Abstract
Background
Hirschsprung disease (HSCR) is a congenital intestinal disease characterised by functional obstruction of the colon. Herein, we investigated the role and mechanism of the gene GFRA4 in HSCR.
Methods
GFRA4 expression in the ganglionic and aganglionic segment tissues in patients with HSCR and healthy colon tissues were detected using qRT-PCR, western blot, and immunohistochemistry. Cell proliferation, cycle distribution, apoptosis, changes in mitochondrial membrane potential, and differentiation were assessed in mouse enteric neural crest stem cells (ENCSCs) using the CCK-8 assay, EdU staining, flow cytometry, JC-1 probe, and immunofluorescence, respectively. GSEA analysis was performed to screen the signaling pathways regulated by GFRA4.
Results
GFRA4 was downregulated in aganglionic segment tissues compared to control and ganglionic segment tissues. GFRA4 overexpression promoted proliferation and differentiation, and inhibited apoptosis in ENCSCs, while GFRA4 down-regulation had the opposite result. GFRA4 activated the hedgehog pathway. GFRA4 overexpression enhanced the expression of key factors of the hedgehog pathway, including SMO, SHH, and GLI1. However, GFRA4 down-regulation reduced their expression. An antagonist of hedgehog pathway, cyclopamine, attenuated the effect of GFRA4 overexpression on proliferation, differentiation, and apoptosis of ENCSCs.
Conclusion
GFRA4 promotes proliferation and differentiation but inhibits apoptosis of ENCSCs via the hedgehog pathway in HSCR.
Impact
-
This study confirms that GFRA4 improves the proliferation and differentiation of ENCSCs via modulation of the hedgehog pathway.
-
This study for the first time revealed the role and the mechanism of the action of GFRA4 in HSCR, which indicates that GFRA4 may play a role in the pathological development of HSCR.
-
Our findings may lay the foundation for further investigation of the mechanisms underlying HSCR development and into targets of HSCR treatment.
Similar content being viewed by others
Introduction
As an alimentary tract disease, Hirschsprung disease (HSCR) is clinically characterised by intestinal obstruction, abdominal distension, delayed meconium excretion, and vomiting.1 The incidence of HSCR has been reported to be approximately 1/5000 in newborn children (male: female = 4:1).2 Research has shown that the primary cause of HSCR is the failure of proliferation and migration of enteric neural crest cells (ENCCs) to the distal part of the colon during embryonic development.3 At present, the most common therapeutic treatment for HSCR is surgical removal of the abnormally innervated intestine.4 However, postoperative intestinal dysfunction is common.5 Thus, probing the nosogenesis and identifying potential therapeutic targets for HSCR treatment remains an urgent unmet need.
Previous studies have indicated that HSCR pathogenesis is related to the levels of glial cell-derived neurotrophic factor (GDNF) and rearranged during transfection (RET).6,7 Presently, four GDNF receptor alpha (GFRA) proteins (GFRA1, GFRA2, GFRA3, and GFRA4) have been identified to bind GDNF.8 Unlike the other GFRA family members, GFRA4 is smaller in size and lacks the first Cys-rich domain.9 Moreover, GFRA4 has been shown to activate RET to promote neuronal survival and differentiation.10 Interestingly, GFRA4 expression was shown to be downregulated in glioma primary cells compared with normal brain primary cells.11 Based on these findings, we surmised that GFRA4 may play an important role in HSCR pathogenesis. However, there has been little research on the influence and underlying role of GFRA4 on the pathogenesis of HSCR.
In the present study, we detected GFRA4 expression in HSCR tissues and investigated the biological function of GFRA4 in mouse enteric neural crest stem cells (ENCSCs). Moreover, we found that GFRA4 affected the biological function of ENCSCs by regulation of the hedgehog signaling pathway. Our findings may lay the foundation for further investigation of the mechanisms underlying HSCR development and into targets of HSCR treatment.
Methods
Human specimen collection
In this study, 20 HSCR specimens and 20 normal colon specimens were collected from patients at our hospital. The resected HSCR tissues include ganglionic and aganglionic segments which were taken from the most proximal and most distal margin of the resected pull-through specimen, respectively. Normal colon tissues collected from imperforate anus patients after colostomy were used as controls. After surgery, specimens were stored at −80 °C. Patients and their guardians all signed an informed consent form to participate. This study was authorised by the Ethics Committee of Shandong Provincial Hospital Affiliated to Shandong First Medical University. Ganglionic and aganglionic segment tissues were identified by histopathological analysis of hematoxylin and eosin (H&E)-stained slides.
Quantitative real-time polymerase chain reaction (qRT-PCR)
RNA isolater Total RNA Extraction Reagent (Vazyme, China) was utilised to isolate total RNA from ENCSCs and patient tissue. PrimeScript™ RT Reagent Kit with gDNA Eraser (Takara, Japan) was used to synthesise complementary DNA. SYBR Green PCR Mix Kit (Takara) was used to perform qRT-PCR reactions. The primers were as follows: human GFRA4 (forward: 5′-GTCACCCCTAACTACGTGGAC-3′, reverse: 5′-CGGTTCCTGGTAAAGAGCCC-3′), human GAPDH (forward: 5′-TGCAACCGGGAAGGAAATGA -3′, reverse: 5′- GCCCAATACGACCAAATCAGA -3′), and mouse SMO (forward: 5′-AGCCTTTGCGCTACAACGTG-3′, reverse: 5′-TTCCGGAGGCCGGACCA-3′), mouse SHH (forward: 5′-GGTGATCCTTGCTTCCTCGC-3′, reverse: 5′-T TTGTCTTTGCACCTCTGAGTC-3′), mouse GLI1 (forward: 5′- TTGCAGCCAGGAGTTCGATT-3′, reverse: 5′- TCCGACAGCCTTCAAACGTG-3′), and mouse GAPDH (forward: 5′- CAGGAGAGTGTTTCCTCGTCC -3′, reverse: 5′- GATGGGCTTCCCGTTGATGA -3′). GAPDH was used as an internal control for normalization. The relative mRNA expression was calculated using the 2−∆∆CT method.
Immunohistochemical staining
After fixation in 4% paraformaldehyde, specimens were embedded in paraffin. After dewaxing with xylene and rehydration in graded alcohols, to retrieve antigens, sections (5-μm thick) were treated with citrate buffer (pH = 6.0) at 95 °C for 40 min. Next, sections were treated with 3% hydrogen peroxide solution for 10 min at room temperature (RT). Subsequently, sections were incubated with 5% bovine serum albumin solution for 1 h at RT. The primary polyclonal GFRA4 antibody (1:100; Bioss, China) was incubated with sections at 4 °C overnight. After washing three times with phosphate buffer saline (PBS), sections were incubated with the secondary antibody (1:50; Beyotime Biotechnology, China) for 1 h at RT, after which they were treated with diaminobenzidine (DAB) solution (Beyotime Biotechnology) to develop colour before being counterstained in hematoxylin. Finally, the sections were observed under a light microscope.
Cell culture and treatment
Mouse ENCSCs were purchased from Procell Life Science&Technology Co., Ltd. (China). Cells were cultured in serum-free culture medium containing 15% chick embryo extract, 20 ng/mL FGF, 20 ng/mL EGF, 35 ng/mL Retinoic acid, 1% N2, 2% B27, and 50 mM β-mercaptoethanol in poly-D-lysine-coated and fibronectin-coated wells at 37 °C and 5% CO2 and enriched by multiple replatings as described previously.12 ENCSCs at passage 3 were harvested for immunofluorescent staining using anti-Nestin (ab6142, Abcam).
Shanghai Genechem Co., Ltd. (China) provided the overexpression plasmid of the GFRA4, empty plasmid (vector), siRNAs [siGFRA4-1 (sense: 5’-GCAAGCUCUUUACAAGGAACC-3’, anti-sense: 5’- UUCCUUGUAAAGAGCUUGCGG-3’), siGFRA4-2 (sense: 5’-CGCGUUGUCUGCGCGUCUACG-3’, anti-sense: 5’-UAGACGCGCAGACAACGCGGG-3’)] and its negative control (siNC; sense: 5’-UUCUCCGAACGUGUCACGUTT-3’, anti-sense: 5’-ACGUGACACGUUCGGAGAATT-3’). Lipofectamine 2000 reagent (Invitrogen) was used for transfection following the manufacturer’s instructions. Cells were treated with 5 μM cyclopamine (Selleck) for 24 h to inhibit the hedgehog pathway.
Cell Counting Kit-8 (CCK-8) assay
Transfected cells were cultured for 0, 24, 48 h, and 72 h. Subsequently, cells in each well were treated with 10 μl CCK-8 solution (Beyotime Biotechnology) for 1 h at 37 °C. Finally, the optical density (OD) at 450 nM was measured using a microplate reader (Bio-Tek).
5-Ethynyl-2’-deoxyuridin (EdU) staining
After the indicated treatment, cells were incubated with EdU (50 μM; Guangzhou RiboBio Co., Ltd) for 2 h at 37 °C before being fixed in 4% formaldehyde. Cells were then incubated with 1 × Apollo reaction mixture for 30 min, and then stained with Hoechst 33342 for 30 min. Finally, EdU positive cells were analyzed under a fluorescent microscope (Nikon, Japan).
Flow cytometry analysis
For cell cycle analysis, cells were suspended with 300 μL of ice-cold PBS. Subsequently, cells were fixed in 70% ethanol for 2 h at 4 °C and treated with propidium iodide (PI, Beyotime Biotechnology). For cell apoptosis analysis, cells were resuspended in binding buffer, and then incubated with Annexin V-FITC (Beyotime Biotechnology) and PI for 15 min at RT in the dark. Finally, a FACScan flow cytometer (Becton Dickinson) was used to analyze cell cycle distribution and apoptosis.
JC-1 detection of mitochondrial membrane potential
According to the instructions of the mitochondrial membrane potential kit (Beyotime Biotechnology), transfected cells were incubated with 1 ml JC-1 dye buffer for 20 min at 37 °C. After centrifugation at 600 g for 3 min, cells were washed with 1 × JC-1 dye buffer. Ultimately, the red/green fluorescence intensity was measured under a fluorescent microscope.
Immunofluorescence
Cells were stabilised using 4% paraformaldehyde. After washing with PBS three times, cells were permeated with 0.3% Triton X-100 (Sigma) for 15 min. Subsequently, 5% BSA (Beyotime Biotechnology) was used to block cells for 1 h at RT. Next, cells were incubated with primary antibodies (anti-TUJ1 and anti-GFAP; Cell Signaling Technology) overnight at 4 °C, and then incubated with secondary antibody (Cell Signaling Technology) for 2 h at RT. DAPI (Beyotime Biotechnology) was used to counterstain cells. Finally, TUJ1 and GFAP positive cells were observed under a fluorescence microscope (Nikon Eclipse Ni-U, Japan).
Western blot
Proteins were extracted from tissues or cells using RIPA buffer (Beyotime Biotechnology). Next, 30 μg proteins were separated using SDS-PAGE and transferred onto a PVDF membrane. Membranes were blocked with 5% non-fat dried milk. After washing with TBST for three times, the membrane was incubated with primary antibodies against GFRA4 (Bioss.), Bcl-2, Cleaved Caspase3, TUJ1, GFAP (Cell Signaling Technology), Bax, SMO, SHH, GLI1, and GAPDH (Proteintech) at 4 °C overnight, and then incubated with secondary antibodies (Cell Signaling Technology) at RT for 2 h. Finally, proteins were visualised using an enhanced chemiluminescence (ECL) Kit (Merck), and ImageJ software was utilized to quantify protein expression.
Bioinformatic analysis
The signal pathways that could be regulated by GFRA4 were analyzed using gene set enrichment analysis (GSEA) based on the Wiki Pathways gene sets dataset. To rank gene sets associated with risk, R package cluster Profiler was used in GSEA.
Statistical analysis
All data from three independent experiments were presented as the mean ± standard deviation. GraphPad Prism software was used for statistical analyses using Student’s t-test or analysis of variance followed by Turkey’s post-test. P < 0.05 indicated statistical significance.
Results
Human HSCR specimens show low expression of GFRA4
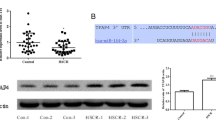
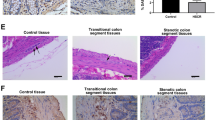
In this study, the expression of GFRA4 in the ganglionic and aganglionic segment tissues in patients with HSCR and normal colon tissues were detected. qRT-PCR data and western blotting results showed that GFRA4 mRNA and protein level expression was decreased significantly in aganglionic specimens compared with control and ganglionic segment tissues (P < 0.01, Fig. 1a, b). Next, immunohistochemistry staining of GFRA4 was performed, revealing a low-level expression of GFRA4 in aganglionic specimens as compared to the control and ganglionic segment tissues (Fig. 1c). H&E staining of ganglion cells was subsequently used to identify the ganglionic and aganglionic segment tissues from patients with HSCR.
a The mRNA level of GFRA4 in the ganglionic and aganglionic segment tissues in 20 patients with HSCR and 20 normal colon tissues were detected using qRT-PCR. b The protein level of GFRA4 in randomly selected control, ganglionic and aganglionic segments (4 in each group) were detected using western blot. c H&E staining of ganglion cells was used to identify the ganglionic and aganglionic segment tissues from patients with HSCR. GFRA4-positive cells were detected using immunohistochemical staining. **P < 0.01.
GFRA4 improves the proliferation of ENCSCs
To further investigate the biological function of GFRA4, functional gain and loss experiments were performed in ENCSCs. As shown in Fig. 2a, b, compared with the vector group, GFRA4 overexpression increased cell viability (P < 0.01), but GFRA4 low-expression (siGFRA4-1 and siGFRA4-2) decreased cell viability compared with the siNC group (P < 0.05). EdU assay further confirmed that GFRA4 overexpression increased the number of EdU-positive cells (P < 0.01, Fig. 2c), while GFRA4 low-expression reduced the number of EdU-positive cells (P < 0.01, Fig. 2d). Furthermore, GFRA4 overexpression reduced the number of cells in the G0/G1 phase and increased the number of cells in the S phase (P < 0.01, Fig. 2e), but GFRA4 low-expression led to the opposite results (Fig. 2f).
GFRA4 inhibits mitochondria-dependent apoptosis in ENCSCs
Next, we investigated the effect of GFRA4 on ENCSC apoptosis. As shown in Fig. 3a, b, GFRA4 overexpression reduced the apoptosis rate (P < 0.01), but GFRA4 low-expression promoted ENCSC apoptosis (P < 0.05). Furthermore, GFRA4 overexpression promoted Bcl-2 expression and inhibited the expression of Bax and cleaved caspase3 (P < 0.05, Fig. 3c), while GFRA4 low-expression resulted in the opposite results (Fig. 3d). When apoptosis occurs, JC-1 is released from mitochondria due to mitochondrial membrane potential depolarization, leading to a green fluorescence. Our results showed that GFRA4 overexpression increased the red/green fluorescence ratio in ENCSCs (P < 0.01, Fig. 3e), but GFRA4 low-expression reduced the red/green fluorescence ratio (P < 0.01, Fig. 3f).
GFRA4 improves the differentiation of ENCSCs
We assessed the change of TUJ1 (a neuronal marker) and GFAP (a common marker of glial cells) to explore the influence of GFRA4 on ENCSC differentiation. GFRA4 overexpression increased the TUJ1- and GFAP-positive cells (P < 0.01, Fig. 4a, c), while GFRA4 low-expression reduced the TUJ1- and GFAP-positive cells (P < 0.01, Fig. 4b, d). Western blotting confirmed the results of immunofluorescence (Fig. 4e–h).
GFRA4 activates the hedgehog signaling pathway in ENCSCs
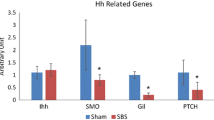
GSEA analysis showed that GFRA4 regulate a list of signaling pathways, including the hedgehog signaling pathway, longevity regulation pathway, GnRH signaling pathway, etc (Fig. 5a). Herein, we selected the hedgehog signaling pathway for further investigation as it ranked first in the enrolment score. Subsequently, the change of key factors of hedgehog signaling pathway was assessed. As shown in Fig. 5b, c, GFRA4 overexpression promoted the mRNA expression of SMO, SHH and GLI1 (P < 0.01), but the lower expression of SMO, SHH and GLI1 mRNA was observed in the si-GFRA4-1 and si-GFRA4-2 groups (P < 0.01). Importantly, similar results were presented in western blot analysis (Fig. 5d, e).
GFRA4 improves proliferation and differentiation but inhibits apoptosis of ENCSCs via activation of the hedgehog signaling pathway
To clarify the relationship of hedgehog signaling pathway and GFRA4 in ENCSCs, we treated cells with cyclopamine (an antagonist of the Hedgehog signaling pathway). EdU staining results showed that GFRA4 overexpression increased the number of EdU-positive cells compared with vector group (P < 0.01, Fig. 6a), while cyclopamine attenuated the promotive role of GFRA4 overexpression. Meanwhile, compared with GFRA4 overexpression group, cyclopamine increased the number of cells in the G0/G1 phase and apoptosis, and decreased the number of cells in S phase (P < 0.05, Fig. 6b, c). Furthermore, cyclopamine reduced the TUJ1- and GFAP-positive cells (P < 0.05, Fig. 6d, e).
a Proliferation of ENCSCs was assessed using EdU assay. b Flow cytometry was conducted to investigate the cell cycle distribution in ENCSCs. c Apoptosis of ENCSCs was tested using flow cytometry. d TUJ1-positive cells were detected using immunofluorescence. e GFAP-positive cells were detected using immunofluorescence. *P < 0.05, **P < 0.01.
Discussion
As the most common disorder of the enteric nervous system (ENS) at birth, HSCR is usually diagnosed by anorectal manometry, barium enema, and biopsy of the rectum.13 The development of HSCR has been reported to be regulated by a multitude of genes, including many coding and non-coding genes.14,15,16 In the present study, we investigated the role of GFRA4 expression in HSCR. We found that GFRA4 was expressed at a lower level in aganglionic tissues than that in normal and ganglionic colon tissues. These findings indicate that GFRA4 may play a role in the pathological development of HSCR.
Perturbation of ENCC migration, proliferation, differentiation, survival, or apoptosis has been shown to be involved in the pathogenesis of HSCR. ENCCs cannot invade, proliferate, or migrate to the hindgut during the 5th to 12th week of embryogenesis.17 Although ENCCs can reach the distal intestine, they cannot proliferate or survive.18,19 Cell apoptosis usually occurs during development, and it plays a role as a homeostatic mechanism to maintain the number of cells in tissues.13 Apoptotic cell death is a normal process within early ENS development and proper apoptosis of neural crest cells has been shown to be conducive to functional ENS.20,21 In the present study, GFRA4 overexpression was found to promote cell proliferation, increased the number of cells in the S phase, and inhibit cell apoptosis. Additionally, GFRA4 low-expression led to the opposite results. We previously found that microRNA-483-5p low-expression promoted cell proliferation and reduced cell early apoptosis by targeting GFRA4 in 293 T and SH-SY5Y cells.22 Further research showed that overexpression of GFRA4 promoted cell proliferation and cell cycle progression, but inhibited apoptosis in SH-5YSY cells.23 A decrease of the mitochondrial membrane potential is considered the earliest event in the apoptosis cascade.24 A substantial change in the mitochondrial membrane potential leads to activation of the caspase protease family, subsequently leading to an apoptosis cascade reaction.25 Our results showed that GFRA4 overexpression inhibited the damage of mitochondrial membrane potential, enhanced Bcl-2 expression, and reduced Bax and cleaved caspase3 expression. These results demonstrated that GFRA4 promoted cell proliferation and inhibited cell apoptosis in HSCR.
Aganglionic bowel is commonly observed in patients with HSCR and often requires surgical resection.26 In patients with HSCR, stem cell therapy has been studied as a potential method to generate ganglia and restore gastrointestinal neuromuscular function.27 Prior studies have shown that injected neural crest stem cells migrate to the myenteric plexus of the aganglionic colon where they produce a glial network in vivo.28,29 Kato et al.previously reported that GFAP was strongly expressed in the normal gut and ganglionic segments in six cases of HSCR.30 In the present study, we detected the markers of differentiated neurons (TUJ1) and gliocytes (GFAP), and found that GFRA4 overexpression enhanced TUJ1 and GFAP expression. These data suggested that GFRA4 promoted the differentiation of ENCSCs into neurons and glial cells in HSCR.
To further explore the mechanism of GFRA4 in HSCR, we used GSEA software to analyze the downstream regulatory pathways of GFRA4, subsequently finding that GFRA4 activated the hedgehog signaling pathway. The hedgehog signaling pathway participate in many biological processes, including embryogenesis, proliferation, and tissue regeneration.31 Lau et al. previously reported that activation of the hedgehog pathway promoted development of cultured human pluripotent stem cells into ENCCs.32 Other studies indicated that the differentiation of ENCCs was regulated by hedgehog signaling during the development of ENS, and that aberrant activation of the hedgehog pathway caused an abnormal ratio of neuron to glia ratio in mice.33,34 In the present study, GFRA4 overexpression promoted the expression of key factors of the hedgehog signaling pathway (SMO, SHH, and GLI1), while knockdown of GFRA4 reduced the expression of SMO, SHH, and GLI1. Importantly, cyclopamine, an antagonist of the hedgehog signaling pathway, reversed the role of GFRA4 overexpression on cell proliferation, apoptosis, and differentiation of ENCSCs. Taken together, these results indicate that GFRA4 improved proliferation and differentiation, while reducing the apoptosis of ENCSCs via the hedgehog signaling pathway.
Overall, the present study showed that GFRA4 was down-regulated in aganglionic HSCR tissues. Further, GFRA4 promoted proliferation and differentiation but reduced apoptosis in ENCSCs, which was related to the hedgehog signaling pathway.
Data availability
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
References
Butler Tjaden, N. E. & Trainor, P. A. The developmental etiology and pathogenesis of Hirschsprung disease. Transl. Res. 162, 1–15 (2013).
Muise, E. D. & Cowles, R. A. Rectal biopsy for Hirschsprung’s disease: a review of techniques, pathology, and complications. World J. Pediatr.: WJP 12, 135–141 (2016).
Gunadi et al. Long-term functional outcomes of patients with Hirschsprung disease following pull-through. BMC Pediatr. 22, 022–03301 (2022).
Xia, R. P. et al. Circ-ITCH overexpression promoted cell proliferation and migration in Hirschsprung disease through miR-146b-5p/RET axis. Pediatr. Res. 92, 1008–1016 (2022).
Kyrklund, K. et al. ERNICA guidelines for the management of rectosigmoid Hirschsprung’s disease. Orphanet J. Rare Dis. 15, 020–01362 (2020).
Soret, R. et al. Genetic Background Influences Severity of Colonic Aganglionosis and Response to GDNF Enemas in the Holstein Mouse Model of Hirschsprung Disease. Int. J. Mol. Sci. 22, 13140 (2021).
Jain, S. et al. Organotypic specificity of key RET adaptor-docking sites in the pathogenesis of neurocristopathies and renal malformations in mice. J. Clin. Invest. 120, 778–790 (2010).
Souza, R. P. et al. Genetic association of the GDNF alpha-receptor genes with schizophrenia and clozapine response. J. Psychiatr. Res. 44, 700–706 (2010).
Enokido, Y. et al. GFR alpha-4 and the tyrosine kinase Ret form a functional receptor complex for persephin. Curr. Biol. 8, 1019–1022 (1998).
Yang, J., Runeberg-Roos, P., Leppänen, V. M. & Saarma, M. The mouse soluble GFRalpha4 receptor activates RET independently of its ligand persephin. Oncogene 26, 3892–3898 (2007).
Lee, K. et al. Proteome-wide discovery of mislocated proteins in cancer. Genome Res. 23, 1283–1294 (2013).
Ngan, E. S. et al. Hedgehog/Notch-induced premature gliogenesis represents a new disease mechanism for Hirschsprung disease in mice and humans. J. Clin. Investig. 121, 3467–3478 (2011).
Du, C. et al. Apoptotic neuron-secreted HN12 inhibits cell apoptosis in Hirschsprung’s disease. Int J. Nanomed. 11, 5871–5881 (2016).
Xie, H. et al. Long none coding RNA HOTTIP/HOXA13 act as synergistic role by decreasing cell migration and proliferation in Hirschsprung disease. Biochem Biophys. Res Commun. 463, 569–574 (2015).
Wu, F. et al. MPGES-1 derived PGE2 inhibits cell migration by regulating ARP2/3 in the pathogenesis of Hirschsprung disease. J. Pediatr. Surg. 54, 2032–2037 (2019).
Chen, G. et al. MicroRNA-939 inhibits cell proliferation via targeting LRSAM1 in Hirschsprung’s disease. Aging 9, 2471–2479 (2017).
Lake, J. I. & Heuckeroth, R. O. Enteric nervous system development: migration, differentiation, and disease. Am. J. Physiol. Gastrointest. Liver Physiol. 305, 2 (2013).
Langer, J. C. Hirschsprung disease. Curr. Opin. Pediatr. 25, 368–374 (2013).
Shu, X. et al. Treatment of aganglionic megacolon mice via neural stem cell transplantation. Mol. Neurobiol. 48, 429–437 (2013).
McKeown, S. J., Stamp, L., Hao, M. M. & Young, H. M. Hirschsprung disease: a developmental disorder of the enteric nervous system. Wiley Interdiscip. Rev. Dev. Biol. 2, 113–129 (2013).
Wallace, A. S. et al. Inhibition of cell death results in hyperganglionosis: implications for enteric nervous system development. Neurogastroenterol. Motil. 21, 768–e49 (2009).
Wang, G. et al. Downregulation of microRNA-483-5p Promotes Cell Proliferation and Invasion by Targeting GFRA4 in Hirschsprung’s Disease. DNA Cell Biol. 36, 930–937 (2017).
Wang, G. et al. Demethylation of GFRA4 Promotes Cell Proliferation and Invasion in Hirschsprung Disease. DNA Cell Biol. 37, 316–324 (2018).
Yang, S. et al. Sesamin induces A549 cell mitophagy and mitochondrial apoptosis via a reactive oxygen species-mediated reduction in mitochondrial membrane potential. Korean J. Physiol. Pharm. 24, 223–232 (2020).
Zhang, B., Bian, W., Pal, A. & He, Y. Macrophage apoptosis induced by aqueous C60 aggregates changing the mitochondrial membrane potential. Environ. Toxicol. Pharm. 39, 237–246 (2015).
Thomas, A. L. et al. Autologous Transplantation of Skin-Derived Precursor Cells in a Porcine Model. J. Pediatr. Surg. 55, 194–200 (2020).
Rollo, B. N. et al. Enteric Neural Cells From Hirschsprung Disease Patients Form Ganglia in Autologous Aneuronal Colon. Cell Mol. Gastroenterol. Hepatol. 2, 92–109 (2015).
Cooper, J. E. et al. In Vivo Transplantation of Enteric Neural Crest Cells into Mouse Gut; Engraftment, Functional Integration and Long-Term Safety. PLoS One 11, e0147989 (2016).
Cooper, J. E. et al. In vivo transplantation of fetal human gut-derived enteric neural crest cells. Neurogastroenterol. Motil. 29, 6 (2017).
Kato, H. et al. Immunocytochemical characterization of supporting cells in the enteric nervous system in Hirschsprung’s disease. J. Pediatr. Surg. 25, 514–519 (1990).
Ingham, P. W. Hedgehog signaling. Curr. Top. Develop. Biol. 149, 1–58 (2022).
Lau, S. T. et al. Activation of Hedgehog Signaling Promotes Development of Mouse and Human Enteric Neural Crest Cells, Based on Single-Cell Transcriptome Analyses. Gastroenterology 157, 1556–1571 (2019).
Ellis, T. et al. Patched 1 conditional null allele in mice. Genesis 36, 158–161 (2003).
Fattahi, F. et al. Deriving human ENS lineages for cell therapy and drug discovery in Hirschsprung disease. Nature 531, 105–109 (2016).
Funding
Project ZR2020MH048 supported by Shandong Provincial Natural Science Foundation and the Natural Science Foundation of Tibet Autonomous Region (No.XZ202101ZR0004G)
Author information
Authors and Affiliations
Contributions
Conception and design: F.F.Z.; Perform research: F.F.Z., L.J.Z., and M.Y.C.; Data analysis and interpretation: B.Z.M., F.G., and G.W.; Manuscript writing: All authors; Final approval of manuscript: All authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, F., Cui, M., Zhang, L. et al. GFRA4 improves the neurogenic potential of enteric neural crest stem cells via hedgehog pathway. Pediatr Res (2024). https://doi.org/10.1038/s41390-024-03158-8
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41390-024-03158-8
- Springer Nature America, Inc.