Abstract
Antibiotic resistance is one of the greatest problems in modern medicine and a global threat to health care. Aminoglycoside phosphotransferases (APHs) currently pose a serious threat to antimicrobial therapy; therefore, research on the functions and obtainment of 3D structures of aminoglycoside phosphotransferases is an important and urgent issue that will allow the development of approaches to overcome resistance to aminoglycoside antibiotics. Soil actinobacteria of the Streptomyces genus contain the largest number of aph genes; these genes can be transferred to them from antibiotic producing strains. The review analyzes the current data on the actinobacteria of the Streptomyces genus as a reservoir of drug resistance genes, as well as approaches to the identification of aph genes associated with resistance to aminoglycoside antibiotics on the example of the model strain S. rimosus ATCC 10970 (oxytetracycline producer). The data on the development of test systems for the screening of inhibitors and potential drugs are discussed. The inhibition of proteins that provide a natural level of bacterial resistance to a number of aminoglycoside antibiotics may help overcome multidrug resistance in pathogenic actinobacteria and expand the range of drugs used due to the synergistic effect of the antibiotic with the ARH inhibitor compound.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
INTRODUCTION
Antibiotic resistance is one of the greatest problems in modern medicine and a global threat to health care. According to the data of the Centers for Disease Control and Prevention, at least 2 million people are infected with antibiotic resistant bacteria, and at least 7.23 million people die annually from diseases caused by antibiotic resistant microorganisms (Hossion and Sasaki, 2013). The World Health Organization (WHO) asked the UN to discuss this problem and to adopt appropriate recommendations. The problem is determined not only by the catastrophic spread of clinical strains of multidrug-resistant (MDR) bacteria, but also by the spread of MDR bacteria in food, farm animals, and plants, as well as in soil and water sources (Link et al., 2007). Thus, it is relevant to study soil bacteria as the main reservoir and a possible source of drug-resistance genes (Gibson et al., 2015).
Aminoglycosides represent a large group of biologically active, secondary metabolites (Davies and Wright, 1997). Since the discovery of this group of antibacterial drugs, they have been widely used as a therapeutic agent in the treatment of various severe infectious diseases caused by gram-negative microorganisms (Hermann, 2007). However, the range of the application of aminoglycoside antibiotics is narrowing due to the emergence of bacteria that are resistant to them (Wright et al., 1998). Nevertheless, aminoglycosides are currently among the most widely used antibiotics in clinical practice due to their high efficiency and low cost (Forge and Schacht, 2000; Block and Blanchard, 2019).
There are several main mechanisms of resistance to aminoglycosides: the modification of ribosomes, the target of aminoglycosides; a decrease in membrane permeability and active transport from the cell; enzymatic modification of the antibiotic; and biofilm formation. However, of all the known mechanisms of resistance to aminoglycosides, modification by enzymes is the most widespread (Wright and Thompson, 1999).
Enzymes that modify aminoglycosides catalyze the modification reactions of various –OH and –NH2 groups of the 2-dioxystreptamine core or sugar residues. These include nucleotidyltransferases (adenylyltransferases), phosphotransferases, and acetyltransferases (Wright, 2011). Acetyltransferases modify the amino group of the aminoglycoside molecule, and phosphotransferase and adenylyltransferase affect hydroxyl group. The effect of aminoglycoside-modifying enzymes (acetylation, phosphorylation, and adenylation, respectively) leads to a change in the structure of the antibiotic molecule such that it is unable bind to the bacterial ribosome, as a result of which protein synthesis is not inhibited and the bacterial cell remains viable (Zarate et al., 2018).
Enzymatic modification of the antibiotic molecule by phosphotransferases is a common mechanism of aminoglycoside resistance in bacterial strains (Frase et al., 2012). This is why the study of the structure and functions of aminoglycoside phosphotransferases is an important and urgent issue that will allow the development of approaches to overcome resistance to aminoglycoside antibiotics.
AMINOGLYCOSIDE ANTIBIOTICS
Aminoglycosides (aminocyclitols) represent a large group of water-soluble antibiotics with a wide antimicrobial spectrum of activity (Wright and Thompson, 1999; Chandrika and Garneau-Tsodikova, 2018). The aminoglycoside-antibiotic molecule contains two or more amino sugars that are linked by glycosidic bonds to the aminocyclitol ring (Busscher et al., 2005). Spectinomycin also belongs to the aminoglycosides, in the molecule of which there are no bonds between the aminocyclitol ring and the amino sugar (Bryskier, 2005; Veyssier and Bryskier, 2005).
Aminoglycoside antibiotics possess a bactericidal effect, the mechanism of which consists of the binding of the aminoglycoside to the decoding site of the ribosome and the disruption of protein synthesis (Sutcliffe, 2005; Hermann, 2005, 2007).
The aminoglycoside antibiotics include streptomycin, kanamycin, neomycin, gentamicin, tobramycin, amikacin, etc. There are three generations (Table 1) of aminoglycoside antibiotics (Reshedko, 1999; Hotta and Kondo, 2018).
The newest aminoglycoside antibiotic to date is plazomicin, a semisynthetic derivative of sisomycin. A drug based on plazomicin is currently in clinical trials (Serio et al., 2017; Shaeer et al., 2019).
The antibacterial action of aminoglycosides is mediated by their chemical structure. Since aminoglycoside antibiotics contain a significant number of positively charged groups, they have high affinity to negatively charged molecules, such as nucleic acids (Jana and Deb, 2006).
Ribosomes are one of the main targets of the effect of antibiotics in the bacterial cell. Most antibiotics used in clinical practice inhibit the elongation stage during protein synthesis: aminoglycosides, chloramphenicol, lincosamides, macrolides, oxazolidinones, streptogramins, and tetracyclines. Despite the large size of ribosomes, antibiotics interact only with several sites. Antibiotics that interact with the 30S subunit bind either the P site or the A site (Ogle et al., 2003; Vicens and Westhof, 2003; Ogle and Ramakrishnan, 2005; Zaher and Green, 2009; Wilson, 2014).
The main target of aminoglycoside antibiotics is a small subunit of the bacterial ribosome (30S), which includes 21 proteins and 16S rRNA. Aminoglycosides bind to 16S ribosomal RNA, more precisely, to the A site, which leads to a disruption of translation during protein synthesis (Fourmy et al., 1996; Carter et al., 2000; Ramirez and Tolmasky, 2010). The model of the molecular interaction of aminoglycoside antibiotics with the A site was studied in detail via X-ray diffraction analysis (Magnet and Blanchard, 2005).
SOIL BACTERIA OF THE Streptomyces GENUS AS PRODUCERS OF AMINOGLYCOSIDES
The largest number of antibiotics (at least 70%) widely used in practice refers to substances formed by soil actinobacteria (the Actinomycetales order). Soil bacteria belonging to the Streptomyces genus are producers of the most important aminoglycoside antibiotics (Block and Blanchard, 2019). Table 2 presents producers of various aminoglycosides (Wright et al., 1998).
Actinobacteria are one of the largest bacterial phyla and are widespread in aquatic and terrestrial ecosystems. Most of the representatives of this group of bacteria are saprophytes, microorganisms that live in the soil, but they are found in fresh and salt water, as well as in air. They are usually present in soil at densities of 106–109 bacterial cells per gram of soil, while streptomycetes account for more than 95% of all actinomycete strains isolated from soil (Barka et al., 2015). Some actinobacteria are causative agents of human and animal diseases: Actinomyces israelii is the causative agent of actinomycosis. Actinomyces meyeri, Actinomyces neuii, and Actinomyces turicensis are causative agents of diseases localized in various parts of the body, mouth, skin, and mucous membranes (Kononen and Wade, 2015).
Streptomyces is the largest genus of actinomycetes. It includes gram-positive aerobic bacteria that form a network of branched filaments, substrate mycelium, and aerial mycelium. Currently, there are about 843 species and 38 subspecies of the Streptomyces genus (LPSN, http://www.bacterio.net/streptomyces.html). The cell wall of streptomycetes contains alanine, glutamic acid, glycine, and LL-2 and 6‑diaminopimelic acids (LL-DAP). Their DNA is characterized by a high content of guanine and cytosine, and the content of G + C in the most widely studied species, e.g., Streptomyces coelicor, reaches 72.1% (Bentley et al., 2002). Their main habitats are soil and sea water (Siti et al., 2017). The Streptomyces genus is the largest genus capable of synthesizing antibiotics, and it has been used since the 1940s–1950s in the industrial production of antibiotics (Newman and Cragg, 2007).
Although streptomycetes are considered primarily to be free-living, terrestrial, soil-forming bacteria, some species are symbionts with fungi, insects, plants, and animals (Seipke et al., 2012), and some strains inhabit marine soils (Fiedler et al., 2005). Several species of streptomycetes are plant pathogens that cause disease by affecting roots and tubers (Zhang and Loria, 2017). The most economically important disease caused by members of the Streptomyces genus is the potato scab, which is characterized by surface lesions of potato tubers (Loria et al., 2006). Another pathogenic species, S. ipomoeae, causes soil rot on sweet potatoes (Tomihama et al., 2016). Other pathogenic streptomycete species include S. europaeiscabiei, S. stelliscabiei, S. luridiscabiei, S. puniciscabiei, S. niveiscabiei, S. reticuliscabiei, and S. caviscabies (Goyer et al., 1996; Bouchek-Mechiche et al., 2000; Park et al., 2003). Numerous studies of representatives of this genus showed that they can be found in almost all ecosystems of our planet, while they successfully compete with representatives of other phylogenetic groups, dominating in the microbial population, due to their unusual metabolism and secondary metabolites (Berdy, 2012).
Many representatives of the Streptomyces genus are well studied, since they are commercially significant producers of secondary metabolites and hydrolytic enzymes. One of these strains is Streptomyces lividans, a classical object of molecular genetic research that is also used to obtain homologous and heterologous hydrolytic enzymes for industrial use. Due to the commercial value of the strain, its metabolic pathways and the mechanisms of the secretion of some substances were studied in great detail (Gullón and Mellado, 2018).
The development of technologies related to genome sequencing and the analysis of data libraries, as well as the calculation resources of computer devices, has accelerated and simplified the identification of numerous clusters of genes for the biosynthesis of natural medicinal compounds (BGCs) in the streptomycete genomes. However, most of these BGCs are silent or are expressed in the original strains at a low level, which makes it relevant to study them with genome editing. Numerous strategies, including those using CRISPR/CRISPR-Cas technologies, are being developed. This method of genome editing has a higher accuracy than other methods and a higher efficiency for genome editing in various model organisms, including streptomycetes (Tao et al., 2014; Cobb et al., 2015).
In addition, streptomycete strains are promising sources of new antibiotics against multidrug/extensively drug-resistant strains, including methicillin-resistant Staphylococcus aureus. Streptomycetes were already sources of such antibiotics, in particular, vancomycin. Over the years, the number of new antibiotic compounds has decreased significantly, which resulted in the fact that fewer drug compounds reached the stage of clinical trials. Since the discovery of streptomycin in 1944, studies have identified more than 10 400 biologically active substances in Streptomyces. At the same time, the search for new antibiotics has slowed, and the rate of resistance formation to them has increased, which makes the search for new antibiotics and their producers from unusual habitats urgent (Berdy, 2012). For this purpose, strains from poorly studied regions are primarily considered: mangroves, deserts, and marine and freshwater reservoirs, as well as endophytic forms. The microorganisms that inhabit these regions face difficult environmental conditions, high salinity, high temperatures, and low humidity. These streptomycete strains have a unique metabolism and form unique, biologically active compounds (Kemung et al., 2018).
CLASSIFICATION OF AMINOGLYCOSIDE PHOSPHOTRANSFERASES
Aminoglycoside phosphotransferases are enzymes that modify aminoglycoside antibiotics via phosphorylation of their hydroxyl groups in the presence of ATP as a cofactor (Smith and Baker, 2002). It was previously thought that only ATP is the donor of the phosphate moiety, but it was recently found that several phosphotransferases use GTP instead of ATP (Shakya and Wright, 2010; Shi and Berghuis, 2012; Smith et al., 2012). More than 30 phosphotransferases have been identified from clinical strains and strains producing aminoglycoside antibiotics; the identity of their amino acid-sequences varies from 20 to 40%.
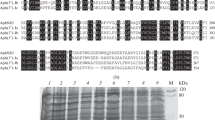
It was found that the enzymes of the aminoglycoside-phosphotransferase family are similar in structure and function to eukaryotic serine-threonine protein kinases and, according to the modern classification, are classified as kinases that modulate bacterial resistance to antibiotics (Kim and Mobashery, 2005; Morar and Wright, 2010). Kinases that modify antibiotics include aminoglycoside phosphotransferases (APH) and macrolide phosphotransferases (MPH). Figure 1 shows a phylogenetic analysis of kinases that modify aminoglycoside antibiotics (Shakya et al., 2011).
Depending on the hydroxyl-group position of the antibiotic modified by the enzyme, there are seven subfamilies of aminoglycoside phosphotransferases: APH(2"), APH(3'), APH(3"), APH(4), APH(6), APH(9), and APH(7") (Vakulenko and Mobashery, 2003; Ramirez and Tolmasky, 2010; Shakya et al., 2011). Table 3 presents the information on these subfamilies (Vakulenko and Mobashery, 2003; Ramirez and Tolmasky, 2010; Shi et al., 2013).
The subfamily of aminoglycoside-3'-phosphotransferases is the most widespread and studied; this subfamily includes eight isozymes. The Aph(3')-V enzymes were found in actinobacteria: in the neomycin-producing strain Streptomyces fradiae (Va), in Streptomyces ribosidificus (Vb), and in Micromonospora chalcea (Vc) (Thompson and Gray, 1983; Hoshiko et al., 1988 ; Salauze et al., 1991; Wright and Thompson, 1999).
In the streptomycin-producing strain Streptomyces griseus, two streptomycin phosphotransferases, APH(3")-Ia and APH(6)-Ia, were found (Heinzel et al., 1988; Collins et al., 2007).
In recent years, a large number of actinobacterial genomes have been sequenced. During the annotation of genomes in actinobacteria of the Streptomyces genus, 4–14 genes of aminoglycoside phosphotransferases were found. They determine the initial (natural) level of resistance to aminoglycoside antibiotics. The functions of genes annotated as aph in the sequenced genomes are poorly understood at present and require further research to test whether these hypothetical enzymes are true aph resistance genes (Anderson et al., 2002).
In a study of the spectrum of antibiotic resistance in 110 strains of the Streptomyces genus, it was found that S. rimosus ATCC 10970 (an oxytetracycline producer) is the only one among those studied that is resistant to most aminoglycoside antibiotics at a concentration of 10–20 μg/mL (Danilenko et al., 1977).
Streptomyces rimosus AMINOGLYCOSIDE PHOSPHOTRANSFERASES
Aminoglycoside Phosphotransferase Aph(3')-VIII
During the multistage selection of S. rimosus ATCC 10970 cells resistant to increasing doses of kanamycin (from 10 to 4 × 104 μg/mL), a strain with aminoglycoside-3'-phosphotransferase activity and chromosomal DNA rearrangements was obtained. The antibiotic resistance determinant of the S. rimosus ATCC 10970 strain, designated as Kmr, was found; it was amplified in a DNA fragment of 10.3 × 106 bp and characterized by genetic instability. The frequency of the transition Kmr ↔ Kms is 1 × 10–3. This type of instability is widespread among actinomycetes and is typical for most signs that are not related to primary metabolism (Potekhin and Danilenko, 1985).
The Kmr determinant was cloned into the Streptomyces lividans 66 strain as a part of the vector plasmid SLP1.2; the phenomenon of the amplification of this determinant within the constructed hybrid plasmids pSU1–pSU13 was studied in detail. It was shown that the subcloning of this chromosomal DNA fragment in the described plasmids makes Streptomyces lividans cells resistant to kanamycin, neomycin, and paromomycin. Crude, cell-free extracts of such transformants demonstrate aminoglycoside phosphotransferase activity, the substrate specificity of which corresponds to the spectrum of antibiotic resistance. The nucleotide sequence of the DNA fragment containing the Kmr determinant was determined, and the open reading frame for the aminoglycoside 3'-phosphotransferase gene was found. Based on the nucleotide sequence and substrate specificity of the encoded enzyme, the aph gene from S. rimosus ATCC 10970 was assigned to a new class of genes and designated as aph(3')-VIII, and its product was designated as APH(3')-VIII (Potekhin and Danilenko, 1985; Starodubtseva et al., 1985; Danilenko et al., 1997).
During further research, the aph(3')-VIII gene was cloned into the unicellular green alga Chlamydomonas reinhardtii as a part of the plasmid vector pSU973. It was demonstrated that nuclear transformants of this alga carrying the vector pSU973:aph(3')-VIII are highly resistant to paromomycin (300–500 µg/mL). Analysis of the total proteins of the transformants via polyacrylamide gel electrophoresis under nondenaturing conditions showed that a protein with a molecular weight of ~30 kDa exhibits aminoglycoside phosphotransferase activity (Sizova et al., 1996, 2001).
The nucleotide sequence of the aph(3')-VIII gene was clarified and deposited in GenBank under the number AAG11411. This sequence is 804 bp in length and encodes an APHVIII protein with a length of 267 amino acids, a molecular weight of 29.1 kDa, and an isoelectric point of 4.59. Comparison of the amino-acid sequences of APH(3')-VIII and aminoglycoside phosphotransferases from strains that produce aminoglycoside antibiotics made it possible to detect significant differences and to assign this enzyme to a separate group (Sizova et al., 2002).
To determine the role of the aph(3')-VIII gene from the S. rimosus ATCC 10970 strain, it was also cloned into E. coli BL21 (DE3) as part of the plasmid vector pSI10. Earlier, chromosomal mutations affecting the level of cell resistance to aminoglycosides were found in S. lividans, which may be localized in the gene encoding the APH(3')-VIII protein activator (Potekhin and Danilenko, 1985). In this case, the role of protein-activity activators in Streptomyces can be played by serine-threonine protein kinases of the eukaryotic type. To test the hypothesis that APHVIII is activated via phosphorylation by protein kinases, the heterologous expression of the aph(3')-VIII gene was studied in transformed prokaryotic cells that do not contain eukaryotic protein kinases (E. coli) and in eukaryotic cells (Chlamydomonas reinhardtii). It was demonstrated that the Chlamydomonas transformants have a higher level of resistance to paromomycin at a lower content of the APH(3')-VIII protein. This phenomenon may be associated with protein activation via phosphorylation by serine-threonine protein kinases. Comparison of the amino-acid residues APH(3')-VIII and serine-threonine protein kinases of actinomycetes revealed a local similarity of 38 amino acids in the conserved region involved in ATP binding. A bioinformatic search for potential phosphorylation regions found four regions for serine-threonine protein kinases, including Ca2+-dependent ones (Sizova et al., 2002).
The aminoglycoside-phosphotransferase activity was determined radiochemically in a crude cell-free extract of S. lividans TK64 containing the plasmid pSU951:aph(3')-VIII. Plasmid-free S. lividans TK64 was used as a negative control, and an extract of E. coli cells transformed with the pSV2neo plasmid that produced streptomycin transferase was used as a positive control. After the incubation of aminoglycoside antibiotics with 32Р-γATP with the cell-free extract, radiolabeled phosphorylated antibiotics were isolated via thin-layer chromatography with polyethyleneimine-cellulose, and the level of radioactivity of the obtained samples was measured. The APH(3')-VIII enzyme was shown to have phosphotransferase activity against the aminoglycoside antibiotics kanamycin, neomycin, and paromomycin (Danilenko et al., 1997).
The aminoglycoside-phosphotransferase activity of APH(3')-VIII in crude cell extracts of Chlamydomonas reinhardtii was also studied via autoradiography, which found result enzymatic activity against paromomycin and neomycin (Sizova et al., 1996).
The substrate specificity of APH(3')-VIII was determined via isolation of the water-soluble fraction of proteins from E. coli transformed with the vector pET16b:aph(3')-VIII. A protein fraction with a molecular weight of about 30 kDa was detected via gel electrophoresis under denaturing conditions. The transformants were resistant to paromomycin at a concentration of up to 60 μg/mL (Sizova et al., 2002).
In further studies, APH(3')-VIII containing His10 from E. coli extracts was isolated and purified with the use of columns with Ni-NTA agarose under native conditions, after which SDS-PAGE was performed. Fractions containing a 31.5-kDa protein were purified via chromatography (Elizarov et al., 2005). The kanamycin kinase activity of the enzyme was analyzed in a reaction mixture containing 0.3 μg Aph(3')-VIII, 1.2 mg/mL kanamycin, 7.5 mM [γ-32P]ATP, 4 mM NaHCO3, 20 mM Tris-HCl pH 7.8, 10 mM MgCl2, 60 mM KCl, and 3 mM DTT. At the same time, a high enzymatic activity against the aminoglycoside antibiotic kanamycin was demonstrated via autoradiography (Elizarov et al., 2012).
Actinomycetes represent a promising model for research on the interaction between eukaryotic protein kinases and aminoglycoside phosphotransferases in the regulation of cellular processes and drug resistance. Via immunoprecipitation with antibodies to APH(3')-VIII and in vitro labeling, it was found that endogenous protein kinases in S. rimosus extracts actively phosphorylate APH(3')-VIII at two serine residues. Moreover, the amount of phosphate introduced in APH(3')-VIII is 1.84 times higher in the presence of Ca2+. Further analysis showed that the phosphorylation of APH(3')-VIII is catalyzed by serine-threonine protein kinases (STPKs) with molecular weights of 55 and 74 kDa. At the same time, the activity of the 55‑kDa kinase depends on calcium and calmodulin. It was found that kanamycin phosphotransferase activity of the phosphorylated protein APH(3')-VIII is 3.72 times higher than that of the unmodified enzyme. The studied protein kinases are involved in the regulation of kanamycin resistance in S. rimosus cells, which can be modulated through changes in the activity of specific ligand-dependent STPKs (Elizarov et al., 2005).
Bioinformatic analysis and molecular modeling were used to identify four potential phosphorylation sites of APH(3')-VIII: S95, S146, S160, and S215. For further identification of phosphorylation sites, mutant variants of the aph(3')-VIII gene were obtained from point mutations, with Ser → Ala substitutions in the identified positions. Comparative analysis of the kanamycin kinase activity of the nonphosphorylated and phosphorylated forms of the initial and mutant variants of the APH(3')-VIII protein showed that Ca2+-dependent phosphorylation of Ser146 in APH(3')-VIII leads to a six- to sevenfold increase in the kanamycin kinase activity of APH(3')-VIII. Thus, Ser146, which is located in the enzyme-activation loop, is critical for its activity. It was also shown that APH(3')-VIII is an enzyme activated by protein kinases (Elizarov et al., 2012).
A full length model of the APH(3')-VIII structure was created with the Swiss-Model server for modeling based on the structures of the enzymes APH(3')-II (identifier 1ND4) and APH(3')-III (identifier 1L8T), which are available in the PDB database. Molecular modeling made it possible to identify (Fig. 2) phosphorylation site Ser146 in the enzyme-activation loop (Elizarov et al., 2012).
Analysis of the molecular dynamics of the complex of nonphosphorylated APHVIII with kanamycin, ATP, and two Mg2+ ions revealed changes in the enzyme structure due to weakening of the contact between the C- and N-terminal domains. The mobility of domains determines the release of ATP from the substrate bound to the C-terminal lobe and the interaction of phosphate with the N-terminal lobe, which leads to the catalytically inactive form APH(3')-VIII (Elizarov et al., 2012).
X-ray crystallography was used to obtain the 3D structure of APH(3')-VIII at a resolution of 2.15 Å; the identification number in PDB is 4H05 (Fig. 3). Analysis of the obtained structure and comparison with the already existing structures of aminoglycoside-3'-phosphotransferases allowed the identification of the Ser146 region in the enzyme-activation loop. It was shown that there was a change in the conformation in this region that occurs upon interaction with the substrate (Boyko et al., 2016).
APH(3')-VIII and its comparison with the structures of other aminoglycoside-(3')-phosphotransferases (Boyko et al., 2016). (a) APHVIII, monomer; (b) comparison of APHVIII and APH-II in a complex with kanamycin A (ID PDB: 1ND4); (c) comparison of APHVIII and APH-III in a complex with ADP and kanamycin A (PDB ID: 1L8T); (d) magnified view of the substrate binding region. The region of the flexible loop NPL in the region of 21–27 amino-acid residues (numbering according to APHVIII) is indicated by the arrow, and the loop between β4 and β5-folded layers is also indicated. APH-I in the complex with ADP (PDB ID: 4EJ7).
Thus, aminoglycoside-3'-phosphotransferase of a new type, APHVIII, was identified and characterized in the S. rimosus ATCC 10970 strain (Potekhin and Danilenko, 1985; Sizova et al., 2002), and the 3D structure of the enzyme was determined, PDB code 4H05 (Boyko et al., 2016).
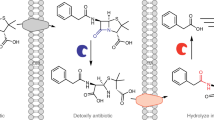
Modulation of the APH(3')-VIII activity by serine-threonine protein kinases is used in practice to create highly efficient test systems for the selection of STPK inhibitors with the APH(3')-VIII protein construct and target human and bacterial STPKs. The principle of operation of test systems of this type is based on the fact that phosphorylation of the APH(3')-VIII enzyme, which inactivates aminoglycoside antibiotics, by protein kinases increases the resistance of bacterial cells to kanamycin. In contrast, protein kinase inhibitors make bacterial cells more sensitive to kanamycin. These properties of the test system allow a biotarget-directed search for protein kinase inhibitors as potential drugs of a new generation. The first test system of this type was a system developed based on the strain Streptomyces lividans ТK24 (66) APH+ (Danilenko et al., 2008). One such system is the E. coli/aphVIII/ pk25 test system, which is used to prescreen Pk25 inhibitors of the Streptomyces lividans strain and its structural homologs. In particular, this test system can be used to prescreen STPK inhibitors of a number of pathogenic microorganisms, such as PknA and PknJ of M. tuberculosis strains, StkP of S. pneumonia, and some human STPKs, including PKA, CaMKI, and Pac2 (Becker et al., 2010).
Another test system of this type is the E. coli/aphVIII/ pim-1 system. Figure 4 shows the principle and features of this system. Site-directed mutagenesis in the region of Ser146 allowed this site to be optimized for the most efficient phosphorylation by protein kinase Pim-1. STPKs of the Pim family positively regulate the cell cycle and play an important role in the pathogenesis of tumors of the blood system, supporting cell proliferation. Inhibitors of STPKs of the Pim family are potential drugs for the treatment of leukemia and lymphomas (Zhukova et al., 2011).
Another system of this type is the recently developed E. coli/aphVIII/gsk3β test system. This system includes the gene for the catalytic domain of the GSK3β protein kinase and the APH(3')-VIII gene, a substrate of phosphorylation. To optimize the functioning of the test system, two APH(3')-VIII modifications were obtained in the region of the phosphorylation site Ser146. The targeted selection of GSK3β-protein kinase inhibitors is a promising direction in the drug therapy for type-2 diabetes, Alzheimer’s disease, and chronic inflammatory diseases (Alekseeva et al., 2018).
Aminoglycoside Phosphotransferase APH(3”)-Id
Analysis of the genome of S. rimosus ATCC 10970 (Pethick et al., 2013) made it possible to identify 14 genes annotated as aph, including aphVIII. These 14 aph genes were designated aphSR1–14. Based on phylogenetic similarities, only three of the 14 genes, aphSR5 (aphVIII), aphSR3, and aphSR2, were assigned to the known subfamilies APH(3'), APH(3''), and APH(7''), respectively.
The aph(3'')-Id gene, which encodes streptomycin phosphotransferase, in S. rimosus ATCC 10970 was first identified for Streptomyces, which are not producers of aminoglycoside antibiotics; the APH(3")-Id enzyme was biochemically characterized (Alekseeva et al., 2019). The aph genes, which belong to the APH(3'') subfamily, are streptomycin phosphotransferases. It is known that resistance to streptomycin mediated by aminoglycoside phosphotransferases is the result of the action of two classes of enzymes, APH(3') and APH(6) (Wright and Thompson, 1999). Phylogenetic analysis of phosphotransferases from clinical bacterial strains and producers of aminoglycoside antibiotics (Shakya et al., 2011) and 14 APH S. rimosus ATCC10970 showed that AphSR3 belongs to the APH(3'') subfamily. The aph(3'') genes were described earlier: aph(3'')-Ia in streptomycin producer Streptomyces griseus (Heinzel et al., 1988), aph(3'')-Ib in the plasmid RSF1010 Escherichia coli (Scholz et al., 1989), and aph(3'')-Ic in Mycobacterium fortuitum (Ramón-García et al., 2006). Multiple sequence alignments showed that AphSR3 has all conserved domains typical for the APH(3'') subfamily (Heinzel et al., 1988; Wright and Thompson, 1999; Ramón-García et al., 2006). The aphSR3 gene was designated aph(3'')-Id. Analysis of the resistance to aminoglycoside antibiotics showed that the aph(3'')-Id gene determines the resistance of E. coli cells to streptomycin. The APH(3'')-Id (AphSR3) protein was first isolated in native conditions. Evaluation of the phosphotransferase activity of the APH(3'')-Id protein in vitro with two methods showed that the APH(3'')-Id protein is enzymatically active and capable of catalyzing phosphorylation of the substrate, the aminoglycoside antibiotic streptomycin. The ability of the newly identified streptomycin phosphotransferase to undergo autophosphorylation in vitro is unique to this class of enzymes. This property is well known for eukaryotic serine-threonine and tyrosine protein kinases (Hashimoto et al., 2008), as well as for bacterial serine-threonine protein kinases of the eukaryotic type (Damle and Mohanty, 2014). Autophosphorylation is an important mechanism for the regulation of the biological activity of protein kinases. However, there are no literature data on the autophosphorylation of aminoglycoside phosphotransferases. It should be noted that the autophosphorylation of serine/threonine and tyrosine is a unique property for this group of enzymes (Alekseeva et al., 2019).
Aminoglycoside Phosphotransferase AphSR2
A study on the effect of serine-threonine protein kinases on increased resistance to aminoglycoside antibiotics during the cocloning of the aphSR2 gene in E. coli and STPK genes pkSR1 and pkSR2 localized in the same cluster of the Streptomyces rimosus ATCC 10970 genome was described. When these genes were cocloned in E. coli, it was found that aphSR2 causes neomycin resistance, which is modeled by pkSR1. It was shown that cloning of the pkSR1 gene in E. coli confers resistance to neomycin and hygromycin. E. coli containing pkSR1 possesses increased resistance to neomycin, and coexpression of the genes aphSR2 and pkSR1 results in a twofold increase in the resistance level. The presented data are the second example of the effect of STPK on modulation of the level of aminoglycoside-antibiotic resistance in Streptomyces bacteria. It was shown that APHSR2 is the second aminoglycoside phosphotransferase of streptomycetes and, in particular, of S. rimosus, for which the data show that the level of resistance is increased by STPK and is an accumulative result of their coexpression. In practical terms, the data can be used to study the distribution and features of the functions of genes that determine natural resistance to aminoglycoside antibiotics in actinobacteria of the Streptomyces genus (Rudakova et al., 2020).
CONCLUSIONS
The results of many years of our own research and analysis of the literature indicate, it would seem, that aminoglycoside phosphotransferases currently pose a serious threat for antimicrobial therapy (Shakya and Wright, 2010; Ribeiro da Cunha et al., 2019). Classical representatives of aph, which are common in clinical strains of bacteria, cause resistance to antibiotics. They can be divided into several groups according to their substrate specificity: kanamycin phosphotransferases, streptomycin phosphotransferases, gentamycin phosphotransferases, and hygromycin phosphotransferases. The other genes may perform other functions (Shakya et al., 2011; Wright, 2011). To overcome the problem of antibiotic resistance, advances in the next generation of genome sequencing, bioinformatics, and analytical chemistry will be combined.
Aminoglycoside phosphotransferases are well studied in actinobacteria of producers of aminoglycoside antibiotics, but they are insufficiently studied in other actinobacteria of the Streptomyces genus.
Current approaches to the identification of genes responsible for antibiotic resistance include genomic DNA sequencing, followed by gene annotation. Actinobacteria of the Streptomyces genus contain 4–16 aph genes. The functions of genes annotated as aph in sequenced genomes are currently poorly understood and require further research to test whether these hypothetical enzymes are true aph resistance genes (Wright, 2019).
The genes that encode bacterial aminoglycoside phosphotransferases have a common evolutionary ancestor. The classification of the studied aph genes from Streptomyces bacteria based on phylogenetic similarity with previously known aph genes belonging to seven subfamilies of aminoglycoside phosphotransferases makes it possible to identify genes associated with resistance to aminoglycoside antibiotics and, in theory, to predict the spectrum of resistance encoded by this aph gene.
Thus, in the model S. rimosus ATCC 10970 strain with a high level of resistance to aminoglycosides, only three genes, aphSR5 (aph(3')-VIII), aphSR3 (aph(3'')-Id), and aphSR2, belong to the known subfamilies APH(3'), APH(3''), and APH(7'') based on phylogenetic similarity. Aminoglycoside phosphotransferases APH(3') modify the 3'-OH-group in a wide range of aminoglycosides, including kanamycin, neomycin, paromomycin, lividomycin, ribostamycin, butirosin, amikacin, and isepamicin. APH(3'') modify the 3''-ОН-groups of streptomycin, and APH(7'') provide bacterial resistance to hygromycin.
The emergence of drug-resistant bacteria has led to the need to develop new drugs. The obtainment of crystal structures of the studied enzymes allows the in silico docking of APH protein inhibitors belonging to different subfamilies. The inhibition of proteins that provide a natural level of bacterial resistance to a number of aminoglycoside antibiotics could help to overcome multidrug resistance in pathogenic actinobacteria and to expand the range of used drugs due to the synergistic effect of the antibiotic with the APH-inhibitor compound.
REFERENCES
Alekseeva, M.G., Mavletova, D.A., and Danilenko, V.N., Test-system Escherichia coli/aphVIII/gsk3β for selective screening of inhibitor of serine-threonine protein kinase GSK3β, Genetika (Moscow), 2018, vol. 54, no. 13, pp. 14–17.
Alekseeva, M.G., Boyko, K.M., Nikolaeva, A.Y., et al., Identification, functional and structural characterization of novel aminoglycoside phosphotransferase APH(3'')-Id from Streptomyces rimosus subsp. rimosus ATCC 10970, Arch. Biochem. Biophys., 2019, vol. 671, no. 4, pp. 111–122.
Anderson, A.S., Clark, D.J., Gibbons, P.H., and Sigmund, J.M., The detection of diverse aminoglycoside phosphotransferases within natural populations of actinomycetes, J. Ind. Microbiol. Biotechnol., 2002, vol. 29, no. 2, pp. 60–69.
Barka, E.A., Vatsa, P., Sanchez, L., et al., Taxonomy, physiology, and natural products of Actinobacteria,Mol. Biol. Rev., 2015, vol. 80, no. 1, pp. 1–43.
Bekker, O., Alekseeva, M., Osolodkin, D., et al., New test system for screening serine-threonine protein kinase inhibitors: Escherichia coli APHVIII/Pk25, Acta Nat., 2010, vol. 2, no. 3, pp. 126–139.
Bentley, S.D., Chater, K.F., Cerdeno-Tarraga, A.M., et al., Complete genome sequence of the model actinomycete Streptomyces coelicolor A3, Nature, 2002, vol. 417, pp. 141–147.
Berdy, J., Thought and facts about antibiotics: where we are now and where we are heading, J. Antibiot., 2012, vol. 65, no. 8, pp. 385–395.
Block, M. and Blanchard, D.L., Aminoglycosides, StatPearls Online, 2020, p. 621.
Bouchek-Mechiche, K., Gardan, L., Normand, P., and Jouan, B., DNA relatedness among strains of Streptomyces pathogenic to potato in France: description of three new species, S. europaeiscabiei sp. nov. and S. stelliscabiei sp. nov. associated with common scab, and S. reticuliscabiei sp. nov. associated with netted scab, Int. J. Syst. Evol. Microbiol., 2000, no. 50, pp. 91–99.
Boyko, K.M., Gorbacheva, M.A., Korzhenevskiy, D.A., et al., Structural characterization of the novel aminoglycoside phosphotransferase AphVIII from Streptomyces rimosus with enzymatic activity modulated by phosphorylation, Biochem. Biophys. Res. Commun., 2016, vol. 477, pp. 595–601.
Bryskier, A., Antibiotics and antibacterial agents: classifications and structure-activity relationships, in Antimicrobial Agents: Antibacterials and Antifungals, New York: Wiley, 2005, pp. 13–38.
Busscher, G., Rutjes, F., and Delft, F., 2-Deoxystreptamine: central scaffold of aminoglycoside antibiotics, Chem. Rev., 2005, no. 105, pp. 775–791.
Carter, A., Clemons, W., Brodersen, D., et al., Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics, Nature, 2000, no. 407, pp. 340–348.
Chandrika, T.N. and Garneau-Tsodikova, S., Comprehensive review of chemical strategies for the preparation of new aminoglycosides and their biological activities, Chem. Soc. Rev., 2018, vol. 47, no. 4, pp. 1189–1249.
Cobb, R.E., Wang, Y., and Zhao, H., High-efficiency multiplex genome editing of Streptomyces species using an engineered CRISPR/Cas system, ACS Synth. Biol., 2015, vol. 4, pp. 723−728.
Collins, A.C., Ashenafi, M., Saunders, A.A., and Byrnes, W.M., Cloning and expression of streptomycin inactivating enzymes APH(6)-Ia and APH(6)-Id, Cell. Mol. Biol., 2007, vol. 53, no. 3, pp. 74–79.
Damle, N.P. and Mohanty, D., Mechanism of autophosphorylation of mycobacterial PknB explored by molecular dynamics simulations, Biochemistry, 2014, vol. 53, pp. 4715–4726.
Danilenko, V.N., Puzynina, G.G., and Lomovskaya, N.D., Multiple resistance to antibiotics in actinomycetes, Genetika (Moscow), 1977, vol. 13, no. 10, pp. 1831–1842.
Danilenko, V.N., Akopyants, K.E., Sizova, I.A., and Michurina, T.A., Determination of the nucleotide sequence and characterization of the novel aminoglycoside phosphotransferase aphVIII gene from the Streptomyces rimosus strain, Russ. J. Genet., 1997, vol. 33, no. 11, pp. 1264–1272.
Danilenko, V.N., Simonov, A.Y., Lakatosh, S.A., et al., Search for inhibitors of bacterial and human protein kinases among derivatives of diazepines[1,4] annelated with maleimide and indole cycles, J. Med. Chem., 2008, vol. 51, no. 24, pp. 7731–7736.
Davies, J. and Wright, G., Bacterial resistance to aminoglycoside antibiotics, Trends Microbiol., 1997, vol. 5, no. 6, pp. 234–240.
Elizarov, S.M., Sergienko, O.V., Sizova, I.A., and Danilenko, V.N., Dependence of aminoglycoside 3'-phosphotransferase VIII activity on serine/threonine protein kinases in Streptomyces rimosus, Mol. Biol. (Moscow), 2005, vol. 39, no. 2, pp. 226–233.
Elizarov, S.M., Alekseeva, M.G., Novikov, F.N., et al., Identification of phosphorylation sites in aminoglycoside phosphotransferase VIII from Streptomyces rimosus,Biochemistry (Moscow), 2012, vol. 77, no. 11, pp. 1258–1265.
Fiedler, H.P., Bruntner, C., Bull, A.T., et al., Marine actinomycetes as a source of novel secondary metabolites, Antonie Leeuwenhoek, 2005, vol. 87, pp. 37–42.
Forge, A. and Schacht, J., Aminoglycoside antibiotics, Audiol. Neurootol., 2000, vol. 5, pp. 3–22.
Fourmy, D., Recht, M., Blanchard, S., and Puglisi, J., Structure of the A site of Escherichia 16S ribosomal RNA complexed with an aminoglycoside antibiotic, Science, 1996, vol. 27, pp. 1367–1374.
Frase, H., Toth, M., and Vakulenko, S., Revisiting the nucleotide and aminoglycoside substrate specificity of the bifunctional aminoglycoside acetyltransferase(6')-Ie/aminoglycoside phosphotransferase(2'')-Ia enzyme, J. Biol. Chem., 2012, vol. 287, pp. 43262–43269.
Gibson, M.K., Crofts, T.S., and Dantas, G., Antibiotics and the developing infant gut microbiota and resistome, Curr. Opin. Microbiol., 2015, vol. 27, pp. 51–56.
Goyer, C., Faucher, E., and Beaulieu, C., Streptomyces caviscabies sp. nov., from deep-pitted lesions in potatoes in Quebec, Canada, Int. J. Syst. Evol. Microbiol., 1996, vol. 46, pp. 635–639.
Gullón, S. and Mellado, R.P., The cellular mechanisms that ensure an efficient secretion in Streptomyces,Antibiotics, 2018, vol. 7, no. 33, pp. 1–13.
Hashimoto, Y.K., Satoh, T., Okamoto, M., and Takemori, H.J., Importance of autophosphorylation at Ser186 in the A-loop of salt inducible kinase 1 for its sustained kinase activity, Cell. Biochem., 2008, vol. 104, pp. 1724–1739.
Heinzel, P., Werbitzky, O., Distler, J., and Piepersberg, W., Second streptomycin resistance gene from Streptomyces griseus codes for streptomycin-3"-phosphotransferase. Relationships between antibiotic and protein kinases, Arch. Microbiol., 1988, vol. 150, no. 2, pp. 184–192.
Hermann, T., Drugs targeting the ribosome, Curr. Opin. Struct. Biol., 2005, vol. 15, pp. 355–366.
Hermann, T., Aminoglycoside antibiotics: old drugs and new therapeutic approaches, Cell. Mol. Life Sci., 2007, vol. 64, pp. 1841–1852.
Hoshiko, S., Nojiri, C., Matsunaga, K., et al., Nucleotide sequence of the ribostamycin phosphotransferase gene and of its control region in Streptomyces ribosidificus,Gene, 1988, vol. 68, pp. 285–296.
Hossion, A.M. and Sasaki, K., Novel quercetin glycosides as potent anti-MRSA and anti-VRE agents, Recent Pat. Anti-Infect. Drug Discovery, 2013, vol. 8, no. 3, pp. 198–205.
Hotta, K. and Kondo, S., Kanamycin and its derivative, arbekacin: significance and impact, J. Antibiot., 2018, vol. 71, pp. 417–424.
Jana, S. and Deb, J., Molecular understanding of aminoglycoside action and resistance, Microbiol. Biotechnol., 2006, vol. 70, pp. 140–150.
Kemung, H., Tan, L., Khan, T., et al., Streptomyces as a prominent resource of future anti-MRSA drugs, Front. Microbiol., 2018, vol. 9, pp. 2221–2247.
Kim, C. and Mobashery, S., Phosphoryl transfer by aminoglycoside 3'-phosphotransferases and manifestation of antibiotic resistance, Bioorg. Chem., 2005, vol. 33, pp. 149–158.
Kononen, E. and Wade, W.G., Actinomyces and related organisms in human infections, Clin. Microbiol. Rev., 2015, vol. 28, no. 2, pp. 419–442.
Krause, K.M., Serio, A.W., Kane, T.R., and Connolly, L.E., Aminoglycosides: an overview, Cold Spring Harbor Perspect. Med., 2016, vol. 6, no. 6, pp. 1–19.
Li, D., Li, H., Ni, X., et al., Construction of a gentamicin C1a-overproducing strain of Micromonospora purpurea by inactivation of the gacD gene, Microbiol. Res., 2013, vol. 168, no. 5, pp. 263–267.
Link, N.A., Chen, P., Lu, W.J., et al., Collective form of cell death requires homeodomain interacting protein kinase, J. Cell Biol., 2007, vol. 178, no. 4, pp. 567–574.
Loria, R., Kers, J., and Joshi, M., Evolution of plant pathogenicity in Streptomyces,Ann. Rev. Phytopathol., 2006, vol. 44, pp. 469–487.
Magnet, S. and Blanchard, J., Molecular insights into aminoglycoside action and resistance, Chem. Rev., 2005, vol. 105, pp. 477–497.
Morar, M. and Wright, G., The genomic enzymology of antibiotic resistance, Ann. Rev. Genet., 2010, vol. 44, pp. 25–51.
Motkova, M.O., Gladkikh, E.G., and Korobkova, T.P., Stability of the antibiotic formation trait of the tobramycin producer Streptomyces cremeus subsp. Tobramycini,Antibiotiki, 1984, vol. 29, no. 2, pp. 83–85.
Nepal, K.K., Oh, T.J., and Sohng, J.K., Heterologous production of paromamine in Streptomyces lividans TK24 using kanamycin biosynthetic genes from Streptomyces kanamyceticus ATCC 12853, Mol. Cells, 2009, vol. 27, no. 5, pp. 601–608.
Newman, D.J. and Cragg, G.M., Natural products as sources of new drugs over the last 25 years, J. Nat. Prod., 2007, vol. 70, no. 3, pp. 461–477.
Ogle, J.M. and Ramakrishnan, V., Structural insights into translational fidelity, Ann. Rev. Biochem., 2005, vol. 74, pp. 129–177.
Ogle, J.M., Carter, A.P., and Ramakrishnan, V., Insights into the decoding mechanism from recent ribosome structures, Trans. Biochem. Sci., 2003, vol. 28, pp. 259–266.
Ota, Y., Tamegai, H., Kudo, F., et al., Butirosin-biosynthetic gene cluster from Bacillus circulans,J. Antibiot., 2000, vol. 53, no. 10, pp. 1158–1167.
Park, D.H., Yu, Y.M., Kim, J.S., et al., Characterization of Streptomycetes causing potato common scab in Korea, Plant Dis., 2003, vol. 87, pp. 1290–1299.
Pethick, F.E., MacFadyen, A.C., Tang, Z., et al., Draft genome sequence of the oxytetracycline-producing bacterium Streptomyces rimosus ATCC 10970, Genome Announce., 2013, vol. 1, no. 2, pp. 1–2.
Potekhin, Ya.A. and Danilenko, V.N., The determinant of kanamycin resistance of Streptomyces rimosus: amplification in the chromosome and reversed genetic instability, Mol. Biol. (Moscow), 1985, vol. 19, no. 3, pp. 805–811.
Ramirez, M. and Tolmasky, M., Aminoglycoside modifying enzymes, Drug Resist. Updates, 2010, vol. 13, pp. 151–171.
Ramón-García, S., Otal, I., Martín, C., et al., Novel streptomycin resistance gene from Mycobacterium fortuitum,Antimicrob. Agents Chemother., 2006, vol. 50, no. 11, pp. 3920–3922.
Reshed’ko, G., The importance of enzymatic modification of aminoglycosides in the development of resistance in bacteria, Klin. Mikrobiol., Antimikrob.Khimioter., 1999, vol. 1, no. 1, pp. 40–50.
Ribeiro da Cunha, B., Fonseca, L.P., and Calado, C.R., Antibiotic discovery: where have we come from, where do we go? Antibiotics, 2019, vol. 8, no. 2, p. e45.
Rudakova, N.N., Alekseeva, M.G., Zakharevich, N.V., et al., Aminoglycoside phosphotransferase AphSR2 from Streptomyces rimosus ATCC 10970: dependence of antibiotic resistance on serine-threonine protein kinases PkSR1 and PkSR2, Russ. J. Genet., 2020, vol. 56, no. 1, pp. 112–117.
Salauze, D., Perez-Gonzalez, J.A., Piepersberg, W., and Davies, J., Characterization of aminoglycoside acetyltransferase-encoding genes of neomycin-producing Micromonospora chalcea and Streptomyces fradiae,Gene, 1991, vol. 101, pp. 143–148.
Scholz, P., Haring, V., Wittmann-Liebold, B., et al., Complete nucleotide sequence and gene organization of the broad-host-range plasmid RSF1010, Gene, 1989, vol. 75, no. 2, pp. 271–288.
Seipke, R.F., Kaltenpoth, M., and Hutchings, M.I., Streptomyces as symbionts: an emerging and widespread theme? FEMS Microbiol. Rev., 2012, vol. 36, pp. 862–876.
Serio, A., Magalhães, M., Blanchard, J.S., and Connolly, L., Aminoglycosides: mechanisms of action and resistance, in Antimicrobial Drug Resistance, Dordrecht: Springer, 2017, pp. 213–229.
Shaeer, K.M., Zmarlicka, M.T., Chahine, E.B., and Piccicacco, N., Plazomicin: a next-generation aminoglycoside, Pharmacotherapy, 2019, vol. 39, no. 1, pp. 77–93.
Shakya, T. and Wright, G., Nucleotide selectivity of antibiotic kinases, Antimicrob. Agents Chemother., 2010, vol. 54, pp. 1909–1913.
Shakya, T.A., Stogios, P., Waglechner, N., et al., Small molecule discrimination map of the antibiotic resistance kinome, Chem. Biol., 2011, vol. 18, pp. 1591–1601.
Shi, K. and Berghuis, A., Structural basis for dual nucleotide selectivity of aminoglycoside 2''-phosphotransferase IVa provides insight on determinants of nucleotide specificity of aminoglycoside kinases, Biol. Chem., 2012, vol. 287, pp. 13094–13102.
Shi, K., Caldwell, S., Fong, D., and Berghuis, A., Prospects for circumventing aminoglycoside kinase mediated antibiotic resistance, Front. Cell. Infect. Microbiol., 2013, vol. 3, no. 22, pp. 1–17.
Siti, J.A., Mohd, B., Syarul, N.B., et al., Discovery of antimalarial drugs from Streptomycetes metabolites using a metabolomic approach, J. Trop. Med., 2017, vol. 2017, pp. 1–7.
Sizova, I.A., Lapina, T.V., Frolova, O.N., et al., Stable nuclear transformation of Chlamydomonas reinhardtii with a Streptomyces rimosus gene as the selective marker, Gene, 1996, vol. 181, pp. 13–18.
Sizova, I., Fuhrmann, M., and Hegemann, P.A., Streptomyces rimosus aphVIII gene coding for a new type phosphotransferase provides stable antibiotic resistance to Chlamydomonas reinhardtii,Gene, 2001, vol. 277, pp. 221–229.
Sizova, I.A., Khegemann, P., Furmann, M., and Danilenko, V.N., Streptomyces rimosus aminoglycoside 3'-phosphotransferase VIII: comparisons with aminoglycoside 3'-phosphotransferases of aminoglycoside-producing strains and with eukaryotic protein kinases, Mol. Biol. (Moscow), 2002, vol. 36, no. 1, pp. 18–25.
Smith, C.A. and Baker, E.N., Aminoglycoside antibiotic resistance by enzymatic deactivation, Curr. Drug Targets: Infect. Disord., 2002, vol. 2, pp. 143–160.
Smith, C., Toth, M., Frase, H., et al., Aminoglycoside-2'' phosphotransferase-IIIa (APH(2'')-IIIa) prefers GTP over ATP: structural templates for nucleotide recognition in the bacterial aminoglycoside-2'' kinases, Biol. Chem., 2012, vol. 287, pp. 12893–12903.
Starodubtseva, L.I., Taisova, A.S., and Danilenko, V.N., Amplification of the determinant of kanamycin resistance Kmr in the composition of constructed hybrid plasmids in Streptomyces lividans strain, Antibiot. Med. Biotekhnol., 1985, no. 5, pp. 565–572.
Sutcliffe, J., Improving on nature: antibiotics that target the ribosome, Curr. Opin. Microbiol., 2005, vol. 8, pp. 534–542.
Takahashi, Y. and Nakashima, T., Actinomycetes, an inexhaustible source of naturally occurring antibiotics, Antibiotics, 2018, vol. 7, no. 45, p. 45.
Tao, W., Yang, A., Deng, Z., and Sun, Y., CRISPR/Cas9-based editing of Streptomyces for discovery, characterization, and production of natural products, Front. Microbiol., 2014, vol. 9, pp. 1660–1668.
Thapa, L.P., Oh, T.J., Liou, K., and Sohng, J.K., Biosynthesis of spectinomycin: heterologous production of spectinomycin and spectinamine in an aminoglycoside-deficient host, Streptomyces venezuelae YJ003, J. Appl. Microbiol., 2008, vol. 105, no. 1, pp. 300–308.
Thompson, C.J. and Gray, G.S., Nucleotide sequence of a streptomycete aminoglycoside phosphotransferase gene and its relationship to phosphotransferases encoded by resistance plasmids, Proc. Natl. Acad. Sci. U.S.A., 1983, vol. 80, pp. 5190–5194.
Tomihama, T., Nishi, Y., Sakai, M., and Ikenaga, M., Draft genome sequences of Streptomyces scabiei S58, Streptomyces turgidiscabies T45, and Streptomyces acidiscabies a10, the pathogens of potato common scab, isolated in Japan, Genome Announce., 2016, vol. 4, pp. 1–2.
Vakulenko, S. and Mobashery, S., Versatility of aminoglycosides and prospects for their future, Clin. Microbiol. Rev., 2003, vol. 16, no. 3, pp. 430–450.
Vastrad, B.M. and Neelagund, S.E., Optimization of medium composition for the production of neomycin by Streptomyces fradiae NCIM 2418 in solid state fermentation, Biotechnol. Res. Int., 2014, vol. 2014, pp. 1–11.
Veyssier, P. and Bryskier, A., Aminocyclitol aminoglycosides, in Antimicrobial Agents: Antibacterials and Antifungals, New York: Wiley, 2005, pp. 453–469.
Vicens, Q. and Westhof, E., Molecular recognition of aminoglycoside antibiotics by ribosomal RNA and resistance enzymes: an analysis of X-ray crystal structures, Biopolymers, 2003, vol. 70, pp. 42–57.
Wilson, D., Ribosome-targeting antibiotics and mechanisms of bacterial resistance, Nat. Rev. Microbiol., 2014, vol. 12, pp. 35–48.
Wright, G.D., Molecular mechanisms of antibiotic resistance, Chem. Commun., 2011, vol. 47, no. 14, pp. 4055–4061.
Wright, G.D., Environmental and clinical antibiotic resistomes, same only different, Curr. Opin. Microbiol., 2019, vol. 51, pp. 57–63.
Wright, G.D. and Thompson, P.R., Aminoglycoside phosphotransferases: proteins, structure, and mechanism, Front. Biosci., 1999, vol. 4, pp. D9–D21.
Wright, G., Berghuis, A., and Mobashery, S., Aminoglycoside antibiotics: structures, functions, and resistance, in Resolving the Antibiotic Paradox: Progress in Understanding Drug Resistance and Development of New Antibiotics, Rosen, B.P. and Mobashery, S., Eds., New York: Plenum, 1998, pp. 27–69.
Zaher, H.S. and Green, R., Fidelity at the molecular level: lessons from protein synthesis, Cell, 2009, vol. 136, pp. 746–762.
Zarate, S.G., De la Cruz Claure, M.L., Benito-Arenas, R., et al., Overcoming aminoglycoside enzymatic resistance: design of novel antibiotics and inhibitors, Molecules, 2018, vol. 23, no. 2, p. E284.
Zhang, Y. and Loria, R., Emergence of novel pathogenic Streptomyces species by site-specific accretion and cis-mobilization of pathogenicity islands, Mol. Plant-Microbe Interact., 2017, vol. 30, no. 1, pp. 72–82.
Zhukova, Yu., Alekseeva, M., Zakharevich, N., et al., Pim family of protein kinases: structure, functions, and roles in hematopoietic malignancies, Mol. Biol., 2011, vol. 45, no. 5, pp. 695–703.
Funding
The work was carried out within the State Task Genetic technologies in biology, medicine, agricultural and environmental activities, project no. 0112-2019-0002 2019.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interests. The authors declare that they have no conflicts of interest.
Statement on the welfare of humans or animals. This article does not contain any studies involving animals performed by any of the authors.
Additional information
Translated by D. Novikova
Rights and permissions
About this article
Cite this article
Rudakova, N.N., Alekseeva, M.G. & Danilenko, V.N. Genes of Aminoglycoside Phosphotransferases in Soil Bacteria of the Streptomyces Genus. Biol Bull Rev 10, 507–519 (2020). https://doi.org/10.1134/S2079086420060055
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S2079086420060055