Abstract
O-specific polysaccharides have been isolated from lipopolysaccharides of the Azospirillum zeae N7, Azospirillum melinis TMCY 0552, and Azospirillum palustre B2 type bacterial strains. Using one- and two-dimensional 1H- and 13C NMR spectroscopy and sugar analysis including the determination of the absolute configurations of monosaccharides, it has been found that the isolated polysaccharides consist of branched tetrasaccharide repeating units of the following structure: →3)-α-L-Rhap2OAc-(1→2)-[β-D-Glcp-(1→3)]-α-L-Rhap-(1→3)-α-L-Rhap-(1→, which has been previously described for a number of Azospirillum strains assigned to serogroup III. Functions of genes responsible for the biosynthesis of O-antigens have been identified by comparison with the sequences presented in the available databases, and a high level of their homology has been shown.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
INTRODUCTION
Gram-negative alpha-proteobacteria of the Azospirillum genus are widely distributed in associations with wild and cultivated cereals in various climatic zones [1]. The Azospirillum species were first described in 1925 but became widely known only after the rediscovery in the 1970s in Brazil [2]. This discovery became a cornerstone in the study of the phenomenon of associativity and gave impetus to the development of this branch of science. Over 40 years of studies of plant-microbial associations involving azospirilla, ideas about their growth-stimulating effect have evolved from the Additive Hypothesis, i.e., the ability to fix nitrogen and produce phytohormones, to the Multiple Mechanisms Hypothesis., which includes in addition enhanced water and mineral uptake, mitigation of biotic and abiotic stressors, and biocontrol of pathogens [1, 3]. To date, the Azospirillum genus includes 22 species [4], most of which are rhizospheric. However, recently it has been increasingly reported that new species have been isolated from ecological niches uncharacteristic of azospirilla, e.g., sulfide and thermal springs, used pavement, and peat bogs [5]. The high adaptive potential of these bacteria is explained by the redundancy and plasticity of their genome and the high proportion of genes introduced by horizontal transfer [6].
As the most studied among the bacteria, which stimulate plant growth and development, the Azospirillum species are part of biofertilizers and are widely used in several South American countries. Application of azospirilla leads to a significant increase (by 5–30%) in cereal yields in 60–70% of field experiments [3, 7]. To minimize undesirable effects during inoculation with azospirilla, it is necessary to take into account some factors including the state of the native microflora, the level of mineral nutrition of the soil, the variability of plant varieties, and the characteristics of the inoculate strains used in terms of the growth-stimulating effect on plants [8]. Taking strain variability into account, the expansion of fundamental knowledge about the molecular mechanisms of associative interaction of plants and azospirilla is necessary to increase the efficiency of their use in agriculture.
It is known that the initial stages of association formation, such as cell attachment, adsorption, and formation of biofilms on the surface of roots, occur with the participation of glycopolymers, which form the surface of bacterial cells. These glycopolymers are capsular polysaccharides and lipopolysaccharides (LPSs) [9]. LPS, the main structural component of the outer membrane of gram-negative bacteria, is also found in extracellular polymer substances. In the azospirilla culture medium, LPS is in the form of a lipopolysaccharide-protein complex (LPPC) [9], which can be used by bacteria as a carbon source under starvation conditions [10]. The ability of LPS from azospirilla to induce deformation of root hairs [10], enhance peroxidase activity, generate hydrogen peroxide, and increase the length and weight of roots in wheat seedlings has been shown [11]. Their positive effect on the morphogenicity of callus and the yield of explant plants has also been demonstrated [12].
LPS is an amphiphilic macromolecule, which consists of three covalently bound domains, i.e., a hydrophobic lipid A, a hydrophilic polysaccharide including a core oligosaccharide, and an O-specific polysaccharide (OPS) (O-antigen). The diversity of monosaccharides in OPS in combination with various types of bonds between them provides almost limitless possibilities for the structural multiplicity of these biopolymers, which leads to serological variability of strains of the same species. In recent years, more than 20 types of the OPS repeating units have been found for representatives of seven Azospirillum species: A. brasilense, A. lipoferum [13], A. halopraeferens [14], A. dobereinerae [15], A. fermentarium [16], A. formosense [17], and A. rugosum [18]. Most of the OPSs are branched heteropolysaccharides with the exception of A. baldaniorum Sp245, a serologically related group of strains and A. doebereinerae and A. fermentarium type strains. The regularity of the OPS structure of some Azospirillum strains is masked by the presence of several types of repeating units and irregular methylation and acetylation of monosaccharide residues, which makes it difficult to create chemotypic classification schemes based on the OPS structure. In most cases, type Azospirillum strains are characterized by the presence of a unique OPS structure. The exceptions are the A. baldaniorum Sp245(T) strain, which show the structural relationship of OPS with some strains of A. lipoferum, A. brasilense [13], and the A. rugosum DSM 19657(T) (the latter contains two O-specific polysaccharides previously found in A. brasilense Jm125A2) [18].
The biosynthesis of O-antigens has been studied in detail with an example of enterobacteria. The genes of these bacteria, which are responsible for the synthesis of OPS, i.e., the genes for the synthesis of nucleotide precursors of monosaccharides, the genes of glycosyltransferases, and the genes for transmembrane transfer and polymerization of O-units, are generally found on the chromosome as an O-antigen gene cluster [19]. The OPS assembly and synthesis occur in three known pathways, i.e., the Wzx/Wzy-dependent, ABC transporter, or synthase pathways [19]. The structure of the gene cluster responsible for O-antigen biosynthesis has not yet been reported for the Azospirillum species.
The goal of this work is to study the structure of O-antigens of type strains of previously unexplored species of A. zeae N7(T) [20], A. melinis TMCY 0552(T) [21], and A. palustre B2(T) [5] and to analyze the genes involved in the biosynthesis of their OPSs.
RESULTS AND DISCUSSION
The A. zeae N7(T), A. melinis TMCY 0552(T) and A. palustre B2(T) strains were chosen from the Collection of microbial cultures of the IBPPM RAS based on screening of serological specificity of LPS extracts of previously unexplored Azospirillum species. The chosen strains demonstrated immunochemical cross-reaction with antisera to LPPC of the A. lipoferum Sp59b strain. The surface glycopolymers of these strains contain epitopes, which cause serological cross-reaction with the A. lipoferum Sp59b strain. This fact allowed one to attribute the above strains to serogroup III, whose representatives are characterized by the presence of the →3)-α-L-Rhap-(1→3)-α-L-Rhap-(1→2)-α-L-Rhap-(1→3) fragment in OPS [13].
We performed a detailed immunochemical analysis of the LPS samples isolated from the dry biomass of the studied bacteria by water-phenolic extraction. The double radial immunodiffusion test showed the fusion of precipitation bands of antibodies to LPPC of the A. lipoferum Sp59b strain with homologous and studied antigens (Fig. 1a) and the absence of the interaction with LPSs of A. baldaniorum Sp245, A. brasilense Sp7, A. brasilense Jm6B2, and A. brasilense SR80 strains. Interstrain differences in the intensity of the antigen–antibody interaction were observed by ELISA. However, the tendency of the interaction of LPSs of the studied strains with antibodies was similar to that of the homologous antigen (Fig. 1b).
The result of double radial immunodiffusion of the A. palustre B2 (1), A. melinis TMCY 0552 (2), A. zeae N7 (3), and A. lipoferum Sp59b (4) lipopolysaccharide samples with antibodies to lipopolysaccharide-protein complex of A. lipoferum Sp59b (5) (a). The result of enzyme immunoassay of lipopolysaccharide samples of the studied strains with antibodies to the lipopolysaccharide-protein complex of A. lipoferum Sp59b (b).
SDS-PAAG electrophoretic analysis of isolated LPS samples and subsequent staining with silver nitrate demonstrated the prevalence of OPS-containing fractions, which were observed in the upper part of the track, and the presence of highly mobile fractions that contained the core and lipid A in the lower part of the track (Fig. 2). Unlike LPSs of the gamma-proteobacterium Pseudomonas putida TSh-18, which were a mixture of molecules with a wide range of molecular weight differing by one repeating unit, LPSs of azospirilla showed a predominance of LPS fractions with the molecular weight of 20–25 kDa. The presence of high-molecular-weight LPSs in the studied strains indicates the predominance of molecules in the S-forms. Therefore, identical or similar antigenic determinants that cause the intersection with the Sp59b strain can be localized in their OPSs.
We analyzed the composition and physicochemical properties of LPSs and the structure of OPSs of the studied strains to identify the chemical nature of the serological cross-reaction. The GLC analysis of the composition of LPS fatty acids after obtaining the corresponding methyl esters revealed the predominance of 3-hydroxytetradecanoic and 3-hydroxyhexadecanoic acids in all samples (more than 70% of the sum of all identified derivatives). Hexadecanoic, hexadecenoic, and octadecenic acids were also present. Taking into account the conservatism of the structure of lipid A within the bacterial genus, the fatty acid profile of the studied strains was consistent with the previous data for LPS of representatives of other Azospirillum species [15–18].
The LPS samples can form supramolecular complexes (micelles) in aqueous solutions due to the amphiphilic nature. The main driving force of LPS self-aggregation is the hydrophobic interaction between the acyl chains of lipid A. The size of micelles is influenced by the lipid A and OPS structures and the ratio of these components in the LPS sample [22]. Thus, with the similarity of the structure of individual structural components, LPSs can differ significantly in functional activity because they aggregate in aqueous solutions in different ways. The DLS measurement of the size and ζ-potential of micelles, which were generated from the LPS molecules of A. zeae N7(T) and A. palustre B2(T) at a concentration of 2 mg/mL at 37°C in an aqueous medium revealed that both samples formed negatively charged micelles with a size of 27.5 nm and 41.0 nm, respectively, (Table 1). The light scattering intensity (I) of the LPS solution of A. palustre was 1.5 times higher than that for LPS of A. zeae. The electrophoretic analysis did not reveal significant differences in the degree of OSP polymerization of the studied strains. Therefore, the observed differences in the size of their LPS micelles may be caused by the microheterogeneity of the lipid A structure (the ratio of forms with varying degrees of acylation). The evaluation of the relative concentration (Nrel) and the relative mass-volume concentration (Crel) by the formulas described earlier [23] showed that under the studied conditions, the number of micelles formed by LPSs of A. palustre B2(T) and the number of LPSs involved in micelle formation are significantly lower than the number of LPSs of A. zeae N7(T).
OPSs of the studied strains were obtained by mild acid hydrolysis of LPSs followed by gel filtration. Using the GLC method, we analyzed the monosaccharide composition by the content of polyol acetates, which were formed after complete acid hydrolysis of all samples. We managed to identify the presence of Rha and Glc in the OPS composition in a ratio of 3 : 1 (detector response). The GLC analysis of acetylated (S)-2-octyl glycosides allowed for determining the D-configuration of Glc and the L-configuration of Rha residues.
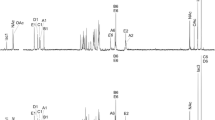
The OPS structure of the studied strains was established using 1D and 2D 1H and 13C NMR spectroscopy. The 1H and 13C NMR spectra of OPSs of the studied strains were almost identical (Fig. 3), which indicated the structural similarity of O-antigens.
The 1H NMR spectrum contained five signals in the low field region at 4.63–5.24 ppm, signals of the methyl groups at 1.26–1.32 ppm, a signal of the O-acetyl group at 2.21 ppm, and signals of protons of monosaccharide rings at 3.31–4.39 ppm. The 13С NMR spectrum contained signals of four anomers at 99.8–105.3 ppm, rhamnose methyl groups at 17.8–18.0 ppm, the O-acetyl group at 22.2 ppm (СН3) and 175.5 ppm (СО), the СН2ОН group at 62.2 ppm, and monosaccharide rings in the region of 70.3–81.3 ppm. The absence of the signals of the monosaccharide rings in the range of 83–88 ppm in the spectrum indicated the pyranose form of monosaccharide residues [24].
The signals in the 1H and 13C NMR spectra were assigned using 2D NMR spectra (homonuclear 1H, 1H COSY, TOCSY, and ROESY experiments and heteronuclear 1H, 13C HSQC and HMBC experiments). Chemical shifts of the signals of the monosaccharide residues are presented in Table 2. Using the intraresidue H, H and H, C correlations and the spin–spin coupling constants 3JH,H, we identified spin–spin systems of four monosaccharides, i.e., A, B, and C with the manno configuration and D with the gluco configuration. The TOCSY spectrum demonstrated the presence of Н1/Н2 and Н2/Н3–Н-6 cross-peaks for the A–C residues and Н1/Н2–H-6 cross-peaks for the D residue. The signals within each spin–spin system were assigned using the COSY spectra.
Abbreviations: LPPC, lipopolysaccharide-protein complex; LPS, lipopolysaccharide; OSP, O-specific polysaccharide.
Н and 13С NMR spectra of О-specific polysaccharide of A. zeae N7(T) (chemical shifts, ppm)The alpha configuration of the A–C residues and the beta configuration of the D residue were determined based on the characteristic chemical shifts of the C5 signals when compared with the literature data [24, 25].
The positions of substitution of monosaccharides were established by the shift in the low field of the C2 and C3 signals of the A residue and the C3 signal of the B and C residues compared with the corresponding unsubstituted monosaccharides [24, 25]. Chemical shifts of C2–C6 of the D residue were close to those of O-methyl-β-Glcp [24] and indicated that the D residue occupied a terminal position in the side chain. The monosaccharide sequence was determined by the ROESY spectra, which demonstrated interresidue correlations between anomeric protons and protons at the linkage carbons: А Н1/В Н3 at δ 5.11/3.74, B H1/C H3 at δ 5.03/4.04, C H1/А H2 at δ 5.24/4.39, and D H1/A H3 at δ 4.63/4.03. We observed the corresponding correlations in the 1Н, 13С HMBC spectra between anomeric protons and carbon atoms at the glycoside bond: А Н1/В С3 at δ 5.11/78.6, B H1/C С3 at δ 5.03/77.0, C H1/А С2 at δ 5.24/78.5, and D H1/A С3 at δ 4.63/81.3.
Our results allowed us to identify the structure of the OPS repeating units in the studied microorganisms, which consisted of three rhamnose residues in the main chain and the glucose residue in the side chain (Fig. 4). The degree of acetylation of the C residue (~75%) was calculated using the integral intensity of the signals of the anomeric proton of the C residue that contained or did not contain the acetyl group (δH 5.13 ppm [26]). This structure of the OPS repeating units is common among azospirilla of serogroup III, for which a different degree of acetylation of the Rha residue has been demonstrated [13]. The observed similarity of the O-antigen structure of different bacterial species can serve as an indirect confirmation of widespread horizontal gene transfer.
To find the relationship of the genes involved in the biosynthesis of O-antigens of the studied strains, we searched for the genes of the L-Rha biosynthesis available in the NCBI database of sequences of the complete azospirila genomes. We have revealed sections of the genomes and arbitrarily designated them as gene clusters or loci that contain genes for the biosynthesis of nucleotide-activated precursors of L-Rha and glycosyltransferase and the ABC transporter genes involved in O-antigen processing. Plasmid localization of the genes of Azospirillum O-antigen biosynthesis may indicate their appearance in the genome because of the horizontal transfer process as well as their involvement in this process, i.e., gene transfer between strains within the same species, different species, genera, and families. The analysis of the closest protein homologs of the O-antigen biosynthesis demonstrated their presence in bacteria of the Rhodospirillaceae, Rhodobacteraceae, and Phyllobacteriaceae families of α-proteobacteria and the Nitrospira family (Table 3). It should be noted that all these bacteria are representatives of either soil microflora or marine ecosystems.
The corresponding loci of the A. oryzae KACC 14407, A. lipoferum Sp59b, and A. palustre B2 strains (the identity of repeating units of surface glycans for the last two strains has been found, and the structure of A. oryzae OPS has not yet been established) demonstrated a high similarity of organization and considerable difference from the cluster of the A. brasilense Sp7 strain (Fig. 5). The analyzed genes were sequentially located on the antisense DNA strands within the detected loci of A. oryzae KACC 14407, A. lipoferum Sp59b, and A. palustre B2 strains. The genes of L-Rha biosynthesis and OPS processing (wzm and wzt) in the A. brasilense Sp7 strain were significantly remote from each other, and open reading frames with unspecified functions and the genes of several hypothetical glycosyltransferases were located between them. The genes of two glycosyltransferases and the wzm gene had the opposite reading direction.
Comparative analysis revealed a high degree of homology of the L-Rha rfbA–rfbD biosynthesis genes and wzm and wzt processing genes (93.5–96.6%) in the A. oryzae KACC 14407, A. lipoferum Sp59b, and A. palustre B2 strains. For comparison, the degree of homology of the functionally related L-Rha rmbA-rmbD biosynthesis genes and wzm and wzt processing genes for the A. brasilense Sp7 type strain was 69.7–77.8%.
Pairwise alignment of the nucleotide sequences of the glycosyltransferase genes included in the gene cluster of the L-Rha synthesis of the A. oryzae KACC 14407, A. lipoferum Sp59b, and A. palustre B2 strains did not show significant similarity with the corresponding genes of the A. brasilense Sp7 strain because of the different OPS structure of these strains and, accordingly, the different specificity of the enzymes responsible for the O-unit assembly. The search for the genes with the maximum degree of similarity for these glycosyltransferases by the megaBLAST algorithm showed that the A. oryzae KACC 14407 strain had the highest degree of homology (95.3%) with the similarly annotated sequence of A. thiophilum BV-S (CP012407.1), whereas the nucleotide sequences for A. lipoferum Sp59b and A. palustre B2 were closest to that for Azospirillum sp. TSH100 glycosyltransferase (CP039640.1) (96.3 and 96.2%, respectively).
Thus, the analysis of the structure of the gene clusters of the L-Rha synthesis for four Azospirillum strains has allowed for the conclusion that the corresponding genes are orthologous, and the A. oryzae KACC 14407, A. lipoferum Sp59b, and A. palustre B2 strains show slight variability in contrast to the more evolutionarily distant A. brasilense Sp7 strain. This fact may indicate the acquisition of these genes during horizontal transfer. The structure of the locus of the A. oryzae KACC 14407 strain and its similarity to that of the loci of the A. lipoferum Sp59b and A. palustre B2 strains suggests that OSP of the A. oryzae strain contains a polysaccharide identical with tetrasaccharide units established in this work. To confirm this assumption, it is necessary to perform appropriate immunochemical tests or analysis of the OPS structure by chemical and physicochemical methods.
EXPERIMENTAL
Cultivation of bacteria. The A. melinis TMCY 0552 (IBPPM 547), A. zeae N7 (IBPPM 550), and A. palustre B2(T) (IBPPM 633) strains are provided by the Collection of Rhizosphere Microorganisms of the Institute of Biochemistry and Physiology of Plants and Microorganisms of the Russian Academy of Sciences (Saratov, Russia). Bacteria were cultured in a liquid malate-salt medium with vitamins [27] at a temperature of 30°С and stirring on a vibrating stand until the end of the exponential growth phase. The cells were precipitated by centrifugation, resuspended in 0.15 M NaCl solution, and the capsule material was washed off the surface by mechanical stirring for five days with a daily change of the washing solution.
Isolation of LPS and OPS. LPSs were isolated from acetone-dried capsule-free cells with a hot 45% aqueous phenol solution without layer separation [28]. Protein impurities were precipitated from the LPS solution by the addition of 40% CCl3COOH to the final pH value of 2.7. The solutions were dialyzed against distilled water, concentrated on a Laborota 4000 rotary evaporator (Heidolph, Germany), and lyophilized on a Bench Top VirTis lyophilizer (United States). LPSs were degraded with 2% CH3COOH at 100°С for 4 h. OPSs from the supernatant were separated by gel chromatography on a column with Sephadex G-50 Fine (GE Healthcare, United States) in 0.025 M pyridine acetate buffer with the detection on a differential flow refractometer (Knauer, Germany). The fraction of O-specific polysaccharide of a high molecular weight was concentrated and lyophilized.
Electrophoresis of LPS samples was performed in 13.5% SDS-PAAG [29]. The components were visualized by staining the gels with a silver nitrate dye [30].
Dynamic light scattering in the solutions of lyophilized LPS samples in deionized water (Milli-Q) at a concentration of 2.0 mg/mL was evaluated using a Malvern Nano-ZS unit (Malvern, Great Britain) in plastic four-sided cuvettes (10 mm) (Sarstedt, Germany). Measurements were carried out at 37°C with fixed focusing of a helium–neon laser (λ = 633 nm in vacuum) in the center of the cuvette (4.65 mm) and a constant diameter of the diaphragm (mounted attenuator, 7). The light scattering intensity at an angle of 173° (the photon count rate, kcps) and the correction function of the scattering intensity fluctuations over time were evaluated. These data were used to assess the most probable modal hydrodynamic diameter (dm) of micelles. The relative values of the numerical concentration (Nrel) and the mass–volume concentration (Srel) of dispersed biopolymer compounds were evaluated by the equation from [23]. The ζ-potential of LPS micelles (2.0 mg/mL) was measured at 37°C using the Malvern Nano-ZS system (Malvern, Great Britain). The measurements were carried out with the default settings recommended by the manufacturer.
Immunochemical studies of LPSs were carried out using polyclonal rabbit antibodies to LPPC of A. lipoferum Sp59b by the double radial immunodiffusion [31] and solid-phase enzyme immunoassay (ELISA) methods. The precipitate in immunodiffusion was stained with Coomassie blue R 250. The interaction of antigens and antibodies in ELISA was detected in polystyrene 96-well plates using goat antirabbit antibodies conjugated with horseradish peroxidase with the addition of hydrogen peroxide and o‑phenylenediamine. The optical density of the studied samples was measured at a wavelength of 490 nm using a Tescan enzyme immunoassay analyzer (Thermo Fisher Scientific, United States).
The analysis of the monosaccharide composition and absolute configurations of sugars after hydrolysis of OPSs with 2M CF3COOH (120°С, 2 h) was carried out by GLC of polyol acetates [32] and acetylated 2‑(S)-octyl glycosides [33] on a Hewlett-Packard 7820A chromatograph on an HP-5 capillary column (Hewlett-Packard, United States) (temperature gradient, from 160°С (1 min) to 290°С; heating rate, 7°C/min).
The composition of fatty acids of LPSs in the form of their methyl esters was evaluated using a GLC-2010 chromatograph (Shimadzu, Japan) equipped with a DB-5 column (Agilent, United States). Methylation was performed by the method described in [34].
NMR spectra were recorded on a DRX-600 spectrometer (Bruker, Germany) in 99.96% D2O at 30°С with trimethylsilylpropanoate-d4 as the internal standard (δC, –1.6; δH, 0.0). The samples were lyophilized twice from 99.9% D2O. Two-dimensional spectra were recorded using standard mathematical software from Bruker (Germany); the TOPSPIN 2.1 program was used for data collection and processing. In the TOCSY and ROESY experiments, the mixing time was 150 and 200 ms, respectively.
Analysis of genes of biosynthesis of O-antigens. The genes of the L-Rha biosynthesis were extracted from the genome-wide sequences of A. brasilense Sp7 (GenBank: AH013753.2), A. oryzae KACC 14407 (CP054615.1), A. lipoferum Sp59b (VTTN01000010.1), and the available preliminary data of the full-genome sequence of A. palustre B2 (GCF_002573965.1 (ASM257396v1 assembly). The functions of the identified gene sequences have been predicted by aligning the corresponding and known protein sequences from GenBank, which are involved in the O-antigen biosynthesis in other bacteria, using the BLASTn analysis [35]. The three- and four-letter designations of the A. brasilense Sp7 genes correspond to the GenBank annotation. Three-letter (wzm and wzt) and four-letter designations (rfbA–rfbD) were assigned to the genes of A. oryzae KACC 14407, A. lipoferum Sp59b, and A. palustre B2 in accordance with their annotations and the results of pairwise alignments of their nucleotide sequences. The image of the gene clusters of the studied Azospirillum strains was determined using the Easyfig visualizer version 2.2.5 [36]. The homology of the nucleotide sequences of the genes was evaluated by pairwise alignments of the corresponding sequences using the BLASTn program.
CONCLUSIONS
The glycans of the cell surface of rhizobacteria play an important role at all stages of the existence of a cell population, both during life in the soil and rhizosphere and the development of symbiotic relationships with plants. Lipopolysaccharides are structural components of the bacterial cell wall, which can be exported to the environment. Polysaccharides are involved in the formation of the outer layer of the cell surface of gram-negative bacteria, thus protecting the cell from the adverse effects of the extracellular environment. In the case of symbiotic microorganisms, they play an important role in the interaction with eukaryotic cells of the host organism.
Progress in studying the structural features of the LPS compositions (including their OPSs) of gram-negative bacteria is largely provided by their role in the development of pathophysiological processes, which accompany bacterial infections of humans and animals. Lipopolysaccharides trigger the immune response of animals and humans and are recognized by their antibodies. This property of LPSs is successfully used in clinics to identify and classify pathogenic bacteria. Collections of O-antigen-specific antisera are used to classify gram-negative organisms in serological testing, which is also effective for nonpathogenic microorganisms, e.g., soil diazotrophs of the Azospirillum genus.
Nucleotide sequences of gene clusters of the O‑antigen biosynthesis can be used as genetic markers for identification of E. coli strains [37]. Bacteria of the Azopirillum genus have not been studied sufficiently in this regard. In this paper, we analyzed the OPS structures of representatives of three Azospirillum species, which were previously unexplored in this regard, and identified gene clusters in their genomes responsible for the biosynthesis of O-antigens with a high level of identity. Our approach can be very efficient for further molecular serodiagnosis of azospirilla by gene clusters of their O-antigens, given the fact that the phenomenon of molecular mimicry is very characteristic of representatives of this genus [13]. It should be noted that the identity of the O-antigen structures does not lead to unification of the surface properties of these microorganisms probably due to the variety of exposed biomacromolecules or because of differences in the micelle formation of amphiphilic LPS molecules in aqueous solution, which we revealed in this study.
Notes
Abbreviations: LPPC, lipopolysaccharide-protein complex; LPS, lipopolysaccharide; OSP, O-specific polysaccharide.
REFERENCES
Cassán, F., Coniglio, A., López, G., Molina, R., Nievas, S., Le Noir, ., de Carlan, C., Donadio, F., Torres, D., Rosas, S., Olivera, P.F., de Souza, E., Díaz Zorita, M., de-Bashan, L., and Mora, V., Biol. Fertil. Soils, 2020, vol. 56, pp. 461–479. https://doi.org/10.1007/s00374-020-01463-y
Döbereiner, J., Marriel, I.E., and Nery, M., Can. J. Microbiol., 1976, vol. 22, pp. 1464–1473. https://doi.org/10.1139/m76-217
Bashan, Y. and Bashan, L.E., Adv. Agron., 2010, vol. 108, pp. 77–136. https://doi.org/10.1016/S0065-2113(10)08002-8
Genus Azospirillum, in List of Prokaryotic Names with Standing in Nomenclature (LPSN). https://bacterio.net/genus/azospirillum.
Tikhonova, E.N., Grouzdev, D.S., and Kravchenko, I.K., Int. J. Syst. Evol. Microbiol., 2019, vol. 69, pp. 2787–2793. https://doi.org/10.1099/ijsem.0.003560
Wisniewski-Dyé, F., Borziak, K., Khalsa-Moyers, G., Alexandre, G., Sukharnikov, L.O., Wuichet, K., Hurst, G.B., McDonald, W.H., Robertson, J.S., Barbe, V., Calteau, A., Rouy, Z., Mangenot, S., Prigent-Combaret, C., Normand, P., Boyer, M., Siguier, P., Dessaux, Y., Elmerich, C., Condemine, G., Krishnen, G., Kennedy, I., Paterson, A.H., González, V., Mavingui, P., and Zhulin, I.B., PLoS Genet., 2011, vol. 7, art. ID e1002430. https://doi.org/10.1371/journal.pgen.1002430
Bomfim, C.A., Coelho, L.G.F., Vale, H.M.M., de Carvalho, MendesI., Megias, M., Ollero, F.J., Dos, Reis., and Junior, F.B., Braz. J. Microbiol., 2021, vol. 52, pp. 2215–2232. https://doi.org/10.1007/s42770-021-00618-9
Cassán, F. and Diaz-Zorita, M., Soil Biol. Biochem., 2016, vol. 103, pp. 117–130. https://doi.org/10.1111/j.1574-6968.1998.tb13150.x
Skvortsov, I.M. and Ignatov, V.V., FEMS Microbiol. Lett., 1998, vol. 165, pp. 223–229. https://doi.org/10.1111/j.1574-6968.1998.tb13150.x
Sigida, E.N., Fedonenko, Y.P., Shashkov, A.S., Toukach, P.V., Shelud’ko, A.V., Zdorovenko, E.L., Knirel, Y.A., and Konnova, S.A., Int. J. Biol. Macromol., 2019, vol. 126, pp. 246–253. https://doi.org/10.1016/j.ijbiomac.2018.12.229
Hernánde-Esquivel, A.A., Castro-Mercado, E., and García-Pineda, E., J. Plant Growth Regul., 2021, vol. 40, pp. 1903–1911. https://doi.org/10.1007/s00344-020-10241-x
Tkachenko, O.V., Burygin, G.L., Evseeva, N.V., Fedonenko, Y.P., Matora, L.Y., Lobachev, Y.V., and Shchyogolev, S.Y., Plant Cell Tiss. Organ Cult., 2021, vol. 147, pp. 147–155.
Fedonenko, Y.P., Sigida, E.N., Konnova, S.A., and Ignatov, V.V., Russ. Chem. Bull., 2015, vol. 64, pp. 1024–1031. https://doi.org/10.1007/s11172-015-0971-x
Sigida, E.N., Fedonenko, Y.P., Shashkov, A.S., Arbatsky, N.P., Zdorovenko, E.L., Konnova, S.A., Ignatov, V.V., and Knirel, Y.A., Beilstein J. Org. Chem., 2016, vol. 12, pp. 636–642. https://doi.org/10.3762/bjoc.12.62
Sigida, E.N., Fedonenko, Y.P., Shashkov, A.S., Zdorovenko, E.L., Konnova, S.A., and Knirel, Y.A., Carbohydr. Res., 2019, vol. 478, pp. 54–57. https://doi.org/10.1016/j.carres.2019.04.009
Sigida, E.N., Fedonenko, Y.P., Shashkov, A.S., Konnova, S.A., and Ignatov, V.V., Carbohydr. Res., 2018, vol. 465, pp. 40–43. https://doi.org/10.1016/j.carres.2018.06.003
Sigida, E.N., Shashkov, A.S., Zdorovenko, E.L., Konnova, S.A., and Fedonenko, Y.P., Carbohydr. Res., 2020, vol. 494, art. ID 108060. https://doi.org/10.1016/j.carres.2020.108060
Sigida, E.N., Kokoulin, M.S., Dmitrenok, P.S., Grinev, V.S., Fedonenko, Y.P., and Konnova, S.A., Russ. J. Bioorg. Chem., 2020, vol. 46, pp. 60–70. https://doi.org/10.1134/S1068162020010112
Samuel, G. and Reeves, P., Carbohydr. Res., 2003, vol. 338, pp. 2503–2519.
Mehnaz, S., Weselowski, B., and Lazarovits, G., Int. J. Syst. Evol. Microbiol., 2007, vol. 57, pp. 2805–2809. https://doi.org/10.1099/ijs.0.65128-0
Peng, G., Wang, H., Zhang, G., Hou, W., Liu, Y., Wang, E.T., and Tan, Z., Int. J. Syst. Evol. Microbiol., 2006, vol. 56, pp. 1263–1271. https://doi.org/10.1099/ijs.0.64025-0
D’Errico, G., Silipo, A., Mangiapia, G., Vitiello, G., Radulescu, A., Molinaro, A., Lanzetta, R., and Paduano, L., Phys. Chem. Chem. Phys., 2010, vol. 12, pp. 13574–13585. https://doi.org/10.1039/c0cp00066c
Burygin, G.L., Sigida, E.N., Fedonenko, Y.P., Khlebtsov, B.N., and Shchyogolev, S.Y., Biophysics (Moscow), 2016, vol. 61, pp. 547–557. https://doi.org/10.1134/S0006350916040059
Bock, K. and Pedersen, C., Adv. Carbohydr. Chem. Biochem., 1983, vol. 41, pp. 27–66.
Lipkind, G.M., Shashkov, A.S., Knirel, Y.A., Vinogradov, E.V., and Kochetkov, N.K., Carbohydr. Res., 1988, vol. 175, pp. 59–75. https://doi.org/10.1016/0008-6215(88)80156-3
Choma, A., Komaniecka, I., and Sowinski, P., Carbohydr. Res., 2009, vol. 344, pp. 936–939. https://doi.org/10.1016/j.carres.2009.02.021
Konnova, S.A., Makarov, O.E., Skvortsov, I.M., and Ignatov, V.V., FEMS Microbiol. Lett., 1994, vol. 118, pp. 93–99.
Westphal, O. and Jann, K., Methods Carbohydr. Chem., 1965, vol. 5, pp. 83–91.
Hitchcock, P.J. and Brown, T.M., J. Bacteriol., 1983, vol. 154, pp. 269–277.
Tsai, C.M. and Frasch, C.E., Anal. Biochem., 1982, vol. 119, pp. 115–119.
Ouchterlony, O. and Nilsson, L.-A., in Handbook of Experimental Immunology, Weir, D.M., Ed., Oxford: Blackwell, 1978, pp. 19.16–19.23.
Sawardeker, J.S., Sloneker, J.H., and Jeanes, A., Anal. Chem., 1965, vol. 37, pp. 1602–1603.
Leontein, K., Lindberg, B., and Lonngren, J., Carbohydr. Res., 1978, vol. 62, pp. 359–362.
Mayer, H., Merkofer, T., Warth, C., and Weckesser, J., J. Endotox. Res., 1996, vol. 3, pp. 345–352. https://doi.org/10.1177/096805199600300409
Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J., Nucleic Acids Res., 1997, vol. 25, pp. 3398–3402.
Sullivan, M.J., Petty, N.K., and Beatson, S.A., Bioinformatics, 2011, vol. 27, pp. 1009–1010.
Liu, B., Furevi, A., Perepelov, A.V., Guo, X., Cao, H., Wang, Q., Reeves, P.R., Knirel, Y.A., Wang, L., and Widmalm, G., FEMS Microbiol. Rev., 2020, vol. 44, pp. 655–683. https://doi.org/10.1093/femsre/fuz028
ACKNOWLEDGMENTS
The authors thank Doctor, Professor A.S. Shashkov for recording NMR spectra of A. zeae and A. melinis. The experiments were performed using the scientific and technical basis of the Center for Collective Use “Symbiosis” at the Institute of Biochemistry and Physiology of Plants and Microorganisms of the Russian Academy of Sciences.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
COMPLIANCE WITH ETHICAL STANDARDS
This article does not contain any studies with the use of humans or animals as objects of research.
Conflict of Interests
The authors state that there is no conflict of interest.
Additional information
Translated by A. Levina
Abbreviations: LPPC, lipopolysaccharide-protein complex; LPS, lipopolysaccharide; OSP, O-specific polysaccharide.
Corresponding author: phone: +7 (8452) 97-04-44.
Rights and permissions
About this article
Cite this article
Sigida, E.N., Grinev, V.S., Zdorovenko, E.L. et al. O-Antigens of Azospirillum zeae N7(T), Azospirillum melinis TMCY 0552(T), and Azospirillum palustre B2(T): Structure Elucidation and Analysis of Biosynthesis Genes. Russ J Bioorg Chem 48, 519–528 (2022). https://doi.org/10.1134/S1068162022030177
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1068162022030177