Abstract
Background
Signal regulatory protein alpha (SIRPα), expressed in the macrophage membrane, inhibits phagocytosis of tumor cells via CD47/SIRPα interaction, which acts as an immune checkpoint factor in cancers. This study aimed to clarify the clinical significance of SIRPα expression in hepatocellular carcinoma (HCC).
Methods
This study analyzed SIRPα expression using RNA sequencing data of 372 HCC tissues from The Cancer Genome Atlas (TCGA) and immunohistochemical staining of our 189 HCC patient cohort. The correlation between SIRPα expression and clinicopathologic factors, patient survival, and intratumor infiltration of immune cells was investigated.
Results
Overall survival (OS) was significantly poorer with high SIRPα expression than with low expression in both TCGA and our cohort. High SIRPα expression correlated with lower recurrence-free survival (RFS) in our cohort. High SIRPα expression was associated with higher rates of microvascular invasion and lower serum albumin levels and correlated with greater intratumor infiltration of CD68-positive macrophages and myeloid-derived suppressor cells (MDSCs). Multivariate analysis showed that SIRPα expression and high infiltration of CD8-positive T cells and MDSCs were predictive factors for both RFS and OS. Patients with high SIRPα expression and infiltration of CD8-positive T cells and MDSCs had significantly lower RFS and OS rates. In spatial transcriptomics sequencing, SIRPα expression was significantly correlated with CD163 expression.
Conclusions
High SIRPα expression in HCC indicates poor prognosis, possibly by inhibiting macrophage phagocytosis of tumor cells, promoting MDSC infiltration and inducing antitumor immunity. Treatment alternatives using SIRPα blockage should be considered in HCC as inhibiting macrophage antitumor immunity and MDSCs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Primary liver cancer is the fourth most common type of cancer in the world. Hepatocellular carcinoma (HCC) is caused by liver cirrhosis, hepatitis B/C virus infection, and alcoholic or nonalcoholic steatohepatitis. Although liver resection is an effective and safe treatment for patients with HCC, the possibility of recurrence after the procedure remains high.1,2
Cancer immunotherapy, such as immune checkpoint inhibition, has emerged as an essential therapeutic option for various cancer types. Tumors have a variety of immune escape mechanisms,3,4,5 and the activation of T cells in HCC is insufficient. Therefore, combination therapy targeting both innate and adaptive responses is essential and has shown remarkable clinical efficacy in HCC.6
Signal regulatory protein alpha (SIRPα) is a transmembrane protein, activating Src-homology-2 domain-containing protein tyrosine phosphatase 1/2 (SHP-1/2) through its cytoplasmic region via ligand interaction (CD47).7,8 Signal regulatory protein alpha is especially abundant in hematopoietic cells such as macrophages, neutrophils, and dendritic cells.9,10 In contrast, CD47 is ubiquitously expressed on the surface of various cells, including HCC cells.11,12 The interaction between SIRPα and CD47 serves as a suppressive signal for phagocytosis. Therefore, this signaling is termed the “don't-eat-me” or antiphagocytic signal.13,14
Tumor cells evade antitumor immunity through immune checkpoint signaling. Therefore, therapies targeting this pathway have been considered as immunologic approaches to cancer treatment. Indeed, preclinical research showed that blocking the SIRPα-CD47 pathway promoted macrophage-mediated phagocytosis in vivo and exhibited robust synergistic antitumor efficacy in a mouse model.15,16
This study aimed to examine the clinical significance of SIRPα expression in HCC tumors using RNA sequencing (RNA-seq) data from a public database and immunohistochemical data from a public database of patients who underwent surgical resection for HCC. We clarified the association between SIRPα expression and clinicopathologic factors and determined the prognostic value of SIRPα expression in patients with HCC. We also investigated the relationship between SIRPα expression and tumor-infiltrating immune cells in HCC tissues. Finally, we examined the correlation between SIRPα expression and tumor-infiltrating immune cells as a prognostic marker.
Methods
Patients
The study enrolled 189 patients with HCC who underwent initial liver resection at the Department of Surgery and Science of Kyushu University Hospital between July 2004 and December 2015. The surgical technique details and patient selection criteria for liver resection in HCC have been previously reported.17 Outpatient follow-up visits were carried out for 1–3 months after discharge. Dynamic computed tomography was performed upon a suspected recurrence. Clinical information and follow-up data were obtained from medical records. This study was approved by the ethics committee of Kyushu University (approval code 2021-467) and conducted in accordance with the Declaration of Helsinki of 1996.
Immunohistochemical Staining
Immunohistochemical staining for CD8,5 CD33,4 and CD6818 was performed as previously described. For SIRPα, immunohistochemical staining was performed on 4-μm formalin‐fixed and paraffin-embedded sections. The sections were first deparaffinized. After inhibition of endogenous peroxidase activity for 30 min with 10% hydrogen peroxidase in methanol, the sections were pretreated with Target Retrieval Solution (Dako, Glostrup, Denmark) in a decloaking chamber at 121 °C for 15 min, then incubated with monoclonal antibodies at 4 °C overnight. Immune complexes were detected using an EnVision Detection System (Dako, Carpinteria CA, USA). Finally, the sections were incubated in 3,3'‐diaminobenzidine, counterstained with hematoxylin, and mounted.
The primary antibodies used were SIRPα (clone D613M, 1:200 dilution; Cell Signaling Technology, Beverly, MA, USA) and a CD33 mouse antibody (PA0555, no dilution; Leica Biosystems, Wetzlar, Germany). The stained slides were scanned using a NanoZoomer digital slide scanner (Hamamatsu Photonics K.K., Shizuoka, Japan). Cells exhibited plasma membrane staining for SIRPα. Spleens were used as positive controls for SIRPα. Immunohistochemical data for SIRPα, CD33, CD68, and CD8 staining were evaluated by three experienced researchers (T.T., K.K., and S.I.) who were blinded to the clinical status of the patients. The final assessments were achieved by consensus.
Gene Analysis with a Visium Platform
Gene analysis with a Visium platform (10x Genomics, Pleasanton, CA, USA) was performed as previously reported. Briefly, a single HCC sample was embedded in optimal slicing temperature compound (TissueTek Sakura) in a 10-mm × 10-mm cryomold at − 80 °C and sectioned at a thickness of 10 μm. Tissue was permeabilized for 3 min, which was defined as the optimal time in tissue optimization time course experiments. Visium Spatial Gene Expression Libraries were prepared according to the Visium Spatial Gene Expression Reagent Kits User Guide. Libraries were sequenced on the NovaSeq 6000 System (Illumina, Tokyo, Japan) in a paired-end mode at sufficient sequence depth. Raw FASTQ files and histology images were processed using Space Ranger Software v1.1.0 (https://support.10xgenomics.com/spatial-gene-expression/software/pipelines/latest/©nstallation). To visualize spatial expression using histologic images, the raw Visium files for each sample were read into Loupe Browser software v4.0.0 (https://support.10xgenomics.com/spatial-gene-expression/software/downloads/latest). We obtained mean sequence read counts of 288,702 and identified median genes of 3300 per spot. The median-normalized average of gene in cluster is calculated as the mean of observed unique molecular identifier (UMI) counts normalized by the size factor for each cell in the representative cluster.
Statistical Analysis
Standard statistical analyses were used to evaluate descriptive statistics such as medians, frequencies, and percentages. Continuous variables without a normal distribution were compared using the Mann–Whitney U test. Logistic regression analysis was performed to identify variables associated with SIRPα expression. Categorical variables were compared using the chi-square test or Fisher’s exact test. Survival data were used to establish a univariate Cox proportional hazard model. Covariates significant at a p value lower than 0.05 were included in the multivariate Cox proportional hazards model. The cumulative overall survival (OS) and recurrence-free survival (RFS) rates were calculated using the Kaplan–Meier method, and differences between curves were evaluated using the log-rank test. Differences were considered statistically significant at a p value lower than 0.05. All statistical analyses were performed using the JMP15 software (SAS Institute Inc., Cary, NC, USA).
Results
Signal Regulatory Protein Alpha Expression was a Prognostic Factor of HCC in the TCGA Dataset
We investigated whether SIRPα affects the prognosis of patients with liver hepatocellular carcinoma (LIHC). The association between SIRPα expression and HCC patient prognosis was examined using a public dataset.
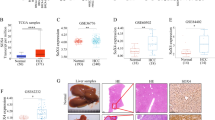
First, we estimated the mRNA expression level of SIRPα in 372 patients with LIHC using The Cancer Genome Atlas (TCGA) dataset. Unlike other cancer types, the expression of SIRPα mRNA in cancer tissue was significantly lower than in noncancerous tissue (respective median values: 10.7 [range, 6.80–13.2] vs 11.2 [range, 10.5–12.5]; p < 0.0001; Fig. 1A). However, diverse patterns of SIRPα mRNA expression have been observed in cancer tissues compared with that in noncancerous tissues. We divided 343 patients into high-expression (n = 172) and low-expression (n = 171) groups according to their SIRPα mRNA expression levels. The OS in the high-expression group was significantly lower than in the low-expression group (respectively, 37.3% vs 59.2%; p = 0.0305, log-rank test; Fig. 1B).
Overall survival rates in the signal regulatory protein alpha (SIRPα) high-expression group were worse than in the low-expression group using a public dataset of liver hepatocellular carcinoma (LIHC). A The mRNA expression level of signal regulatory protein alpha (SIRPα) in tumor tissue compared with normal tissue using the Cancer Genome Atlas. The median mRNA expressions (Log2FPKM) were 10.7 (range, 6.80–13.2) in tumors and 11.2 (range, 10.1–12.5) in normal tissues (p < 0.0001). Overall survival rates for high-expression (n = 172), and low-expression (n = 171) groups of SIRPα were analyzed by the Kaplan-Meier method. Statistical analysis was performed by the log-rank test. Signal regulatory protein alpha (SIRPα) expression was the indicator of poor prognosis of hepatocellular carcinoma (HCC) in the current cohort. B Immunohistochemical staining of SIRPα expression in patients with HCC. C Image of a high SIRPα expression pattern. D Image of a low SIRPα expression pattern. E Recurrence-free survival and F overall survival of high (n = 56), and low (n = 133) expression of SIRPα were analyzed by the Kaplan–Meier method. Statis analysis was undertaken by the log-rank test
Signal Regulatory Protein Alpha Expression and Clinicopathologic Factors
Our cohort of 189 patients with HCC had 135 (71.4%) male patients. The median age of the patients was 68 years (range, 61–76 years; 25–75% quantile). Among the 189 patients, 27 (14.2%) and 118 (62.4%) were positive for hepatitis B surface antigen and hepatitis C virus antibody expression, respectively. The median observation period was 4.11 years (range, 2.28–6.32 years; 25–75% quantile).
Figure 1C, D show representative immunohistochemical staining of SIRPα in HCC tissues. The staining exhibited SIRPα in the intratumor cytoplasm and plasma membrane of mononuclear cells. The median protein expression level of SIRPα was 15% (range, 25–75%; quantile, 3–35%). The patients were divided into high-expression (n = 56) and low-expression (n = 133) groups according to the SIRPα staining.
The associations between SIRPα protein expression and clinicopathologic factors are shown in Table 1. The serum albumin level was lower in the SIRPα high-expression group than in the low-expression group (p = 0.0004). Tumors with microvascular invasion were more frequent in the SIRPα high-expression group than in the low-expression group (p = 0.0097). The primary disease of the patients did not differ significantly.
Survival analysis according to SIRPα expression using the Kaplan-Meier method in our patient cohort showed that both RFS and OS were significantly poorer in the SIRPα high-expression group than in the low-expression group (p < 0.0001 and p = 0.0022, respectively; Fig. 1E, F).
SIRPα High Expression was Associated with Tumor-Infiltrating Immune Cells
We examined whether SIRPα is involved in the regulation of antitumor immunity in HCC. We used immunohistochemistry (IHC) analysis to evaluate the association between SIRPα expression and the number of tumor-infiltrating immune cells such as cytotoxic T cells, macrophages, and myeloid-derived suppressor cells (MDSCs). Studies show that CD8, CD68, and CD33 are used as cell surface markers for cytotoxic T cells, macrophages, and MDSCs, respectively.4,5,19 Representative images of CD8-positive T cells, CD68-positive macrophages, and MDSC staining are shown in Fig. 2A–C.
Signal regulatory protein alpha (SIRPα) expression was correlated with tumor infiltration of macrophage and myeloid-derived suppressor cells (MDSCs). Representative immunohistochemical staining of A CD8-positive T cells, B CD68-positive macrophages, and C myeloid-derived suppressor cells (MDSCs) in patients with hepatocellular carcinoma (HCC) is shown. D The median number of intratumor CD8-positive T cells was 12.3 (range, 0.667–114) in the high-SIRPα group and 13.0 (range, 0.333–105) in the low-SIRPα group (p = 0.7115). E The median number of intratumor CD68-positive macrophages was 131 (range, 26.3–223) in the high-SIRPα group and 95.0 (range, 19.0–244) in the low-SIRPα group (p = 0.0007). F The median number of intratumor MDSCs was 99.3 (range, 6.67–210) in the high-SIRPα group and 74.7 (range, 1.33–2173) in the low-SIRPα group (p = 0.0266). *p < 0.05. **p < 0.001
Although the number of tumor-infiltrating CD8-positive T cells did not differ significantly between the SIRPα high- and low-expression groups (Fig. 2D; p = 0.7115), the SIRPα high-expression group had a significantly higher number of tumor-infiltrating macrophages and MDSCs than the low-expression group (CD68-positive macrophages [p = 0.0007], MDSCs [p = 0.0266]; Fig. 2E, F). Multivariate analysis showed that high SIRPα expression was significantly associated with the number of tumor-infiltrating CD68-positive macrophages (odds ratio [OR], 3.81; 95% confidence interval [CI] 1.65–8.84; p = 0.0018) (Table S1).
We analyzed the contribution of CD8-positive T cells, CD68-positive macrophages, and MDSCs to patient survival. All were significant prognostic factors for RFS and OS with HCC (RFS: CD8-positive T cells [p < 0.0001], CD68-positive macrophages [p < 0.0001], MDSCs [p < 0.0001]; OS: CD8-positive T cells [p < 0.0001], CD68-positive macrophages [p = 0.0116], MDSCs [p = 0.00028]; Fig. 3A–F).
Kaplan–Meier curves showing the survival of patients with hepatocellular carcinoma (HCC) according to the number of tumor-infiltrating CD8-positive T cells, CD68-positive macrophages, or myeloid derived suppressor cells (MDSCs). Recurrence-free survival rates for all the patients according to A high and low intratumor CD8-positive T cell infiltration, B CD68-positive macrophage infiltration, and C MDSC infiltration are shown. Overall survival rates for all the patients according to D high and low CD8-positive T cell infiltration, E CD68-positive macrophage infiltration, and F MDSCs infiltration are shown
Uni- and Multivariate Analysis of Prognostic Factors for RFS and OS
Tables 2 and 3 lists the uni- and multivariate analyses results associated with RFS and OS for the patients with surgically resected HCC. In the multivariate analysis, Cox proportional hazard regression models showed that high expression of SIRPα in tumors was associated with significantly worse RFS and OS (RFS: hazard ratio [HR], 2.78 [95% CI 1.78–4.33, p < 0.0001]; OS: HR, 2.45 [95% CI 1.16–5.20, p = 0.0194]). Low intratumor CD8-positive T cell infiltration in tumors was associated with significantly worse RFS (HR, 2.02; 95% CI 1.27–3.19; p = 0.0028) and OS (HR, 4.08; 95% CI 1.72–9.67; p = 0.0014). High intratumor MDSC infiltration also was associated with significantly worse RFS (HR, 2.42; 95% CI 1.41–4.17; p = 0.0014) and OS (HR, 2.52; 95% CI 1.00–6.35; p = 0.0493). In addition, the prognostic factors for RFS were liver fibrosis (HR, 1.72; 95% CI 1.03–2.89; p = 0.0338) and intrahepatic metastasis (HR, 1.82; 95% CI 10.1–3.26; p = 0.0449). In this analysis, CD68-positive macrophages were not significantly associated with RFS or OS.
Examination of SIRPα Expression in M2 Macrophage
We evaluated the distribution of SIRPα expression using the Visium platform. Figure 4A, B show the distributions of SIRPα and CD163, respectively. The distributions of SIRPα and CD163 were similar. Compared with CD163-negative cells, a significantly higher percentage of CD163-positive cells exhibited SIRPα expression (Fig. 4C).
Stratification of SIRPα Expression and MDSC and CD8-Positive T Cell Infiltration in HCC
Next, we evaluated the significance of SIRPα expression and MDSC and CD8+ T cell infiltration in predicting OS and RFS. High SIRPα expression, low CD8 infiltration, and high MDSC infiltration each was assigned a prognostic risk score of 1 point, and the patients were divided into four groups (0–3 points). The four groups were found to differ significantly in both RFS (p < 0.0001, log-rank test) and OS (p < 0.0001, log-rank test) (Fig. 5A, B). The differences in clinicopathologic characteristics between the patients with the highest risk scores and the others are shown in Table S2. Poor differentiation and smaller tumor size were observed in the group with the highest risk factor score compared with the others (p = 0.0109 and 0.0352, respectively). In addition, the serum albumin level was lower in the group with the highest risk score than in the other groups (p = 0.0007).
Kaplan–Meier curves classified with the numbers of risk factors, including signal regulatory protein alpha (SIRPα), myeloid-derived suppressor cells (MDSCs), and CD8-positive T cells. Kaplan–Meier curves for A recurrence-free survival (RFS) and B overall survival (OS) for the patients with hepatocellular carcinoma (HCC) according to the numbers of risk factors are shown
Discussion
The current study analyzed SIRPα expression in the TCGA and our HCC cohorts. In both cohorts, we identified SIRPα expression as an indicator of poor prognosis in HCC patients compared with the conventional markers such as alpha fetoprotein and des-gamma-carboxy pro-thrombin. Although SIRPα was associated with tumor-infiltrating macrophages and MDSCs, the correlation coefficient was small, with r2 below 0.1 for both the number of intratumor CD68-positive macrophages and SIRPα expression (r2 = 0.065 for the number of intratumor MDSCs and r2 = 0.038 for SIRPα expression). Therefore, the relationship was weak as a confounding factor, and multivariate analysis was used to examine these factors.
A previous study reported that SIRPα expression was an effective prognostic marker in esophageal and intrahepatic cholangiocarcinomas.10,20 In HCC, CD47, the ligand of SIRPα, was positive in 21.7% of the patients and associated with a shorter RFS for the patients who underwent surgical resection.12 However, the clinical significance of SIRPα expression in HCC remained unclear. Therefore, the current study is the first to determine the effect of SIRPα expression on HCC prognosis.
In our cohort, SIRPα expression was associated with microvascular invasion and poor prognosis, indicating that SIRPα may partially contribute to patient survival by enhancing tumor invasiveness.
Suppression of SIRPα expression in dendritic cells also enhanced T cell activation through antigen presentation and suppressed tumor growth in a mouse model.21
Several preclinical studies have shown that blocking the SIRPα-CD47 pathway using anti-SIRPα antibody, SIRPα-Fc fusion proteins, or anti-CD47 antibody, which act as effective decoy receptors, has an antitumor effect against several solid organ or hematopoietic cancers.22,23,24,25,26,27 Several clinical trials for blocking the SIRPα-CD47 pathway have been performed.28,29 Our results provide a basis for further investigations of the therapeutic effects of SIRPα-CD47 inhibition in HCC.
The results of the current study showed that SIRPα expression was significantly correlated with the infiltration of CD68-positive macrophages, indicating that these might have high SIRPα expression. In HCC, the number of tumor-infiltrating CD68-positive macrophages is reported to be a prognostic marker for OS.30 In the multivariate analysis, SIRPα expression was found to be a prognostic factor for both RFS and OS, whereas the number of CD68-positive macrophages was not. This result suggested that macrophage polarization may be more important than the number of tumor-infiltrating macrophages.
Macrophages in malignant tumors are classified into two subtypes. On the one hand, M1 macrophages are involved in efficient antigen presentation and pathogen killing, secreting large amounts of pro-inflammatory molecules and promoting type 1 T helper cells and antitumor immunity. On the other hand, M2 macrophages have high phagocytic activity but secrete small amounts of pro-inflammatory molecules, which are involved in type 2 T helper responses and suppress the antitumor immune response.31,32 In HCC, tumor cells promote M2 polarization in a paracrine manner, and tumor-associated macrophages (TAMs) predominantly consist of M2 macrophages.33,34 Moreover, M2 macrophages promote neovascularization, cancer spreading, and suppression of innate and adaptive immune responses.32 Expression of SIRPα correlates with M2 macrophage differentiation and regulates anti-tumor immunity.10 Actually, in the current study, SIRPα expression was significantly upregulated in CD163-positive cells compared with CD163-negative cells.
Our IHC data also showed that SIRPα expression was correlated with MDSC infiltration. We previously reported that high intratumor MDSC infiltration was correlated with poor RFS and OS for patients with HCC,4 with MDSCs enhancing tumor proliferation and resistance to immunotherapy.35 In addition, MDSCs express SIRPα, and SIRPα blockage reduces the number of MDSCs in a mouse model. Hence, SIRPα blockage may be useful in suppressing MDSCs.
Recently, a clinical trial (983P phase 1 dose-escalation study) of BI 765063, a selective SIRPα inhibitor, was performed in patients with advanced solid tumors and in patients with HCC, indicating a growing interest in therapies that inhibit this signaling. Several clinical trials also are currently evaluating the efficacy of CD47 blockage (Hu5F9-G4, TTI-621) for solid tumor,36 although no clinical trials are dealing primary with HCC. In addition, humanized anti-CD47 antibodies, evaluated in a current phase 1 clinical trial, are reported to be well-tolerated for patients with advanced solid and hematologic malignancies when used in initial and maintenance doses.37
Interest in anti-CD47 antibodies also is growing, and further validation of their efficacy is needed. Therefore, we believe that examination of the SIRPα immune status in patients with HCC is essential to the development of more effective cancer therapies. To the best of our knowledge, no report has described a method to evaluate SIRPα expression by liquid biopsy in solid tumors. However, it will be an important subject of future research to find a method to evaluate SIRPα expression using a peripheral blood-based platform to determine the therapeutic effect of immunotherapy before treatment. In addition, programmed death 1 (PD-1) blockage suppressed intratumor SIRPα and CD47 expression and deteriorated intratumor MDSC invasion in mouse model experiments of head and neck squamous cell carcinoma.38 In our study, we were unable to examine the correlation with SIRPα expression and immunotherapy because our cases did not include patients treated with preoperative immunotherapy. In HCC patients, the relationship between PD-1 and CTLA-4 antibody therapy and SIRPα expression should be verified in the future.
One limitation of the current study was its retrospective single-center design. Further analyses from multiple institutions are required. Additionally, several types of cells in the tumor microenvironment of HCC, such as macrophages and MDSCs, express SIRPα. It is unknown whether these cells suppress the immune reaction via SIRPα. Therefore, further experiments are required to confirm this hypothesis. In conclusion, SIRPα is associated with a poor prognosis in HCC through M2 macrophage and MDSC accumulation.
References
Itoh S, Shirabe K, Taketomi A, et al. Zero mortality in more than 300 hepatic resections: validity of preoperative volumetric analysis. Surg Today. 2012;42:435–40.
Itoh S, Morita K, Ueda S, et al. Long-term results of hepatic resection combined with intraoperative local ablation therapy for patients with multinodular hepatocellular carcinomas. Ann Surg Oncol. 2009;16:3299–307.
Koyama S, Akbay EA, Li YY, et al. Adaptive resistance to therapeutic PD-1 blockade is associated with upregulation of alternative immune checkpoints. Nat Commun. 2016;7:10501.
Tomiyama T, Itoh S, Iseda N, et al. Myeloid-derived suppressor cell infiltration is associated with a poor prognosis in patients with hepatocellular carcinoma. Oncol Lett. 2022;23:93.
Itoh S, Yoshizumi T, Yugawa K, et al. Impact of immune response on outcomes in hepatocellular carcinoma: association with vascular formation. Hepatology. 2020;72:1987–99.
Finn RS, Qin S, Ikeda M, et al. Atezolizumab plus bevacizumab in unresectable hepatocellular carcinoma. N Engl J Med. 2020;382:1894–905.
Ding X, Wang J, Huang M, et al. Loss of microglial SIRPα promotes synaptic pruning in preclinical models of neurodegeneration. Nat Commun. 2021;12:2030.
Barclay AN, van den Berg TK. The interaction between signal regulatory protein alpha (SIRPα) and CD47: structure, function, and therapeutic target. Immunology. 2014;32:25–50.
Murata Y, Kotani T, Ohnishi H, et al. The CD47-SIRP signalling system: its physiological roles and therapeutic application. J Biochem. 2014;155:335–44.
Koga N, Hu Q, Sakai A, et al. Clinical significance of signal regulatory protein alpha (SIRPα) expression in esophageal squamous cell carcinoma. Cancer Sci. 2021;112:3018–28.
Brown EJ, Frazier WA. Integrin-associated protein (CD47) and its ligands. Trends Cell Biol. 2001;11:130–5.
Kim H, Bang S, Jee S, et al. Clinicopathological significance of CD47 expression in hepatocellular carcinoma. J Clin Pathol. 2021;74:111–5.
Matozaki T, Murata Y, Okazawa H, et al. Functions and molecular mechanisms of the CD47–SIRPα signalling pathway. Trends Cell Biol. 2009;19:72–80.
Vaeteewoottacharn K, Kariya R, Pothipan P, et al. Attenuation of CD47-SIRPα signal in cholangiocarcinoma potentiates tumor-associated macrophage-mediated phagocytosis and suppresses intrahepatic metastasis. Transl Oncol. 2019;12:217–25.
Wang H, Sun Y, Zhou X, et al. CD47/SIRPα-blocking peptide identification and synergistic effect with irradiation for cancer immunotherapy. J Immunother Cancer. 2020;8:e000905.
Lin GHY, Chai V, Lee V, et al. TTI-621 (SIRPαFc), a CD47-blocking cancer immunotherapeutic, triggers phagocytosis of lymphoma cells by multiple polarized macrophage subsets. PLoS ONE. 2017;12:e0187262.
Itoh S, Shirabe K, Matsumoto Y, et al. Effect of body composition on outcomes after hepatic resection for hepatocellular carcinoma. Ann Surg Oncol. 2014;21:3063–8.
Itoh S, Yoshizumi T, Kitamura Y, et al. Impact of metabolic activity in hepatocellular carcinoma: association with immune status and vascular formation. Hepatol Commun. 2021. https://doi.org/10.1002/hep4.1715.
Iseda N, Itoh S, Yoshizumi T, et al. ARID1A deficiency is associated with high programmed death ligand 1 expression in hepatocellular carcinoma. Hepatol Commun. 2020. https://doi.org/10.1002/hep4.1659.
Yang H, Yan M, Li W, et al. SIRPα and PD1 expression on tumor-associated macrophage predict prognosis of intrahepatic cholangiocarcinoma. J Transl Med. 2022;20:140.
Liu Q, Wen W, Tang L, et al. Inhibition of SIRPα in dendritic cells potentiates potent antitumor immunity. Oncoimmunology. 2016;5:e1183850.
Yanagita T, Murata Y, Tanaka D, et al. Anti-SIRPα antibodies as a potential new tool for cancer immunotherapy. JCI Insight. 2017;2:e89140.
Ring NG, Herndler-Brandstetter D, Weiskopf K, et al. Anti-SIRPα antibody immunotherapy enhances neutrophil and macrophage antitumor activity. Proc Natl Acad Sci. 2017;114:E10578–85.
Billerhart M, Schönhofer M, Schueffl H, et al. CD47-targeted cancer immunogene therapy: secreted SIRPα-Fc fusion protein eradicates tumors by macrophage and NK cell activation. Mol Ther Oncolytics. 2021;23:192–204.
Zhao XW, van Beek EM, Schornagel K, et al. CD47-signal regulatory protein-α (SIRPα) interactions form a barrier for antibody-mediated tumor cell destruction. Proc Natl Acad Sci. 2011;108:18342–7.
Zhang X, Fan J, Wang S, et al. Targeting CD47 and autophagy elicited enhanced antitumor effects in non-small cell lung cancer. Cancer Immunol Res. 2017;5:363–75.
Chao MP, Alizadeh AA, Tang C, et al. Therapeutic antibody targeting of CD47 eliminates human acute lymphoblastic leukemia. Cancer Res. 2011;71:1374–84.
Kotecki N, Champiat S, Delord J-P, et al. 983P Phase I dose escalation study in patients (pts) with advanced solid tumours receiving first-in-class BI 765063, a selective signal-regulatory protein α (SIRPα) inhibitor, in combination with ezabenlimab (BI 754091), a programmed cell death protein 1 (PD-1) inhibitor. Ann Oncol. 2021;32:S841–2.
Oronsky B, Carter C, Reid T, et al. Just eat it: a review of CD47 and SIRP-α antagonism. Semin Oncol. 2020;47:117–24.
Ding W, Tan Y, Qian Y, et al. Clinicopathologic and prognostic significance of tumor-associated macrophages in patients with hepatocellular carcinoma: a meta-analysis. PLoS ONE. 2019;14:e0223971.
Qian B-Z, Pollard JW. Macrophage diversity enhances tumor progression and metastasis. Cell. 2010;141:39–51.
Bronte V, Murray PJ. Understanding local macrophage phenotypes in disease: modulating macrophage function to treat cancer. Nat Med. 2015;21:117–9.
Tian X, Wu Y, Yang Y, et al. Long noncoding RNA LINC00662 promotes M2 macrophage polarization and hepatocellular carcinoma progression via activating Wnt/β-catenin signaling. Mol Oncol. 2020;14:462–83.
Sica A, Invernizzi P, Mantovani A. Macrophage plasticity and polarization in liver homeostasis and pathology. Hepatology. 2014;59:2034–42.
Kumar V, Patel S, Tcyganov E, et al. The nature of myeloid-derived suppressor cells in the tumor microenvironment. Trends Immunol. 2016;37:208–20.
Alvey C, Discher DE. Engineering macrophages to eat cancer: from “marker of self” CD47 and phagocytosis to differentiation. J Leukocyte Biol. 2017;102:31–40.
Sikic BI, Lakhani N, Patnaik A, et al. First-in-human, first-in-class phase I trial of the Anti-CD47 antibody Hu5F9-G4 in patients with advanced cancers. J Clin Oncol. 2019;37:946–53.
Yu GT, Bu LL, Huang CF, et al. PD-1 blockade attenuates immunosuppressive myeloid cells due to inhibition of CD47/SIRPα axis in HPV negative head and neck squamous cell carcinoma. Oncotarget. 2015;6:42067–80. https://doi.org/10.18632/oncotarget.5955
Acknowledgment
This study was supported by the Japan Society for the Promotion of Science (JSPS) KAKENHI (Grant No. JP-19K09198) and the Research Grant of the Princess Takamatsu Cancer Research Fund.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Disclosure
There are no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Tomiyama, T., Itoh, S., Iseda, N. et al. Clinical Significance of Signal Regulatory Protein Alpha (SIRPα) Expression in Hepatocellular Carcinoma. Ann Surg Oncol 30, 3378–3389 (2023). https://doi.org/10.1245/s10434-022-13058-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1245/s10434-022-13058-y