gene structure and protein structure has been identified, some SNP sites were discovered. Besides, the expression pattern and enzyme activity of NnSSSIII were also analyzed in various organs during different developmental stages. These results would help us to understand its biological functions in lotus and provide essential information for genetic engineering efforts to improve starch content and quality of lotus rhizomes as well.
Material and methods
Plant material and treatments
Nelumbo nucifera. cv. Xianglian, as one of the most widely planted variety in China, was selected in this study. Sprouted seeds were planted in the pots after 3 days of germination and grown in the greenhouse of Wuhan University. Each seedling was grown in individual pots under uniform conditions: 75% humidity, 20–26 °C, 16 h of light and 8 h of dark. Based on the growth time and size of rhizomes, those plants can be divided into three developmental stages: initial stage of rhizome development, the rhizome is elongating; early rhizome development stage, formation of the first internode; middle stage of rhizome development, the swelling of the first internode is apparent; later stage of rhizome-development, the first internode of rhizome is fully expanded. The leaves, petioles, rhizomes and roots were collected at 6:00 p.m. on 6th week, 8th week, 10th week, 12th week and 14th week after germination (Fig. 1), and quick-freezing in liquid nitrogen, stored at −80 °C for next manipulation. Figure 1 showed their growth status, and histological observation of starch accumulation in leaves and rhizomes.
Lotus materials and histological observation in this study. a. Lotus harvested at five different time point are shown above: above: 6th, 8th, 10th, 12th, 14th weeks after germination. b. Histological observation of rhizomes (dyed by 2%KI-0.2%I2) under optics microscope with the magnification of 200×, the starch granule indicated by blue line and descriptive text. c. Histological observation of both leaves (dyed by 2%KI-0.2%I2) under optics microscope with the magnification of 200×, top right corner displayed starch granule with a larger version, the starch granule indicated by blue line and descriptive text
RNA isolation and cDNA synthesis
Total RNA was isolated from leaves, petioles, rhizomes and roots by Plant RNA Extraction kit (TIANGEN, China) and treated with DNase I to remove gDNA (genomic DNA) contamination. RNA quality is examined by 1% agarose gel and bio-photometer (Eppendorf, Germany). For synthesis of the first-strand cDNA, about 1μg RNA was denatured for 5 min at 70 °C with 2uL Oligo (dT) (Promega, America), ice-bathed for 2 min, and then incubated at 42 °C for 1 h by M-MLV Reverse Transcriptase (Promega, America) following the manufacturer’s protocol. The products were stored at −20 °C for later use.
Molecular cloning of NnSSS III
To obtain the full-length cDNA of NnSSSIII, RACE technique was used in this study. Initial fragment of NnSSSIII cDNA was obtained using the primer set based on transcriptome analysis and the conserved regions of SSSIII gene among different plants. SMARTTM RACE cDNA Amplification Kit (Takara, Japan) and Gene Racer TM Kit (Invitrogen, China) were used for 5’-RACE and 3’-RACE respectively. Nested PCR, amplicon purification, sequencing analysis were used to amplification and purify the 5′ ends and 3′-end of the cDNA. Sequence fragments were integrated by DNASTAR to achieve the full-length cDNA. The full-length cDNA was inserted into pGEM-T Vector (Promega), then transferred to E. coli cells (DH5α) for sequence analysis. All the primers were displayed in Table 1.
Characterization of a genetic polymorphism of NnSSSIII cDNA
Genetic polymorphisms of NnSSSIII cDNA from two other breeds of lotus were also analyzed using the gene-specific primers based on the cDNA sequence of Xianglian’s (XSSSIII). The NnSSSIII gene were amplified from cDNAs of cultivated TaiKong (TSSSIII) and wild lotus (WSSSIII). PCR reactions were conducted in 50uL volumes which containing 2uL of total cDNA, 5uL of 10 × PCR buffer, 5uL of 2 mM dNTPs mixture, 3uL of 25 mM MgSO4, 1.6uL of 10pM of each primers, 1uL of 1 U/μL KOD DNA polymerase (TOYOBO, Japan) and 30.8 μL ddH2O with a total volume of 50 μL. Amplification conditions followed the two-step amplification procedure: 94 °C for 2 min, 36 cycles of 98 °C for 10s, (Tm)°C for 30s, and 68 °C for 1 min. The sizes of the PCR products were assessed by electrophoresis and cloned as described above, then sequenced by Sanger sequencing (Augct, China). Aligned sequences by clustalx to find molecular markers.
Bioinformatics analysis
The open-reading frame (ORF) of the NnSSSIII was predicted using ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). The basic parameters of the SSSIII gene was analyzed by NCBI gene (https://www.ncbi.nlm.nih.gov/gene/). The conserved domains were predicted by Pfam sever (http://pfam.xfam.org/search/sequence). All the sequences were aligned using Cluxal-X. Subsequently the phylogenetic NJ (neighbor-joining) tree generated by MEGA6.0v.
Real-time PCR analysis
The experimental design fallowed MIQE (minimum information for publication of quantitative real-time PCR experiments) in this experiment (Bustin et al. 2009). A house-keeping gene, β-actin (GenBank accession no.EU131153) gene was selected as the reference gene in this experiment. The primers of β-actin and SSSIII were designed based on the sequence of CDS by Primer Premier 5.0 and checked by its single melt curve, amplification efficiency and so on (displayed in Table 2). For qRT-PCR (real-time PCR) experiment, the two-step amplification procedure was used: 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. The total PCR reaction mixture contains 2 μL cDNA (1:10 diluted), 0.2 μL forward primer (20 M), 0.2 μL reverse primer (20 M), 10 μL 2 × SYBR mix (TOYOBO, Japan) and 7.6 μL ddH2O with a total volume of 20 μL. The relative gene expression data was calculated using 2-ΔΔCt methods with the guidance of the StepOne software v2.1 (ABI, US).
Enzyme activity assay
SSs utilize ADPG to elongate linear chains by the formation of a-1,4 linkages. Referred to Cheng’s article, the protocol for measuring SSS activity was used to assay the SSS activity of lotus in this study (Cheng et al. 2001). Firstly, total enzyme isolation: about 0.2 g powder of plant material was added into 1 mL HEPES–NaOH (PH7.5) buffer. After proper shaking, the tube was centrifuged at 10000 g for 5 min, and supernatant was extracted to check the enzyme activity of SSS. Next, according to the character of SSS, three steps reaction reacted in this stage.
36uL reaction mixture 1(50 mM HEPES–NaOH (pH 7.4), 1.6 mM ADPG, 0.7 mg amylopectin, 15 mM DTT) was added to the supernatant (20uL), then incubate the total reaction mix at 30 °C for 20 min, bathed in boiling water for 30s, cooling in ice;
20uL reaction mix2 (50 mM HEPES-NaOH (pH 7.4), 4 mM PEP, 200 mM KCl, l0 mM MgCl2, 1.2unit PK) was added follow the same manipulations.
43uL reaction mixture 3 (50 mM HEPES–NaOH (pH 7.4), 10 mM glucose, 20 mM MgC12, 2 mM NADP, 1.4unit HK, 0.35unit G-6-PD) was added to the supernatant after centrifuged at 10000 g for 10 min. Finally, the absorption peak of generated NADPH2 at 340OD was tested by tecan sunrise, and its value was used to estimate the enzyme activity of SSS (Doehlert et al. 1988).
Result
Sequence characterization NnSSSIII
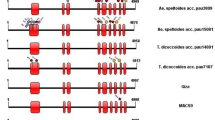
The full-length cDNA sequence is 4070 bp (GenBank accession no. KR062069), containing 79 bp of 5’ UTR (Untranslated Region), the 295 bp of 3’ UTR and an ORF (open reading frame) of 3696 bp which encodes 1231 amino acid residues. The complete genomic structure of NnSSSIII consists of 17 exons and 16 introns which covers a successive region of 70.6 kb. The gene structures are shown in Fig. 2a. The predicted protein was analyzed, three conserved domains were identified, and displayed in Fig. 2b. The most highly conserved amino acid regions present in all other plant species were also present in the SSSIII sequence and showed in Fig. 3c. The sequence alignment that the sequence identity with other plant was higher in both gene and protein levels in NCBI database. In gene level, NnSSSIII showed 83% sequence similarity with Vitis vinifera, and show 76%–83% identity with Most eudicots, under 74% identity with most monocots, respectively. In protein level, NnSSSIII shows 81% identity with Populus trichocarpa which is the highest, 62%–81% identity with most eudicots, is below 67% similarity compared with most monocots.
The gene structure of NnSSSIII (a), functional domains (b) and deduced amino acid sequenceof SSSIIIs (c) and its. a. The green boxes represent exons, thick line represents intros. b. Functional domains of were marked with different colors: CBM53 (green), GT5 (red), GT1 (blue). c. DNAman multiple sequence alignment of the deduced amino acid sequence of NnSSSIII in present study with SSSIIIs protein from other high plants. The functional domains of SSSIIIs were boxed with names on the bottom of sequence
Characterization of genetic diversity in the NnSSSIII gene
Information collecting on genetic diversity within species is particularly important in the efficient use of plant genetic resource. In this study, the cDNA sequences of NnSSSIII were used to determine the genetic diversity among three breeds of lotus. Compared the cDNA nucleotide sequences of the three NnSSSIII genes that showed pretty homology generally. The identities were 99.90% between XSSSIII and TSSSIII, 99.65% between XSSSIII and WSSSIII, and 99.55% between TSSSIII and WSSSIII. Those three NnSSSIII sequences provided a matrix of 4070 bp, which is composed of 3696 bp ORF and 295-bp UTR region. Twenty polymorphic sites were revealed in NnSSSIII gene and demonstrated all the polymorphisms were SNPs. Among of them, twenty SNPs located in the CDS and another one was in the 3’-UTR (Table 3). Thirteen SNPs resulted in nonsynonymous mutation and changed to the amino acid sequence, when other six SNPs were synonymous mutation. Ratio of nonsynonymous/synonymous = 2.17. Selection analysis revealed that Ka/Ks (0.0045/0.0073) <1, which confirmed NnSSSIII gene subjected by purify selection.
Phylogenetic analysis of SSS
As one of a crucial gene in plant starch synthesis pathway, the SSS gene have been widely reported in both dicotyledons and monocotyledons. A NJ tree was constructed by some SSS proteins sequences of plant and their isoforms (Fig. 3). To explore the classification of lotus SSS protein and to elucidate phylogenetic relationship, a NJ tree was constructed using SSS proteins sequences from various monocots and dicots. This NJ phylogenetic tree divided plant SSS proteins into four distinct subgroups: SSSI, SSSII, SSSIII and SSSIV subgroups. As expected, the NnSSSIII protein was classified into the SSSIII subgroup which consist of SSSIII proteins, providing that it is indeed a SSSIII protein of starch biosynthesis pathway in lotus. NnSSSIII and HbSSSIII are closely clustered together, forming a side branch in association with other SSSIII sequences from dicots. The monocot SSSs were clustered into unique subgroups that are distant from dicot SSSs, indicating that divergence event of SSS in dicots predates its divergence from the monocots. Besides, evolutionary distances of SSSI and SSSIII were far away from SSSII and SSSIV, suggesting that SSSI and SSSIII are two ancestral forms of SSS and the origination of SSSII and SSSIV might begin with whole genome duplication.
Expression patterns of SSSIII
The relative expression of NnSSSIII gene was analyzed by qRT-PCR from four tissues (leaves, petioles, rhizomes and roots) of lotus collected after 6th week to 14th week of growth. Distinct special and temporal expression patterns were observed via the result of qRT-PCR (Fig. 4). NnSSSIII was most expressed in leaves, followed by rhizomes, petioles and roots. The highest relative expression in leaves was observed in 8th week lotus, with a value nearly six-fold higher that of rhizomes. However, the expression discrepancy between the leaf and rhizome was minimized (<2 fold) in 14th weeks old lotus. The highest expression of NnSSSIII in a single tissue was observed at intervals between 8th and 10th weeks, and its abundance is decreased in later stages.
Enzyme activity of SSS
As showed in Fig. 5, we also assayed the enzyme activity of SSS in four tissues. It is similar with the expression pattern of NnSSSIII, SSS maintains a high enzyme activity in leaves and a relative low activity in roots throughout all developmental stages. SSS activity reaches the peak in 8th weeks lotus leaves. However, the second highest enzyme activity of SSS was observed in petioles rather than in rhizomes. This is different from the result of qRT-PCR where NnSSSIII has a significant higher expression in rhizomes than in petioles.
Discussion
Starch is a major storage compound which used as a primary storage for metabolism and biosynthesis. As a crucial enzyme in starch biosynthesis, a large number of researches about starch synthase genes have been reported in various kinds of plant species (Ball and Morell 2003). In pea embryo, SSSII is the main soluble starch synthase, related to 60% enzyme activity (Denyer and Smith 1992). In potato, SSSIII was accounts for approximately 80% of the activity in the soluble fraction of the tuber (Marshall et al. 1996). Transcriptomic analysis about rhizome in lotus found that the expression level of NnSSSIII is higher (Yang et al. 2015), this indicates that it might responsible for most synthase activity. The NnSSSIII has isolated and characterized from cDNA libraries by RACE technology, and the length of this gene is over 4000 bp and codes for a large protein of 1231 amino acids.
Over the last decades, many investigations devoted to exploring genomic variation among different germplasms. Twenty SNPs were found according to the NnSSSIII cDNA sequence of three accessions of seed lotus. The interspecific relationships among the three accessions of lotus revealed that Xiang lotus and Taikong lotus show a close relationship, and the wild lotus show higher genomic diversity. In actual production, Xiang lotus and Taikong lotus are cultivated by traditional breeding, and the grain weight of these three accessions was Taikong lotus>Xiang lotus>wild lotus. High nucleotide variation is great significance to adapt to the changeable environment and increase yield of lotus. Ratio of nonsynonymous/synonymous was 2.17 which is higher compared with the whole genomic sequences (1.81) (Huang et al. 2018). Ka/Ks analysis confirmed NnSSSIII subjected by purify selection in natural selection. Although nonsynonymous mutations are affected by natural selection, more nonsynonymous mutations reserved in cultivated seed lotus. It might be the result of artificial selection, further analysis of the trait of starch may reveal some correlation of evolution in the process of seed lotus breeding. It is an important application in exploring the evolution of artificial selection of species. In addition, similar genetic variations in lotus have been well studied. Two SSR molecular markers were identified in AGPL and GBSSI which involved in total starch content, amylose content and amylopectin content (references not shown). Those twenty SNPs would help to define the genetic relationships between the cultivated species, supplied useful information for the study of genetic differentiation, provided molecular markers for the breeding of qualified characteristics (Hu et al. 2015).
The full-length cDNA sequence of NnSSSIII was characterized, which accord with all the main characteristic amino acid residues, motifs, and conserved domains of starch synthase protein family. NnSSSIII showed high homology (60% -70%) with other SSSIII. The phylogenetic analysis indicated NnSSSIII grouped together with plant SSSIII which derived from a common ancestor and preceded the monocot/dicot split. In coding region, there are highly similar sequences. Conserved domains of NnSSSIII protein analysis indicated that the protein contained three conserved regions which is consistent with other plants (Gao et al. 1998; Green et al. 1986). Although SNP analysis showed that NnSSSIII has a high mutation rate and most of SNPs are missense mutations, few mutations occured in functional domain, which indicating the functional region is conservative. In addition, there are no putative transit peptide in the N-terminal of NnSSSIII, but it predicted in many SSSIIIs which have been isolated and identified (Li et al. 2000). The presence of transit peptide may reflect its expression patterns and specific sites of expressed product.
Many studies have shown that the expression of starch synthase related genes fluctuates greatly in different plant tissues (Boraston Alisdair et al. 2004; Cheng et al. 2014; Tenorio et al. 2003). Therefore, the materials of different tissues were harvested in different times and studied the expression patterns of NnSSSIII in the rapid accumulation of starch during rhizomes expanding. The results of qRT-PCR shown that NnSSSIII gene were expressed around tissues. Early in starch accumulation, NnSSSIII was found to higher level expression in leaf, a photosynthetic organ with most chloroplast, compared to the storage tissue (rhizome and root). The SSS were abundantly expressed in the earlier phase of starch accumulate, such an expression pattern was observed in rice, wheat, and Arabidopsis (Busi et al. 2008; Dian et al. 2005; Li et al. 2000; Valdez et al. 2008). As one of the important enzymes in starch synthesis, SSSIII acquired ADP-Glc more convenient when expressed in photosynthetic tissues, which is consistent with NnAGP genes in a high degree, and accelerated amylopectin synthesis to meet the demand of carbon fixation. Lately, the rhizome was abundantly stored starch, and gene expressed in high levels (Fig. 4), the expression discrepancy between the leaf and rhizome was minimized (<2 fold). This similar expression pattern in leaves and rhizome was also observed in NnGBSS, another key enzyme in amylose synthesis (Lu et al. 2012). This spatial and temporal variation from top to bottom may be related to the process of transient starch and storage starch which adapt to different requirements of starch synthesis, to achieve higher efficiency of starch accumulation.
Lotus (Nelumbo nucifera) has been used as an important aquatic economic vegetable for thousands of years in Asia. Its seeds and rhizomes are very popular in the daily diet because its richness in starch. NnSSSIII is considered as one of a key gene controlling starch biosynthesis. Various isoforms of SS play different roles in the synthesis of amylopectin and have specific functions (James et al. 2003). In this study, we cloned the NnSSSIII gene and described its expression patterns, analysis in protein level, and allows for informative studies for more detailed examination in starch synthesis of plants. Also, it is helpful to analyze the function of different isoforms and the regulation mechanism of endosperm starch synthesis to improve taste quality and increase the starch production.
Abbreviations
- SSS:
-
Soluble starch synthase
- AGPase:
-
ADP glucose pyrophosphorylase
- GBSS:
-
Granule-bound starch synthase
- SBE:
-
Starch branching enzyme
- DBE:
-
Starch debranching enzyme
- SP:
-
Starch phosphorylase
- DP:
-
Degree of polymerization
- SNP:
-
Single nucleotide polymorphism
- RACE:
-
Rapid amplification of cDNA ends
- ORF:
-
Open reading frame
- NJ tree:
-
Neighbor-joining tree
- qRT-PCR:
-
Real-time polymerase chain reaction
- MIQE:
-
Minimum information for publication of quantitative real-time PCR experiments
References
Baba T et al (1993) Identification, cDNA cloning, and gene expression of soluble starch synthase in Rice (Oryza sativa L.) immature seeds. Plant Physiol 103:565
Ball SG, Morell MK (2003) From bacterial glycogen to starch: understanding the biogenesis of the plant starch granule. Annu Rev Plant Biol 54:207–233. https://doi.org/10.1146/annurev.arplant.54.031902.134927
Boraston Alisdair B, Bolam David N, Gilbert Harry J, Davies Gideon J (2004) Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem J 382:769
Busi MV et al (2008) Functional and structural characterization of the catalytic domain of the starch synthase III from Arabidopsis thaliana. Proteins: Struct Funct Bioinf 70:31–40. https://doi.org/10.1002/prot.21469
Bustin SA et al (2009) The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55:611–622. https://doi.org/10.1373/clinchem.2008.112797
Cai C, Cai J, Man J, Yang Y, Wang Z, Wei C (2014) Allomorph distribution and granule structure of lotus rhizome C-type starch during gelatinization. Food Chem 142:408–415. https://doi.org/10.1016/j.foodchem.2013.07.091
Cheng F, Jiang D, Wu P, Shi C (2001) The dynamic change of starch synthesis enzymes during the grain filling stage and effects of temperature upon it. Zuo Wu Xue Bao 27:201–206
Cheng N et al (2014) Cloning and characterization of the genes encoding the small and large subunit of the ADP-glucose pyrophosphorylase in lotus (Nelumbo nucifera Gaertn). Acta Physiol Plant 37:1734. https://doi.org/10.1007/s11738-014-1734-2
Denyer K, Smith AM (1992) The purification and characterisation of the two forms of soluble starch synthase from developing pea embryos. Planta 186:609–617. https://doi.org/10.1007/BF00198043
Dian W, Jiang H, Wu P (2005) Evolution and expression analysis of starch synthase III and IV in rice. J Exp Bot 56:623–632. https://doi.org/10.1093/jxb/eri065
Doehlert DC, Kuo TM, Felker FC (1988) Enzymes of sucrose and hexose metabolism in developing kernels of two Inbreds of maize. Plant Physiol 86:1013
Fujita N, Goto S, Yoshida M, Suzuki E, Nakamura Y (2008) The Function of Rice Starch Synthase I Expressed in Escherichia coli. 55. https://doi.org/10.5458/jag.55.167
Gao M, Wanat J, Stinard PS, James MG, Myers AM (1998) Characterization of <em>dull1</em>, a Maize Gene Coding for a Novel Starch Synthase. Plant Cell 10:399
Geng Z, Zongdao C, Yimin W (2007) Physicochemical properties of lotus (Nelumbo nucifera Gaertn.) and kudzu (Pueraria hirsute Matsum.) starches. Int J Food Sci Technol 42:1449–1455. https://doi.org/10.1111/j.1365-2621.2006.01363.x
Green S, Kumar V, Krust A, Walter P, Chambon P (1986) Structural and functional domains of the estrogen receptor. Cold Spring Harb Symp Quant Biol 51:751–758. https://doi.org/10.1101/sqb.1986.051.01.088
Guo HB (2009) Cultivation of lotus (Nelumbo nucifera Gaertn. Ssp. nucifera) and its utilization in China. Genet Resour Crop Evol 56:323–330. https://doi.org/10.1007/s10722-008-9366-2
Hirose T, Terao T (2004) A comprehensive expression analysis of the starch synthase gene family in rice (Oryza sativa L.). Planta 220:9–16. https://doi.org/10.1007/s00425-004-1314-6
Hu J, Gui S, Zhu Z, Wang X, Ke W, Ding Y (2015) Genome-wide identification of SSR and SNP markers based on whole-genome re-sequencing of a Thailand wild sacred Lotus (Nelumbo nucifera). PLoS One 10:e0143765. https://doi.org/10.1371/journal.pone.0143765
Huang L, Yang M, Li L, Li H, Yang D, Shi T, Yang P (2018) Whole genome re-sequencing reveals evolutionary patterns of sacred lotus (Nelumbo nucifera). J Integr Plant Biol 60:2–15. https://doi.org/10.1111/jipb.12606
James MG, Denyer K, Myers AM (2003) Starch synthesis in the cereal endosperm. Curr Opin Plant Biol 6:215–222. https://doi.org/10.1016/S1369-5266(03)00042-6
Leterrier M, Holappa LD, Broglie KE, Beckles DM (2008) Cloning, characterisation and comparative analysis of a starch synthase IV gene in wheat: functional and evolutionary implications. BMC Plant Biol 8:98. https://doi.org/10.1186/1471-2229-8-98
Li Z et al (2000) The structure and expression of the wheat starch synthase III gene. Motifs in the Expressed Gene Define the Lineage of the Starch Synthase III Gene Family. Plant Physiol 123:613
Li L, Zhang X, Shen X, Lei S, Xie K, Gu L, Cao P (2006) Studies on starch RVA profile and starch granule shape in rhizome of Nelumbo nucifera Gaertn. Acta Hort Sin 33:534–538
Lin H-M, Chang Y-H, Lin J-H, Jane J-L, Sheu M-J, Lu T-J (2006) Heterogeneity of lotus rhizome starch granules as revealed by α-amylase degradation. Carbohydr Polym 66:528–536. https://doi.org/10.1016/j.carbpol.2006.04.024
Lu Y, Li L, Zhou Y, Gao Q, Liang G, Chen X, Qi X (2012) Cloning and characterization of the Wx gene encoding a granule-bound starch synthase in Lotus (Nelumbo nucifera Gaertn). Plant Mol Biol Report 30:1210–1217. https://doi.org/10.1007/s11105-012-0430-x
Marshall J, Sidebottom C, Debet M, Martin C, Smith AM, Edwards A (1996) Identification of the major starch synthase in the soluble fraction of potato tubers. Plant Cell 8:1121
Tenorio G, Orea A, Romero JM, Mérida Á (2003) Oscillation of mRNA level and activity of granule-bound starch synthase I in Arabidopsis leaves during the day/night cycle. Plant Mol Biol 51:949–958. https://doi.org/10.1023/A:1023053420632
Valdez HA, Busi MV, Wayllace NZ, Parisi G, Ugalde RA, Gomez-Casati DF (2008) Role of the N-terminal starch-binding domains in the kinetic properties of starch synthase III from Arabidopsis thaliana. Biochemistry 47:3026–3032. https://doi.org/10.1021/bi702418h
Yang M, Zhu L, Pan C, Xu L, Liu Y, Ke W, Yang P (2015) Transcriptomic Analysis of the Regulation of Rhizome Formation in Temperate and Tropical Lotus (Nelumbo nucifera). Sci Rep 5:13059. https://doi.org/10.1038/srep13059
Zhang X, Myers AM, James MG (2005) Mutations affecting starch synthase III in Arabidopsis alter leaf starch structure and increase the rate of starch synthesis. Plant Physiol 138:663–674. https://doi.org/10.1104/pp.105.060319
Zhu F, Bertoft E, Seetharaman K (2014) Distribution of branches in whole starches from maize mutants deficient in starch synthase III. J Agric Food Chem 62:4577–4583. https://doi.org/10.1021/jf500697g
Acknowledgments
This work was supported by the Technology Innovation Project of Hubei Province of China (No.2019ABA108) and National Science and Technology Supporting Program (No. 2012BAD27B01). Fenglin Zhu, Zhongli Hu, Neng Cheng, Ying Diao, all contributed about this research. Neng Cheng designed and performed the experiments, Fenglin Zhu analyzed the results, drew the figureures and wrote the manuscript. Ying Diao and Zhongli Hu provided some scientific advices, correction, analyze the results and approved the final manuscript. All authors have read and approved the manuscript in its final form.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zhu, F., Cheng, N., Sun, H. et al. Molecular cloning and characterization of a gene encoding soluble starch synthase III (SSSIII) in Lotus (Nelumbo nucifera). Biologia 75, 279–288 (2020). https://doi.org/10.2478/s11756-019-00341-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.2478/s11756-019-00341-9