Abstract
In this paper, a mathematical model of COVID-19 has been proposed to study the transmission dynamics of infection by taking into account the role of symptomatic and asymptomatic infected individuals. The model has also considered the effect of non-pharmaceutical interventions (NPIs) in controlling the spread of virus. The basic reproduction number (\(\textbf{R}_{0}\)) has been computed and the analysis shows that for \({\textbf{R}_{0}}<{1}\), the disease-free state becomes globally stable. The conditions of existence and stability for two other equilibrium states have been obtained. Transcritical bifurcation occurs when basic reproduction number is one (i.e. \(R_0=1\)). It is found that when asymptomatic cases get increased, infection will persist in the population. However, when symptomatic cases get increased as compared to asymptomatic ones, the endemic state will become unstable and infection may eradicate from the population. Increasing NPIs decrease the basic reproduction number and hence, the epidemic can be controlled. As the COVID-19 transmission is subject to environmental fluctuations, the effect of white noise has been considered in the deterministic model. The stochastic differential equation model has been solved numerically by using the Euler-Maruyama method. The stochastic model gives large fluctuations around the respective deterministic solutions. The model has been fitted by using the COVID-19 data of three waves of India. A good match is obtained between the actual data and the predicted trajectories of the model in all three waves of COVID-19. The findings of this model may assist policymakers and healthcare professionals in implementing the most effective measures to prevent the transmission of COVID-19 in different settings.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
COVID-19 is a contagious disease that spreads by SARS-COV2 virus among humans via respiratory droplets. After exposure to the virus, usually symptoms start to reveal within 2–14 days. The most common symptoms are sore throat, fever, chills, fatigue, breathing problem and loss of smell and taste [1,2,3]. The first case of COVID-19 was appeared in Wuhan, China in December 2019. After that the COVID-19 cases have increased very rapidly in the world wide resulting in the COVID-19 pandemic [4, 5]. During the early phase of the pandemic, most countries were totally unaware of the rapidity of COVID-19 infection and having poor understanding about the disease outbreaks and its further treatment [6]. This situation got worsen due to lack of widespread testing, unavailability of effective treatment and absence of COVID-19 vaccine [2, 7, 8]. Furthermore, it was also the serious concern to track the prevalence of COVID-19 from those who had no symptoms or mild symptoms [1, 6]. People who had mild or no symptoms come in the category of asymptomatic infection. The asymptomatic cases have played very significant role in rapid spread of infection because of being unaware of their infection and spread it to others rapidly [9, 10]. As per literature, around \(80\%\) of the COVID-19 infection were spread from asymptomatic infected individuals [11]. To fight from these challenges, the public health mitigation strategies like social distancing, wearing of masks, contact tracing, quarantine of infected cases and maintaining the environmental, personal hygienic practices and vaccination were proved to be very effective in reducing the transmission of infection and the number of cases in societies [1, 6, 9, 12,13,14]. The COVID-19 pandemic has had the significant impact on many aspects of society including economy, education system, social behavior etc. [15, 16].
Mathematical modeling has always been used as a very significant tool for answering many concerns of the society emerging in health, social sciences, agriculture, biological sciences and engineering [17, 18]. In history of mankind, many diseases like dengue, malaria, HIV AIDS, influenza have taken the form of epidemic but COVID-19 has taken the form of pandemic and adversely affects the almost all aspects of human life. Many mathematical models have been developed for infectious diseases like dengue, malaria, HIV AIDS, influenza etc [19,20,21,22,23]. However, several research investigations have been done on COVID-19 epidemic to understand the transmission and control [24,25,26,27,28,29,30].
To keep in mind the biology of disease, researchers have considered various aspects that are influencing the transmission as vaccination, symptomatic and asymptomatic infection dynamics [10], effects of various public health interventions under constant environmental conditions [29]. Anggriani et al. [30] proposed the COVID-19 compartmental model with symptomatic and asymptomatic infected cases as different compartments. They have also considered the effect of waning immunity in the model. Further, they performed the parameter estimation for the data of West Java. Jayrold et al. [24] also considered SEIR model for COVID-19 with adding asymptomatic infected compartment. They calibrated the model for several countries and concluded that testing and isolation both are important to eradicate the disease. Serhani and Labbardi [25] proposed the SIR type COVID-19 model considering asymptomatic infected class as well as quarantine and dead compartments. They found that the home containment played a very significant role in controlling the disease transmission. Ali et al. [26] have formulated the SEIR type fractional-order mathematical model for COVID-19 with asymptomatic and symptomatic infected transmission dynamics. They found that COVID-19 epidemic can be controlled by restricting people from migration and strictly enforcing personal measures simultaneously. Some research investigations of COVID-19 have been published considering environmental fluctuations. As, environmental fluctuations affect the growth of epidemic so, it is important to consider stochasticity in the model. Olabode et al. [27] have formulated the SEIR model for COVID-19 using both the deterministic and stochastic modeling. They have considered the CTMC stochastic model and found that disease may not persist even for basic reproduction number (\(R_0>1\)) greater than one. Hussain et al. [28] have proposed the deterministic model along with stochastic differential equation model involving environmental white noise. They analyzed the deterministic model and compared the results of stochastic and deterministic models for the future prediction of the spread of COVID-19.
Considering the above literature reviews in mind, the main motivation of this work is to understand the transmission dynamics of symptomatic and asymptomatic infected individuals in presence of non-pharmaceutical interventions. Since, the combination of lockdown and personal hygiene measures has been proved very effective for controlling the pandemic. The impact of NPIs has been considered in susceptible population. This study has also investigated the potential impact of white noise in the proposed model.The good fit of the model is obtained for all the three waves of COVID-19, India. The obtained results are relevant and applicable to the current pandemic situation. The results of this study also provide valuable insights into the effectiveness of NPIs in controlling the spread of COVID-19 in populations.
The rest of the paper is organized as follows: The Sect. 2 deals with the model formulation and computation of basic reproduction number. In Sect. 3, the deterministic model has been analyzed. Subsequently, the main results have been introduced and proved. In Sect. 4, the role of asymptomatic infected individuals on disease dynamics has been discussed. In Sect. 5, the stochastic differential equation model has been introduced. The Sect. 6 deals with numerical simulations for stochastic and deterministic models. The Sect. 7 has been devoted for parameter estimations of the model parameters by using the COVID-19 data of India. Conclusions have been given in Sect. 8.
2 Deterministic model
The deterministic COVID-19 model is now presented. Let N(t) be the total host population at time t, which is divided into four classes of individuals: susceptible S(t), asymptomatic infected \(I_a(t)\), symptomatic infected \(I_s(t)\) and recovered R(t) such that N(t)=\(S(t)+I_a(t)+I_s(t)+R(t)\). Assuming all parameters are positive, the following model is formulated:
with \({S(0)>0, I_a(0)> 0, I_s(0)> 0, R(0)\ge 0}\) and all other parameters are taken positive.
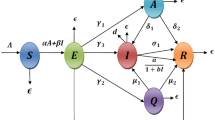
The detailed definition of parameters involved in model (1) are summarized in Table 1. The schematic diagram has been drawn in Fig. 1.
2.1 Positivity & boundedness of solutions
The set \({\Omega =\{(S,I_a,I_s,R)\in R_+^4:0<(S,I_a,I_s,R)\le N\}}\) is positive and attracts all the solutions of the model (1).
Proof
Adding all equations of model (1), it is obtained that:
Further, Eq. (2) can be reduced into
Solving the above differential inequality, for \(t \rightarrow \infty\), the following bound on N(t) is obtained
\(\square\)
2.2 Basic reproduction number (\(R_0\))
The basic reproduction number \(R_0\) is very crucial factor in epidemic models that informs us about disease extinction and persistence. It is defined as the expected number of secondary infectious cases produced by a primary infected individual in susceptible population. Using the next generation matrix approach [31], \(R_0\) is computed as:
Using the next-generation matrix, it gives
where, F is the matrix of new infections from various infected compartments and V is the is the transition matrix from one compartment to another.
At the disease-free state \((I_s=I_a=0)\), the next generation matrix \(FV^{ - 1}\) is computed as:
Thus, the basic reproduction number is obtained by the spectral radius of \(FV^{-1}\), that is obtained as
Let us define,
and
Therefore,
3 Deterministic model analysis
For the model (1), three equilibrium states exist.
3.1 Disease-free state (\(E_0=({\hat{S}}, {\hat{I}}_a, {\hat{I}}_s, {\hat{R}})\))
The disease-free state always exists. It is given as
Theorem 1
The disease-free state is locally asymptotically stable provided \({R_{0}<1}\).
Proof
At the equilibrium point \({E_{0}}\), the Jacobian matrix \(J(E_0)\) for the model (1) is obtained as:
For the matrix \(J(E_0)\), the two of the eigenvalues are having negative real parts given as \(-\alpha -\mu\) and \(-\mu\). However, the other two eigenvalues are \(-\beta _3-k-\mu + \dfrac{\beta _1\omega }{(\alpha +\mu )}\) and \(-\gamma -\mu -\mu _1+\dfrac{\beta _2\omega }{(\alpha +\mu )}\). It is interesting to see that the last two eigenvalues can be written as \((\beta _3+k+\mu )(-1+R_{10})\) and \((\gamma +\mu +\mu _1)(-1+R_{01})\). They are having negative real parts for \(R_{10}<1\) and \(R_{01}<1\). Therefore, for \(R_0<1\), the disease-free state is locally asymptotically stable. \(\square\)
Theorem 2
[32] The disease-free equilibrium is globally asymptotically stable if all eigenvalue of \(A_1\) are real negative and \(A_2\) is a Metzler matrix.
Proof
Let us write the model (1) in the following form:
where, \(x_1=(S,R)\) are non-transmitting compartments and \(x_2=(I_a,I_s)\) are transmitting compartments. Also, \(x_1^*\) is disease-free point.
The subsystem (7) is given as
The matrix \(A_1\) for the subsystem (7) is computed as,
Since \(A_1\) is the lower triangular matrix so the eigenvalues are \(-\alpha -\mu\) and -\(\mu\). As, all eigenvalues are negative, the subsystem is globally asymptotically stable at the disease-free state. Hence, assumptions \(H_1\) and \(H_2\) in [32] are satisfied. Again, the subsystem (8) is given as
The matrix \(A_2\) for the above system is given as
Since all the off-diagonal entries of \(A_2(x)\) are non-negative, \(A_2(x)\) is a Metzler matrix and irreducible for x. Hence, satisfying the assumption \(H_3\) in [32]. The eigenvalues of Metzler matrix are negative for \(\dfrac{\beta _1\omega }{(\alpha +\mu )(\beta _3+k+\mu )}(=R_{10})<1\) and \(\dfrac{\beta _2\omega }{(\alpha +\mu )(\gamma +\mu +\mu _1)}(=R_{01})<1\). Therefore, the Metzler matrix \(A_2\) is stable for \(R_{10}<1\) and \(R_{01}<1\). Hence, it satisfies the assumption \(H_5\) in [32]. Thus, the disease-free state \(E_0\) is globally asymptotically stable for \(R_0<1\). \(\square\)
3.2 Asymptomatic infection free state (\(E_1=(S^*, 0, I_s^*, R^*)\))
Asymptomatic infection free state exists for
The expressions for non-zero state variables at equilibrium point \(E_1\) are obtained as follows:
3.2.1 Stability analysis of asymptomatic infection free state \(E_1\)
Theorem 3
The asymptomatic infection free state is locally asymptotically stable provided \(R_{01}>R_{10}\).
Proof
At the equilibrium point \({E_{1}}\), the Jacobian matrix \(J(E_1)\) for the model (1) is obtained as:
For the matrix \(J(E_1)\), one of the eigenvalues is \(-\mu\). The eigenvalue \(-\dfrac{L(R_{01}-R_{10})}{\beta _2\omega }\) is having negative real parts for \(R_{01}>R_{10}\) where \(L=(\gamma +\mu +\mu _1)(\alpha +\mu )(\beta _3+k+\mu )\). However, the other two eigenvalues are \(-\dfrac{-\beta _2\omega \pm 2(\gamma +\mu +\mu _1)\sqrt{(\gamma +\mu +\mu _1)(\alpha +\mu )(R_{01}-1)}}{2(\gamma +\mu +\mu _1)}\). It is easy to see that all eigenvalues are having negative real parts for \(R_{01}>1\) and \(R_{01}>R_{10}\). Therefore, \(E_1\) state is locally asymptotically stable for \(R_{01}>R_{10}\). \(\square\)
3.3 Endemic state (\(E_2\)=(\(\bar{S}, \bar{I_a}, \bar{I_s}, \bar{R}\)))
Endemic state exists for
The expressions for non-zero state variables at equilibrium point \(E_2\) are obtained as follows:
3.3.1 Local stability
The endemic state is locally asymptotically stable provided \(R_{10}>R_{01}\).
Proof
At the equilibrium point \({E_{2}}\), the Jacobian matrix \(J(E_2)\) for the model (1) is obtained as:
For the matrix \(J(E_2)\), one of the eigenvalues is -\(\mu\). The rest of the eigenvalues are given by following characteristic polynomial:
\(\lambda ^3+ A \lambda ^2+ B\lambda + C=0\) where,
\(G= \beta _1^3\omega (\mu + \mu _1 + \gamma )(\beta _3 + 2\mu + \mu _1 + k + \gamma )+ \alpha (\beta _3 + \mu + k)^2(-\beta _2^2 \beta _3 + \beta _1\beta _2(\beta _3 + \mu + k)- \beta _1^2 (\mu + \mu _1 + \gamma ))-\beta _1^2 (\beta _3^2 (\beta _2\omega + \mu (\mu + \mu _1 + \gamma )) + (\mu + k)(\mu ^3 + \mu ^2 (\mu _1 + k + \gamma ) + \beta _2\omega (2\mu _1 + k + 2\gamma ) + \mu (3 \beta _2\omega + k (\mu _1 + \gamma ))) + \beta _3(2 \mu ^3 + 2 \mu ^2 (\mu _1 + k + \gamma ) + \beta _2\omega (\mu _1 + 2 k + \gamma )+ \mu (3\beta _2\omega + 2 k (\mu _1 + \gamma ))))\)
It is clear from the expressions A, B and C that \(A>0\) for \(R_{10}>R_{01}\). Also, the expressions \(AB-C\) and C are positive for \(R_{10}>1\) and \(R_{10}>R_{01}\). Hence, the endemic state is locally asymptotically stable for equation (16) to be satisfied. \(\square\)
The Fig. 2 has been drawn to show the stability region of various equilibrium states in \(R_{10}-R_{01}\) plane.
4 Impact of asymptomatic infection on disease dynamics: transcritical bifurcation
In this section, the effect of asymptomatic parameter on the qualitative dynamics of the model has been discussed. For this, bifurcation diagram for the parameter \(\beta _3\) with respect to the infection level \({\bar{I}}_s\) has been plotted by keeping rest of the parameters fixed. The values of fixed parameters are provided in the caption of Fig. 3. It is found that as \(\beta _3\) varies from 0 to 0.6, the basic reproduction number \(R_{10}\) remains greater than \(R_{01}\) i.e. \(R_{10}>R_{01}\). Accordingly, endemic state \(E_2\) gets stable. At about \(\beta _3\)=0.63 (say \(\beta _3^*\)), \(R_{10}=R_{01}\), at \(\beta _3^*\) endemic and asymptomatic free states get merge and only asymptomatic free state exists and gets stable. The Fig. 4 has been drawn to show the change in stability at different values of parameter \(\beta _3\) with respect to the variable \(I_a(t)\). For \(\beta _3\)=0.5 (\(<\beta _3^*\)), endemic state gets stable that is shown by the red line in Fig. 4. Further, for \(\beta _3=\beta _3^*\), the states \(E_1\) and \(E_2\) get merge and asymptomatic infection free state \(E_1\) exists and gets stable as shown by blue line in Fig. 4. For \(\beta _3\)=0.7 (\(>\beta _3^*\)), asymptomatic infection free state exists and gets stable as shown in Fig. 4 by pink line.
5 Stochastic differential equation model
The model (1) for COVID-19 is deterministic as all parameters are fixed. However, in real life scenario environmental noise affects the spread of disease. To consider the effect of environmental noise on the system (1), it is assumed that the stochastic perturbations are of white noise type and they are proportional to the variable. The stochastic differential equation models can be obtained by using the modeling techniques given in [33,34,35,36]. Let \(X_i\) (i=1,2,3,4) denotes the random variables for \(SI_aI_sR\) model respectively. To develop the stochastic differential equation model, the expectation \(E^*[\Delta C]\) and variance \(E^*[\Delta C\Delta C^T]\) depend on the associated transition probabilities of the model (1) given in Table 2. The expectation and variance are computed as
\(v_{11}=\omega +\beta _1SI_a+\beta _2SI_s+\alpha S+\mu S\), \(v_{22}=\beta _1SI_a+(\beta _3 + k+\mu )I_a\), \(v_{33}=\beta _2SI_s+\beta _3I_a+(\gamma +\mu +\mu _1)I_s\) and \(v_{44}=\gamma I_s+ \mu R + \alpha S+ k I_a\). The It\(\hat{o}\) SDE model for the system (1) has the following form:
and G is \(4\times 12\) matrix satisfying \(GG^T=V\) and W(t) is a vector of 12 independent Wiener process,
Considering the above facts, the stochastic model is formulated as
\(v_{11}=\omega +\beta _1SI_a+\beta _2SI_s+\alpha S+\mu S\), \(v_{22}=\beta _1SI_a+(\beta _3 + k+\mu )I_a\), \(v_{33}=\beta _2SI_s+\beta _3I_a+(\gamma +\mu +\mu _1)I_s\) and \(v_{44}=\gamma I_s+ \mu R + \alpha S+ k I_a\).
6 Stochastic and deterministic simulations results
The Euler-Maruyama method is one of the numerical methods that can be used to solve the SDEs numerically. The SDEs (20) is solved numerically by using the Euler-Maruyama method as given in equation (21).
Time series solutions of susceptible and asymptomatic infected populations starting from the initial condition (22201, 20, 30, 23176500) converges to the disease-free state (22282, 0, 0, 23177000) for deterministic system (shown by red color) in (a) and (b) respectively. However, the stochastic system shows the convergence around the disease-free state with large fluctuations as shown in blue color
Time series solutions of symptomatic infected and recovered populations starting from the initial condition (22201, 20, 30, 23176500) converges to the disease-free state (22282, 0, 0, 23177000) for deterministic system (shown by red color) in (a) and (b) respectively. However, the stochastic system shows the convergence around the disease-free state with large fluctuations as shown in blue color
The estimated parameters of second wave of COVID-19 (as given in Table 4) have been used to solve the SDEs numerically. For \(\beta _1\)=0.00000348275 and taking rest of the parameters values from Table 4, basic reproduction numbers \(R_{10}\) and \(R_{01}\) are computed as 0.857744 and 0.0145215 respectively. Therefore, \(R_0=0.857744(<1)\). Clearly, disease-free state exists and globally stable by Theorem 1. Using the data given in Table 4, the stochastic differential equations (20) have been solved numerically using the Euler-Maruyama method and solutions have been plotted along with the solutions of deterministic system (1). In Fig. 5a, the stochastic disease dynamics for the 300 paths of S(t) are obtained along with the deterministic solution. One can see that stochastic solutions for S(t) are having large oscillations and they are converging towards the equilibrium value. Again, in Fig. 5b, one can see that time series for \(I_a(t)\) is showing oscillations at the beginning and finally these oscillations get die out with probability one. In Fig. 6a, the time-series of symptomatic infected \(I_s(t)\) is showing strong oscillatory behavior at beginning but after sometime it converges to zero. The Fig. 6b for recovered population R(t) is also showing strongly oscillatory behavior near equilibrium value. The histograms have also been plotted in Figs. 7and 8 to confirm these results. It can be seen from histograms that the distribution of the S(t), \(I_a(t)\), \(I_s(t)\) and R(t) are around the equilibrium values of respective variables.
Further, for \(\beta _1\)=0.00000448275 and \(\beta _2\)= 0.000097028 and taking rest of the parameter values from Table 4, \(R_{10} = 1.1040>1\) and \(R_{01} =2.4707>1\). Clearly, the asymptomatic infection free state \(E_1\) exists by using (11) and is also getting stable by Theorem 3. The deterministic and corresponding stochastic solutions have been plotted in Figs. 9 and 10. It can be seen that the stochastic solutions of S(t) for 300 sample paths are showing strong oscillatory behavior around equilibrium value. However, the stochastic solution for asymptomatic infected individuals \(I_a(t)\) are showing oscillatory behavior at beginning and oscillations converges to zero after sometimes. The stochastic solutions for symptomatic infected \(I_s(t)\) and recovered population R(t) are exhibiting strong oscillatory behavior near equilibrium value. The histograms have also been plotted in Figs. 11 and 12 to confirm the distribution of state variables around equilibrium values.
By changing \(\beta _1=0.00001948275\) and keeping rest of the parameters same as given in Table 4, the \(R_{10} =4.7982\) and \(R_{01} = 0.0145215\). Here, \(R_{10}\) is greater than 1 and also \(R_{10}>R_{01}\). Now, using (16), the endemic state \(E_2\) exists and stable. The time-series for the stochastic dynamics of the system near endemic state along with the deterministic solutions have been plotted in Figs. 13 and 14. It is clear from Figs. 13 and 14 that due to stochasticity, the system exhibits huge oscillations around equilibrium values of model (1). The histograms in Figs. 15 and 16 have also been plotted to show the distribution of system variables.
The important observation from these simulations results is that the presence of stochasticity in the model (1) does not change the dynamics of any equilibrium states. However, stochasticity brings large oscillations around the solution of the system (1).
Time series solutions of susceptible and asymptomatic infected populations starting from the initial condition (8995, 30, 715, 16,63,5889) converges to the asymptomatic infection free state (9018.4, 0, 617, 16,63,7000) for deterministic system (shown by red color) in (a) and (b) respectively. However, the stochastic system shows the convergence around the asymptomatic infection free state with large fluctuations as shown in blue color
Time series solutions of symptomatic infected and recovered populations starting from the initial condition (8995, 30, 715, 16,63,5889) converges to the asymptomatic infection free state (9018.4, 0, 617, 16,63,7000) for deterministic system (shown by red color) in (a) and (b) respectively. However, the stochastic system shows the convergence around the asymptomatic infection free state with large fluctuations as shown in blue color
The histograms show the distribution of individuals across the (a) symptomatic infected and (b) recovered populations, with each bin representing a range of respective population sizes at the asymptomatic infection free state. The x-axis represents the population size, while the y-axis represents the frequency of individuals in each bin
Time series solutions of susceptible and asymptomatic infected populations starting from the initial condition (4631, 7821, 730, 15307500) converges to the endemic state (4643.728, 7914.77, 741.88, 15307611.167) for deterministic system (shown by red color) in (a) and (b) respectively. However, the stochastic system shows the convergence around the endemic state with large fluctuations as shown in blue color
Time series solutions of symptomatic infected and recovered populations starting from the initial condition (4631, 7821, 730, 15307500) converges to the endemic state (4643.728, 7914.77, 741.88, 15307611.167) for deterministic system (shown by red color) in (a) and (b) respectively. However, the stochastic system shows the convergence around the endemic state with large fluctuations as shown in blue color
Histograms show the distribution of individuals across the (a) susceptible and (b) asymptomatic infected populations, with each bin representing a range of respective population sizes at endemic state. The x-axis represents the population size, while the y-axis represents the frequency of individuals in each bin
7 Parameter estimations
In this section, the parameter estimation has been performed for the model (1). The COVID-19 data of India for three waves has been used to estimate the parameters of the model. The data has been taken from the World Health Organization (WHO) site [38]. Since during all the three waves of COVID-19, the consequences were different. Therefore, parameter estimation has been done for all the three waves separately. The parameters \(\beta _1\), \(\beta _2\), \(\beta _3\), \(\alpha\), \(\mu _1\) and \(\gamma\), k and \(\omega\) have been estimated by using the ’fminsearch’ function from the MATLAB Optimization Toolbox that uses the nonlinear least-squares curve fitting technique. The natural death rate of human population has been taken from [37].
The confirmed cases of the first wave of COVID-19 starting from March 3, 2020, to February 2, 2021 have been considered for the model fitting. In Fig. 17, the COVID-19 cases of first wave have been shown by the blue dots and the fitted curve for model (1) has been shown by solid red line. The good match between the data and the predicted solution of the model is obtained. The estimated values of the model parameters for the first wave of COVID-19 are given in Table 3.
The Fig. 18 shows the fitted solution of the model along with confirmed cases of second wave of COVID-19 started from February 3, 2021 to December 14, 2021. The estimated parameters of the model (1) for the second wave of COVID-19 are given in Table 4. The fitted solution of model (1) and the confirmed second wave of COVID-19 cases have been shown by solid red line and blue dots respectively in Fig. 18. The good match is obtained between the data and the predicted solution of the model.
Further, to estimate the parameters of the model (1) for the third wave of COVID-19, the confirmed cases of third wave of COVID-19 started from December 15, 2021 to January 27, 2023 have been considered for the model fitting. The Fig. 19 shows the fitted solution of model (1) and the confirmed third wave of COVID-19 cases by solid red line and blue dots respectively. The estimated parameters of the third wave of COVID-19 are given in Table 5. The good match is obtained between the data and the predicted solution of the model.
It can be concluded from all the estimated values of parameters for the three waves of COVID-19, the transmission rate for asymptomatic infected population is higher than the symptomatic infected population in first and second wave of COVID-19. However, the transmission rate \(\beta _3\) that is responsible for converting the infection from asymptomatic to symptomatic is higher in all three waves of COVID-19. Thus, most of the time, asymptomatic infection has been converted into the symptomatic infection. Disease-induced death rate was very high in second wave as compared to the first and third COVID-19 wave. That estimate is inline with the fact.
8 Conclusions
In this paper, the non-linear model of COVID-19 has been formulated to study the transmission dynamics of infection in deterministic and stochastic framework. For the deterministic model, three equilibrium states exist. The disease-free state always exists and is found to be globally stable for \(R_0<1\). The asymptomatic infection free state exists for \(R_{01}>1\) and is found to be locally stable for \(R_{01}>R_{10}\). However, for \(R_{10}>1\) and \(R_{10}>R_{01}\), the endemic state exists and is found to be locally stable. At \(R_{10}=R_{01}(=1)\), the stability of the asymptomatic infection free state and the endemic state changes, hence transcritical bifurcation occurs at this point. It can be seen from the expressions of \(R_{10}\) and \(R_{01}\) that \(R_{10}\) is associated with the parameters of asymptomatic infection and \(R_{01}\) is dependent on the parameters of symptomatic infection. It can be concluded that whenever the asymptomatic infection cases increase, COVID-19 infection will persist in the population as the endemic state is stable for \(R_{10}>R_{01}\). However, when symptomatic cases will dominate, COVID-19 infection can be eradicated by increasing the non-pharmaceutical interventions (NPIs). It is also important to note that increasing the rate of NPIs can control COVID-19 infection to some extent.
The environmental noise has been added to the deterministic model to make it more realistic. The stochastic differential equation model has been formulated and solved numerically using the Euler-Maruyama method. Stochastic and the corresponding deterministic solutions have been plotted by using the estimated parameters of second wave of COVID-19. It is found that deterministic and the corresponding stochastic solution converge to their respective states. However, the stochastic solutions are showing large fluctuations around the deterministic solutions due to environmental noise.
Parameter estimations have been done by using the COVID-19 data of three waves of India provided by the official sources. It is found from the estimated values of parameters that the value of parameter \(\beta _3\) is quite high as compared to the symptomatic infection transmission parameter \(\beta _2\) in all three COVID-19 waves. This indicates that most of the symptomatic infections were transformed from the asymptomatic ones which are found true in reality. The parameter estimates from the three waves of COVID-19 are consistent with the actual outcomes. As in second wave, the transmision and disease-induced death rates were quite high as compared to the first and third waves of COVID-19 in India [39, 40].
References
World health organization, coronavirus disease (COVID-19) dashboard. https://covid19.who.int/August 2020 (2020)
J.T. Wu, K. Leung, G.M. Leung, Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. The Lancet 395(10225), 689–697 (2020)
S.S. Musa, D. Gao, S. Zhao, L. Yang, Y. Lou, D. He, Mechanistic modeling of the coronavirus disease 2019 (COVID-19) outbreak in the early phase in Wuhan, China, with different quarantine measures. Acta Math. Appl. 43(2), 350–364 (2020)
H. Nishiura, N.M. Linton, A.R. Akhmetzhanov, Initial cluster of novel coronavirus (2019-nCoV) infections in Wuhan, China is consistent with substantial human-to-human transmission. J. Clin. Med. 9, 488 (2020). https://doi.org/10.3390/jcm9020488
A. Gowrisankar, T.M.C. Priyanka, Santo Banerjee, Omicron: a mysterious variant of concern. Eur. Phys. J. Plus 137, 100 (2022)
Centers for disease control and prevention, coronavirus disease (COVID-19). https://www.cdc.gov/coronavirus/2019-nCoV/index.html August 2020 (2020)
S.E. Eikenberry, M. Mancuso, E. Iboi, T. Phan, K. Eikenberry, Y. Kuang, E. Kostelich, A.B. Gumel, To mask or not to mask: modeling the potential for face mask use by the general public to curtail the COVID-19 pandemic. Infect. Dis. Model. 5, 293–308 (2020). https://doi.org/10.1016/j.idm.2020.04.001
E.A. Iboi, C.N. Ngonghala, A.B. Gumel, Will an imperfect vaccine curtail the COVID-19 pandemic in the US? Infect. Dis. Model. 5, 510–524 (2020). https://doi.org/10.1016/j.idm.2020.07.006
Z. Gao, Y. Xu, C. Sun, X. Wang, Y. Guo, S. Qiu, K. Ma, A systematic review of asymptomatic infections with COVID-19. J. Microbiol. Immunol. Infect. 54(1), 12–16 (2021)
X. Kang, Hu. Ye, Z. Liu, S. Sarwar, Forecast and evaluation of asymptomatic COVID-19 patients spreading in China. Results Phys. 34, 105195 (2022)
D.K. Hazra, B.S. Pujari et al., Modelling the first wave of COVID-19 in India. PLoS Comput. Biol. 18(10), e1010632 (2022)
L.X. Hong, A. Lin, Z.B. He, H.H. Zhao, J.G. Zhang, C. Zhang, L.J. Ying, Z.M. Ge, X. Zhang, Q.Y. Han, Q.Y. Chen, Y.H. Ye, J.S. Zhu, H.X. Chen, W.H. Yan, Mask wearing in pre-symptomatic patients prevents SARS-CoV-2 transmission: an epidemiological analysis. Travel Med. Infect. Dis. 36, 101803 (2020). https://doi.org/10.1016/j.tmaid.2020.101803
A.A. Khan, S. Ullah, R. Amin, Optimal control analysis of COVID-19 vaccine epidemic model: a case study. Eur. Phys. J. Plus 137, 156 (2022)
V.R. Saiprasad, R. Gopal, V.K. Chandrasekar et al., Analysis of COVID-19 in India using a vaccine epidemic model incorporating vaccine effectiveness and herd immunity. Eur. Phys. J. Plus 137, 1003 (2022)
H. Inoue, Y. Todo, Has Covid-19 permanently changed online purchasing behavior? EPJ Data Sci. 12, 1 (2023)
X. Gao, X. Shi, H. Guo, Y. Liu, To buy or not buy food online: the impact of the Covid-19 epidemic on the adoption of e-commerce in China. PLoS ONE 15, 0237900 (2022)
H.A. Adekola, I.A. Adekunle, H.O. Egberongbe, S.A. Onitilo, I.N. Abdullahi, Mathematical modeling for infectious viral disease: the COVID-19 perspective. J. Public Aff. 20(4), e2306 (2020). https://doi.org/10.1002/pa.2306
T.V. Porgo, S.L. Norris, G. Salanti, L.F. Johnson, J.A. Simpson, N. Low, M. Egger, C.L. Althaus, The use of mathematical modeling studies for evidence synthesis and guideline development: a glossary. Res. Synth. Methods 10(1), 125–133 (2019). https://doi.org/10.1002/jrsm.1333
H.W. Hethcote, P. Van den Driessche, Some epidemiological models with nonlinear incidence. J Math. Biol. 29, 271–287 (1991)
J. Hui, L. Chen, Impulsive vaccination of SIR epidemic models with nonlinear incidence rate. Discrete Contin. Dyn. Syst. Ser. B 4, 595–605 (2004)
A. Mishra, B. Ambrosio, S. Gakkhar, M.A. Aziz Alaoui, A network model for control of dengue epidemic using sterile insect technique. Math. Biosci. Eng. 15, 441–460 (2018)
A. Mishra, S. Gakkhar, The effects of awareness and vector control on two strains dengue dynamics. Appl. Math. Comput. 246, 159–167 (2014)
A. Mishra, S. Gakkhar, Non-linear dynamics of two-patch model incorporating secondary dengue infection. Int. J. Appl. Comput. Math. 4, 19 (2018). https://doi.org/10.1007/s40819-017-0460-z
J.P. Arcede, R.L. Caga-anan, C.Q. Mentuda, Y. Mammeri, Accounting for symptomatic and asymptomatic in a SEIR-type model of COVID-19. Math. Model. Nat. Phenom. 15, 34 (2020)
M. Serhani, H. Labbardi, Mathematical modeling of COVID-19 spreading with asymptomatic infected and interacting peoples. J. Appl. Math. Comput. 66, 1–20 (2021)
Z. Ali, F. Rabiei, M.M. Rashidi, T. Khodadadi, A fractional-order mathematical model for COVID-19 outbreak with the effect of symptomatic and asymptomatic transmissions. Eur. Phys. J. Plus 137, 395 (2022)
D. Olabode, J. Culp, A. Fisher, A. Tower, D. Hull-Nye, X. Wang, Deterministic and stochastic models for the epidemic dynamics of COVID-19 in Wuhan, China. Math. Biosci. Eng. 18(1), 950–967 (2021)
S. Hussain, E.N. Madi, H. Khan, S. Etemad, S. Rezapour, T. Sitthiwirattham, N. Patanarapeelert, Investigation of the stochastic modeling of COVID-19 with environmental noise from the analytical and numerical point of view. Mathematics 9, 3122 (2021)
S. Bugalia, V.P. Bajiya, J.P. Tripathi, M.T. Li, G.Q. Sun, Mathematical modeling of COVID-19 transmission: the roles of intervention strategies and lockdown. Math. Biosci. Eng. 17(5), 5961–5986 (2020). https://doi.org/10.3934/mbe.2020318
N. Anggriani, M.Z. Ndii, R. Amelia, W. Suryaningrat, M.A.A. Pratama, A mathematical COVID-19 model considering asymptomatic and symptomatic classes with waning immunity. Alex. Eng. J. 61(1), 113–124 (2022)
P. Driessche, J. Watmough, Reproduction numbers and sub-threshold endemic equilibria for compartmental models of disease transmission. Math. Biosci. 180, 29–48 (2002)
J.C. Kamgang, G. Sallet, Global asymptotic stability for the disease free equilibrium for epidemiological models. C. R. Math. 341, 433–438 (2005)
E.J. Allen, L.J.S. Allen, A. Arciniega, P. Greenwood, Construction of equivalent stochastic differential equation models. Stoch. Anal. Appl. 26, 274–291 (2008)
E.J. Allen, Stochastic differential equations and persistence time for two interacting populations. Dyn. Contin. Discret. Impulse Syst. A Math. Anal. 5, 271–281 (1999)
L.J.S. Allen, An introduction to stochastic processes with applications to biology, 2nd edn. (CRC Press, Boca Raton, 2010)
B. Øsendal, Stochastic differential equations: an introduction with applications, 6th edn. (Springer, Berlin, 2003)
Worldometer, COVID-19 Coronavirus Pandemic (2020) https://www.worldometers.info/coronavirus/countries. Accessed 12 May 2020
P. Asrani, M. S. Eapen, M. I. Hassan, S. S. Sohal, Implications of the second wave of COVID-19 in India. Lancet Respir Med. 9(9), e93–e94 (2021). https://doi.org/10.1016/S2213-2600(21)00312-X
S. Singh, A. Sharma, A. Gupta, M. Joshi et al., Demographic comparison of the first, second and third waves of COVID-19 in a tertiary care hospital at Jaipur, India. Lung India 39(6), 525–531 (2022). https://doi.org/10.4103/lungindia.lungindia_265_22
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Awasthi, A. A mathematical model for transmission dynamics of COVID-19 infection. Eur. Phys. J. Plus 138, 285 (2023). https://doi.org/10.1140/epjp/s13360-023-03866-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1140/epjp/s13360-023-03866-w