Abstract
Previously, in the strain Streptomyces rimosus ATCC10970 (producer of oxytetracycline), the aminoglycoside phosphotransferase AphVIII, determining kanamycin, neomycin, and paromomycin resistance, was identified and characterized. Recently, the authors obtained the 3D structure of AphVIII. The 14 aph genes, including gene aphVIII, were annotated when the genome of S. rimosus ATCC10970 was sequenced. In the present study, a new aph(3'')-Id (aphSR3) gene encoding streptomycin phosphotansferase was first identified in the strain of S. rimosus ATCC10970 using bioinformatic and comparative phylogenetic analysis of the aphSR1-aphSR14 genes with the previously known aph genes from clinical isolates and producer strains of aminoglycoside antibiotics belonging to seven subfamilies. When cloning, it was found that the gene aphSR3 (aph(3'')-Id) in Escherichia coli causes resistance to streptomycin at a concentration of 150 μg/mL. The obtained data can be used in practical terms to study the distribution and features of the functions of genes that determine the natural resistance to aminoglycoside antibiotics in actinobacteria of the genus Streptomyces.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
The subject of this study is aminoglycoside phosphotransferase (aph) genes which determine the natural resistance to aminoglycoside antibiotics. They represent a serious threat to antimicrobial therapy and thus are of important epidemiological significance. The multiple drug resistance (MDR) of bacteria to antibiotics is a global problem in the field of medicine, agroindustrial complex, and public health in general and is caused by the spread of bacteria in food and agricultural animals and plants, as well as in soil and water sources. In this connection, the study of the resistivity of soil bacteria as one of the main reservoirs and possible sources of drug resistance genes becomes urgent at the first level [1, 2].
Aminoglycoside phosphotransferase (Aph) (EC 2.7.1.) is a family of enzymes that modify aminoglycoside antibiotics by phosphorylation of their hydroxyl groups in the presence of ATP as a cofactor. Classical representatives of aph, common in clinical strains of bacteria, can be divided according to substrate specificity into the genes encoding kanamycin phosphotransferases, streptomycin phosphotransferases, gentamicin phosphotransferases, and hygromycin phosphotransferase; other genes can perform other functions [3].
Initially, the genes of aminoglycoside phosphotransferases were detected on plasmids and mobile elements in clinical strains of gram-negative and gram-positive bacteria [4, 5]. Aph enzymes of the second type were found in actinobacteria—strains producing aminoglycoside antibiotics [6, 7]. Later, Aph enzymes of the third type, which determine the natural resistance to aminoglycosides in other bacteria, including soil ones, were detected. However, the functions of genes annotated as aph in sequenced genomes have not been sufficiently studied yet [8]. The aph genes are annotated in many genomes, including commensal bacteria of intestinal microbiota, from which they can be transferred to clinically significant strains [9].
A phylogenetic analysis of Aph obtained from clinical strains and strains producing aminoglycoside antibiotics revealed that, depending on the position of the hydroxyl group of the antibiotic modified by the enzyme, seven subfamilies of aminoglycoside phosphotransferases are distinguished: Aph(2''), Aph(3'), Aph(3''), Aph(4), Aph(6), Aph(7''), and Aph(9) [10].
Actinobacteria of the genus Streptomyces contain the largest number of aph genes. To date, the 301 genomic sequence of actinobacteria strains belonging to the genus Streptomyces is represented in the NCBI database (https://www.ncbi.nlm.nih.gov/genome/). The bioinformatic analysis conducted by us showed that from 4 to 16 aminoglycoside phosphotransferases genes are present in the genomes. They determine the initial (natural) level of resistance to these antibiotics. However, studies of the structure and functions of Aph of a new type are not represented in the world literature.
Previously, when studying the resistance spectrum of 110 strains of the genus Streptomyces to aminoglycoside antibiotics, it was found that the ATCC10970 S. rimosus strain (producer of oxytetracycline) is resistant to most aminoglycoside antibiotics at a concentration of 10–20 μg/mL [11, 12]. In the S. rimosus strain, we identified and characterized the aminoglycoside-3'-phosphotransferase of a new type, AphVIII, which determines the resistance to kanamycin, neomycin, and paromomycin [13]. An important feature of AphVIII is its ability to modulate aminoglycoside phosphotransferase activity by phosphorylation of the Ser146 site in the activation loop of the enzyme [14]. We recently obtained a 3D structure of AphVIII with a resolution of 2.15 Å (code PDB 4H05) [15]. 14 aph genes, including the aphVIII gene, were annotated when sequencing the genome of S. rimosus subsp. rimosus ATCC10970 [16].
Within the framework of the current article, bioinformatic and phylogenetic analysis of genes of the Aph ATCC10970 S. rimosus strain, cloning of selected aph genes into E. coli, and study of the spectrum and level of resistance to aminoglycoside antibiotics of E. coli strains containing recombinant plasmids are presented.
Sequences of the aminoglycoside phosphotransferases genes of ATCC10970 S. rimosus strain were obtained from the NCBI database. In the genome of the S. rimosus strain, 14 Aph genes were annotated, named as follows (according to the number of gene loci): SRIM_04805—aphSR1; SRIM_07573—aphSR2; SRIM_08058—aphSR3; SRIM_10001—aphSR4; SRIM_10156—aphSR5; SRIM_11866—aphSR6; SRIM_16495—aphSR7; SRIM_16815—aphSR8; SRIM_16890—aphSR9; SRIM_23171—aphSR10; SRIM_27244—aphSR11; SRIM_32326—aphSR12; SRIM_33711—aphSR13; and SRIM_39853—aphSR14. The aphSR5 amino acid sequence completely coincides (100% identity, blastp) with the previously sequenced and described aphVIII sequence (no. AAG11411 in the GenBank database). AphVIII was previously assigned to the 3'‑APH(3')-VIII subfamily [13]. A comparative analysis of amino acid sequences of 14 Aph according to the LALIGN program (http://www.ch.embnet.org/software/LALIGN_form.html) showed a low degree of identity between them (26–36%).
The comparative phylogenetic analysis of amino acid sequences of the identified gene products with the previously known aph genes from clinical isolates and strains producing aminoglycoside antibiotics belonging to seven subfamilies of aminoglycoside phosphotransferases, Aph(2"), Aph(3'), Aph(3"), Aph(4), Aph(6), Aph(7"), and Aph(9) [10], was conducted using the Clustal Omega (EMBL-EBI) (http://www.ebi.ac.uk/Tools/msa/clustalo/) and MEGA v. 6.0 (http://www.megasoftware.net/) software programs.
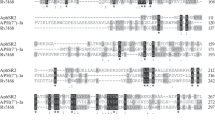
On the basis of the results of the analysis, AphSR5 (AphVIII) can be attributed to the Aph(3') subfamily and AphSR3 can be attributed to the Aph(3") subfamily; this follows from the arrangement of phosphotransferase data on the tree in clusters of these subfamilies and is confirmed by the high bootstrap value of the corresponding nodes of the tree (98–99%). Subsequent analysis of the alignment of the AphSR3 amino acid sequence with the Aph(3")-Ia Streptomyces griseus (streptomycin producer), Aph(3")-Ib Escherichia coli, and Aph(3")-Ic Mycobacterium fortuitum sequences allowed us to mark it as Aph(3")-Id (Fig. 1a).
Characteristics of the aphSR3 (aph(3'')-Id) gene of ATCC10970 S. rimosus strain: (a) comparison of the AphSR3 amino acid sequence with the Aph(3")-Ia, Aph(3")-Ib, and Aph(3")-Ic sequences (conservative residues are shown in black); (b) electrophoresis of soluble fraction of proteins belonging to different E. coli strains: (1–3) BL21(DE3) E. coli strain containing the plasmids (1) pET16b, (2, 3) pET16b:aph(3")-Id; (4–6) BL21(DE3) strain pLysS containing the plasmids (4) pET16b, (5, 6) pET16b:aph(3")-Id; (7–9) BL21(DE3) strain CodonPlus containing the plasmids (7) pET16b, (8, 9) pET16b:aph(3")-Id; M—SM0441protein molecular weight marker (Fermentas, Lithuania).
According to phylogenetic analysis, AphSR2 is located on one branch with Aph(7")-Ia, but bootstrap support for the corresponding node of the tree is low (<60%); nevertheless, the considered sequences contain a number of conserved amino acid residues that distinguish them from other Aph and provide the possibility to assign them to the subfamily Aph(7"). The remaining 11 aminoglycoside phosphotransferases cannot be assigned to any of the subfamilies described above, since they are located on a tree outside their clusters.
Isolation of the aph(3")-Id gene (aphSR3 locus of the SRIM_08058 gene) was carried out from the genomic DNA of S. rimosus strain by the PCR method. DNA amplification was carried out using the Dialat Ltd RSK-100 kit on a PTC-0150 device (MJ Research, Inc.) using oligonucleotides: AphSR3-N (5'-tcgtcatatggtgatcgatctgaccgcattc-3') and AphSR3-C (5'-agccggatcctcatccccaggtcagggggt-3'). Amplification mode: preheating at 95°С for 5 min; then 30 cycles of amplification: 1 min at 94°C, 1 min at 62°C, 1 min at 72°C; final elongation for 10 min at 72°C. The amplified DNA fragment was cloned in the pET16b expression vector (Novagen, United States) into NdeI and BamHI restriction endonuclease sites.
To study the expression of the aph(3'')-Id gene, BL21(DE3) (F–, dcm, ompT, hsdS(\({{{\text{r}}}_{{{{{\text{B}}}^{ - }}}}}\,{{{\text{m}}}_{{{{{\text{B}}}^{ - }}}}}\)), gal λ (DE3)) (Novagen); BL21(DE3) pLysS (F–ompThsdS(\({{{\text{r}}}_{{{{{\text{B}}}^{ - }}}}}\,{{{\text{m}}}_{{{{{\text{B}}}^{ - }}}}}\)) galdcm λ(DE3) [pLysS CamR]) (Stratagene, United States); and BL21(DE3) CodonPlus (F–ompT hsdS (\({{{\text{r}}}_{{{{{\text{B}}}^{ - }}}}}\,{{{\text{m}}}_{{{{{\text{B}}}^{ - }}}}}\)) dcm+ TetRgal λ(DE3) endA Hte [argU ileY leuW CamR]) (Stratagene) E. coli competent cells were transformed using the pET16b:aph(3'')-Id hybrid plasmid and grown in liquid LB medium at 37°C to an optical density of 0.6 (~2 h); then expression was induced by the addition of IPTG to a final concentration of 1.3 mM. Then, the culture was conducted at 28°C for 18 h, after which the cells were pelleted by centrifugation (5000 rpm, 10 min, 4°C) and suspended in a Sample buffer of the following composition: 62.5 mM Tris-HCl, pH 6.8, 5% glycerin, 2% 2-mercaptoethanol, 0.1% SDS, 0.001% bromophenol blue; then the cells were destroyed by heating at 95°C for 10 min and analyzed by 12.5% Laemmli-SDS-PAGE. As the control, protein fractions of E. coli strains containing pET16b plasmid without insert were used.
When cloning the aph(3'')-Id gene in E. coli cells, an additional fraction of the protein with a molecular weight of about 30 kDa, which corresponds to the calculated molecular weight of the Aph(3")-Id protein in total with the molecular weight of the linker protein of pET16b plasmid containing His-Tag (Fig. 1b), was observed. The maximum expression of the aph (3")-Id gene was established in the BL21(DE3) E. coli strain.
At the next stage of the work, the spectrum of resistance to aminoglycoside antibiotics was tested using the diffusion method. Clones of BL21(DE3) E. coli transformants containing the pET16b:aph(3")-Id recombinant plasmid were used for the analysis. Bacteria grown on LB agar medium with ampicillin were inoculated into LB liquid medium with ampicillin and grown in a thermostated shaker for 18 h at 37°C at 250 rpm.
The bacterial suspension was then mixed with a melted LB agar medium containing ampicillin (150 μg/mL) and an IPTG inducer (20 μg/mL) in a 1 : 1 ratio (v/v). After that, 5 mL of the obtained mixture was poured over the top layer onto pre-prepared Petri dishes with LB agar medium with ampicillin and IPTG (10 μg/mL). The resistance spectrum was tested using paper disks with aminoglycoside antibiotics: kanamycin (30 μg/disk), neomycin (30 μg/disk), amikacin (30 μg/disk), streptomycin (10 μg/disk), gentamicin (10 μg/disk), tobramycin (10 μg/disk), sisomicin (10 μg/disk), netilmicin (10 μg/disk), and isepamicin (30 μg/disk). The results were recorded after incubation for 16–18 h at 37°C.
The studies conducted to determine the growth inhibition zone around paper disks showed that the aph(3'')-Id gene determines the resistance of BL21 (DE3) E. coli to streptomycin. The data obtained are consistent with phylogenetic analysis, according to which the gene is assigned to the Aph(3") class.
Testing the level of resistance to streptomycin by the replica method showed that the BL21(DE3) pET16b:aph(3'')-IdE. coli strain is resistant to streptomycin at a concentration of 150 μg/mL.
It should also be noted that cloning of the aphSR2 gene in E. coli (locus of the gene SRIM_07573), designated by us as aph(7'')-Ib was similarly performed. When cloning the aph(7'')-Ib gene in BL21(DE3) E. coli cells, an additional fraction of the protein with a molecular weight of about 39 kDa was observed, which corresponds to the calculated molecular weight of the Aph(7")-Ib protein including the molecular weight of the His-Tag containing pET16b plasmid linker protein. However, studies to determine the growth inhibition zone around paper disks showed that the aph(7'')-Ib gene does not determine the resistance of E. coli to aminoglycoside antibiotics.
Summarizing the data obtained by the authors in earlier published [13–15] and the present works, only two out of the 14 aph genes of the АТСС10970 Streptomyces rimosus strain—aphVIII (aphSR5) and aph(3'')-Id (aphSR3)—exhibit resistance to aminoglycoside antibiotics. Studies are conducted in E. coli using the pET16b and pET32a expression vectors (Novagen) under the T7 promoter to identify the functions of 12 other genes annotated as Aph genes in ATCC10970 S. rimosus.
Thus, a new aph(3'')-Id (aphSR3) gene encoding streptomycin phosphotransferase was identified for the first time by us in the ATCC10970 S. rimosus strain. When cloning the aphSR3 (aph(3'')-Id) gene in E. coli, it was found that it causes resistance to streptomycin at a concentration of 150 μg/mL.
The presented studies offer new opportunities for studying the distribution and specificity of expression of genes that determine the natural resistance to aminoglycoside antibiotics in actinobacteria of the genus Streptomyces.
Within the framework of the further work, the extraction of Aph(3'')-Id recombinant protein, the analysis of phosphotransferase activity and phosphorylation of substrates in vitro, the crystallization of the test protein, and the X-ray diffraction analysis of the obtained crystals are foreseen.
REFERENCES
Brown, D., Antibiotic resistance breakers: can repurposed drugs fill the antibiotic discovery void?, Nat. Rev. Drug. Discov., 2015, vol. 14, no. 12, pp. 821-832. doi 10.1038/nrd4675
Blair, J.M., Webber, M.A., Baylay, A.J., et al., Molecular mechanisms of antibiotic resistance, Nat. Rev. Microbiol., 2015, vol. 13, no. 1, pp. 42-51. doi 10.1038/nrmicro3380
Surette, M. and Wright, G.D., Lessons from the environmental antibiotic resistome, Annu. Rev. Microbiol., 2017, vol. 71, pp. 309-329. doi 10.1146/annurev-micro-090816-093420
Davies, J. and Gray, G., Evolutionary relationships among genes for antibiotic resistance, Ciba Found Symp., 1984, vol. 102, pp. 219-232.
Wright, G.D., Molecular mechanisms of antibiotic resistance, Chem. Commun. (Cambridge), 2011, vol. 47, no. 14, pp. 4055-4061. doi 10.1039/c0cc05111j
Ozen, C., Malek, J.M., and Serpersu, E.H., Dissection of aminoglycoside-enzyme interactions: a calorimetric and NMR study of neomycin B binding to the aminoglycoside phosphotransferase (3')-IIIa, J. Am. Chem. Soc., 2006, vol. 128, no. 47, pp. 15248-15254.
Collins, A.C., Ashenafi, M., Saunders, A.A., et al., Cloning and expression of streptomycin inactivating enzymes Aph(6)-Ia and Aph(6)-Id, Cell. Mol. Biol., 2007, vol. 53, no. 3, pp. 74-79.
Anderson, A.S., Clark, D.J., Gibbons, P.H., et al., The detection of diverse aminoglycoside phosphotransferases within natural populations of actinomycetes, J. Ind. Microbiol. Biotechnol., 2002, vol. 29, no. 2, pp. 60-69.
Kovtun, A.S., Alekseeva, M.G., Averina, O.V., et al., Identification of aminoglycoside phosphotransferases of bacterial isolates in the microbiota of Russians, Vestn. Ross. Gos. Med. Univ., 2017, no. 2, pp. 14-19.
Shakya, T., Stogios, P.J., Waglechner, N., et al., A small molecule discrimination map of the antibiotic resistance kinome, Chem. Biol., 2011, vol. 18, no. 12, pp. 1591-1601. doi 10.1016/j.chembiol.2011.10.018
Danilenko, V.N., Puzynina, G.G., and Lomovskaya, N.D., Multiple resistance to antibiotics in actinomycetes, Genetica (Moscow), 1977, vol. 13, no. 10, pp. 1831-1842.
Potekhin, Ya.A. and Danilenko, V.N., The determinant of kanamycin resistance of Streptomyces rimosus: amplification in the chromosome and reversed genetic instability, Mol. Biol., 1985, vol. 19, no. 3, pp. 18-25.
Sizova, I.A., Khegemann, P., Furmann, M., et al., Streptomyces rimosus aminoglycoside 3'-phosphotransferase VIII: comparisons with aminoglycoside 3'-phosphotransferases of aminoglycoside-producing strains and with eukaryotic protein kinases, Mol. Biol. (Moscow), 2002, vol. 36, no. 1, pp. 18-25. https://doi.org/10.1023/A:1014282003679.
Elizarov, S.M., Alekseeva, M.G., Novikov, F.N., et al., Identification of phosphorylation sites in aminoglycoside phosphotransferase VIII from Streptomyces rimosus, Biochemistry (Moscow), 2012, vol. 77, no. 11, pp. 1258-1265. https://doi.org/10.1134/S0006297912110041.
Boyko, K.M., Gorbacheva, M.A., Korzhenevskiy, D.A., et al., Structural characterization of the novel aminoglycoside phosphotransferase AphVIII from Streptomyces rimosus with enzymatic activity modulated by phosphorylation, Biochem. Biophys. Res. Commun., 2016, vol. 477, no. 4, pp. 595-601. doi 10.1016/j.bbrc.2016.06.097
Pethick, F.E., MacFadyen, A.C., Tang, Z., et al., Draft genome sequence of the oxytetracycline-producing bacterium Streptomyces rimosus ATCC 10970, Genome Announc., 2013, vol. 1, no. 2. e00063-13. doi 10.1128/genomeA.00063-13
ACKNOWLEDGMENTS
This work was carried out with partial financial support from the Russian Science Foundation (project no. 17-04-01106, dated April 6, 2017).
Author information
Authors and Affiliations
Corresponding author
Additional information
Translated by P. Kuchina
Rights and permissions
About this article
Cite this article
Alekseeva, M.G., Rudakova, N.N., Zakharevich, N.V. et al. New Gene of Aminoglycoside Phosphotransferase aph(3'')-Id from Streptomyces rimosus ATCC10970, Encoding Streptomycin Resistance. Russ J Genet 54, 1254–1258 (2018). https://doi.org/10.1134/S1022795418100034
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1022795418100034