Abstract
Great progress has been made in the investigation on mutation and expression of splicing factor. However, little is known on the role of alternative splicing of splicing factors across cancers. Here, we reported a pan-cancer analysis of alternative splicing of splicing factors spanning 6904 patients across 16 cancer types, and identified 167 splicing factors with implications regulating cancer-specific splicing patterns through alternative splicing. Furthermore, we found that abnormal splicing events of splicing factors could serve as potential common regulators for alternative splicing in different cancers. In addition, we developed a splicing-derived neoepitopes database (ASPNs), which provided the corresponding putative alternative splicing-derived neoepitopes of 16 cancer types. Our results suggested that alternative splicing of splicing factors involved in the pre-RNA splicing process was common across cancer types and may represent an underestimated hallmark of tumorigenesis.
Similar content being viewed by others
Introduction
Alternative splicing allows the synthesis of multiple mature mRNAs from a single gene to translate diverse proteins, which is a critical factor in increasing the complexity of protein functions [1,2,3]. Approximately 95% of the genes in the human genome are alternatively spliced [4]. Alternative splicing changes are frequently observed in tumor samples and are recognized as important signatures for tumor progression and therapy [5]. Aberrant regulation of alternative splicing can lead to tumor progression by influencing the expression of genes or isoforms involved in cell proliferation control, apoptosis, DNA damage response, energy metabolism, angiogenesis, and metastasis [6]. For instance, alternative splicing changes of BCL2L1, NUMB, and MET genes affect pathways involved in apoptosis, cell proliferation, and cellular cohesion in lung cancer [7]. Recent studies have shown that alternative splicing is a potential source of neoepitopes [8,9,10,11]. Especially, B-cell acute lymphoblastic leukemia patients have low prevalence of somatic mutations and copy number variations. It is difficult to select a suitable target for the development of efficient immunotherapies. Nevertheless, patients have extensive alternative splicing differences, which can expand the number of suitable targets for immunotherapy [12].
Alternative splicing is regulated by splicing factors [13]. Mutation of splicing factors can generate different splicing patterns which may generate abnormal isoforms in tumor samples [14]. For instance, RBM10 and U2AF1 in lung cancer [15, 16], and SF3B1 in breast cancer [17, 18] mutated. On the other hand, increasing evidences suggest that changes in the expression of splicing factors can lead to abnormal splicing in cancers [19]. The splicing factor SR and hnRNP family are overexpressed in many cancer types and induce alternative splicing changes, promoting cell proliferation, and metastasis [20]. Recently, a study showed that abnormal splicing of splicing factors such as HNRNPA1 and HNRNPC could produce multiple isoforms in Acute Myeloid Leukemia [21]. However, a comprehensive pan-cancer analysis of alternative splicing of splicing factors has not been reported. To illustrate the influence of abnormal splicing of splicing factors to cancer-specific splicing patterns across 16 cancer types, we globally analyzed gene expression and alternative splicing data from The Cancer Genome Atlas (TCGA) project and constructed the splicing-derived neoepitopes database. Our results implicated that abnormal splicing of splicing factors could serve as a regulator of cancer-specific splicing patterns, which were a potential source of neoepitopes.
Results
Landscape of alternative splicing events across cancer types
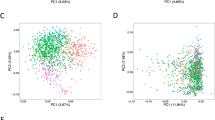
We described alternative splicing landscape across 16 human cancer types from 6904 patients, including 880 matched adjacent normal samples (Table 1 and Methods). Alternative splicing events were divided into seven types, namely Exon Skip (ES), Alternate Donor site (AD), Alternate Acceptor site (AA), Retained Intron (RI), Mutually Exclusive Exons (ME), Alternate Promoter (AP), and Alternate Terminator (AT). To highlight the specificity and commonality of alternative splicing across cancer types, we used t-SNE to visualize the splicing diversity across the full cohort [22]. We observed that cancer from different tissues of origin, such as HNSC and LUSC, were clustered closely together. On the contrary, KICH, KIRP, and KIRC were separated clearly from kidney tissue (Fig. 1a). We also used the same method to visualize the gene expression diversity across the full cohort, and the result showed a similar phenomenon (Supplementary Fig. S1).
a The landscape of alternative splicing for TCGA samples computed on PSI value. Each point represented a sample, colored according to its cancer type. b The DASEs numbers and proportions of splice types across 16 cancer types. DASEs with FDR < 0.05 (t-test and Benjamini–Hochberg) and | ΔPSI | > 0.1. c UpSet plot showed the interactive sets across 16 cancer types of DASEs which were cancer-specific or shared across two cancer types. d Bubble plots showed the GO pathways analyses of DASEs related genes across 16 cancer types. The size of each bubble corresponded to the effect size. The color of each bubble corresponded to the significance of enrichment (well-adopted hypergeometric test). e Bar plots showed the numbers of DASEs that the CDS same (green) or different (blue) across 16 cancer types. f Bar plots showed the distribution of protein domain prediction results.
The difference in the proportion of splicing types was not significant, but we observed that the count of DASEs varied greatly across cancer types from 593 (READ) to 3482 (LUSC) (Fig. 1b). The UpSet graphs were used to display the distribution of DASEs across cancer types (Fig. 1c). In addition to cancer-specific DASEs, we found that DASEs can also share across cancer types. To explore the functions of genes related to DASEs, we utilized metascape to conduct a joint analysis. The results revealed that genes were enriched in cancer development, including cell growth, cell junction assembly, cell part morphogenesis, and regulation of cell adhesion (Fig. 1d).
We expected that not all splicing events will have a potential impact on subsequent protein translation. Among 16 cancer types, the number of DASEs which did not change the CDS was from 133 (READ) to 876 (LUSC) (Fig. 1e). To further clarify the clinical implications of DASEs which did not change the CDS, we used multiple cox regression analysis of these events to determine the association between the PSI value of these events and patients’ overall survival across 16 cancer types (Method). We found survival-related events from DASEs, which did not change the CDS, were detected in different cancer types. There was a big gap in the proportion of splicing types (Supplementary Fig. S2a, b). Besides, we also found other DASEs, such as CCND3_AP_76154 and IL11RA_AP_86208, had opposite survival results in different cancer types (Supplementary Fig. S2c-f).
We translated the spliced and unspliced isoforms of each DASE, which changed the CDS, into amino acid sequences. Furthermore, we used InterProScan to analyze the loss/increase of the spliced protein domain. According to the results, these events which changed protein domain can be divided into four types: complete loss of well-annotated protein domains (CDL), partial loss of protein domains (PDL), the addition of protein domains (DA), and unknown consequence (UC). We found that most events were from the UC group, and these events may alter protein structure. In addition, part of spliced proteins generated by DASEs lose their domain completely (Fig. 1f). Overall, our results indicated that alternative splicing changes recurrently occurred in patients and lead to protein dysfunction.
Alternative splicing of splicing factors across 16 cancer types
Splicing factors amongst the genes related to DASEs had been observed. Therefore, we investigated whether alternative splicing changes of splicing factors could explain the alternative splicing changes in cancers. The differential gene expression of 167 genes encoding splicing factors between normal and tumor sample pairs was analyzed (Supplementary Table S3). We found that few splicing factors in tumor samples undergo splicing and expression changes simultaneously (Fig. 2a). Similar phenomena were found in oncogenes and tumor suppressor genes (Supplementary Fig. S3a, b). The proportion of splicing factors with alternative splicing changes was greater than that with expression changes among some cancer types. Splicing factors that only undergo alternative splicing changes across 16 cancer types were analyzed further. Interestingly, we found that these splicing factors shared in multiple cancer types (Fig. 2b) interacted with each other to form a tightly interconnected network (Fig. 2c). Collectively, splicing factors network analysis indicated that the abnormal splicing of splicing factors can trigger a series of alternative splicing changes in tumor patients.
a Bar plots showed the ratio of splicing factors with only difference in expression (Only DEG), splicing factors with only difference in alternative splicing (Only DASG), and splicing factors with the difference both in expression and alternative splicing (Overlap). b The Circos plot showed the interactive sets of splicing factors with only difference in alternative splicing among 16 cancer types. Each line was colored according to its cancer type. c The interaction network indicated protein-protein interactions (edges) between splicing factors with only difference in alternative splicing (nodes). d Enriched binding motifs of abnormal spliced splicing factors in differentially spliced skipping exon events in each cancer type (P value < 0.05, Wilcoxon’s rank sum test), separated by upstream (left) or downstream (right), and by skipping (top panel) or inclusion (bottom panel). e The heatmap showed the ΔPSI value of DASEs related to splicing factors across cancer types. f The binding motif enrichment analyzes of HNRNPA1 in CHOL and LUSC. The upstream and downstream represented the 250 bp of flanking the exons. The left axis and right axis represented the enrichment score and P value (Wilcoxon’s rank sum test), respectively. The dotted line represents the scores of retained (blue) and skipped (red) exons in tumors, while the black line represents the scores of background from all non-differentially spliced exons. g The unspliced and spliced isoforms of HNRNPA1_ES_212638. Blue and white rectangles were all represented exons. Blue rectangles were represented exons in the CDS region, and white rectangles were represented exons in the UTR region.
To verify this, we used rMAPS [23] to perform binding motifs enrichment analyses of differentially spliced transcripts to determine if they might be targets for the abnormal spliced splicing factors. We focused on the most common alternative splicing types among the typical patterns of alternative splicing (ES, AD, AA, RI, and ME): ES. Compared with adjacent normal samples, we identified that exons were recurrently skipped in tumor samples when the change of PSI value (ΔPSI) < −0.1, while retained when ΔPSI > 0.1. We also identified a control set of non-differentially spliced exons. We observed that the well-conservative binding motifs of these splicing factors were significantly over-represented in the transcript sequences of flanking the skipped/retained exons compared with non-differentially spliced exons (Fig. 2d), and these splicing factors generated many DASEs across cancer types (Fig. 2e). We also found that some motifs showed enrichment in the same cancer type both upstream and downstream and for both inclusion and skipping. The transcript sequences of flanking the skipped/retained exons in the same cancer type were different. These results indicated that the binding preference of these splicing factor might be different in the inclusion and skipping groups. Other motifs showed enrichment only in the same cancer type either inclusion or skipping. Especially compared with the non-differentially skipped exons in CHOL and LUSC, the well-conserved binding motif of HNRNPA1 was significantly over-represented the flanking retained exons in tumor samples. However, HNRNPA1 was not significantly over-represented flanking the retained exons in adjacent normal samples (Fig. 2f). HNRNPA1 serves as a splicing repressor in alternate splicing [12]. As the RNA-binding protein, HNRNPA1 can bind to pre-mRNA in the nucleus and affect pre-mRNA processing, mRNA metabolism and transport [24,25,26]. According to our results, the expression of HNRNPA1 was not significantly different between normal and tumor samples, while HNRNPA1 would produce a nonfunctional protein (HNRNPA1_ES_212638) through alternative splicing. ΔPSI of HNRNPA1_ES_212638 > 0.1 indicated the proportion of exon inclusion in tumor samples within transcript was higher, which would bind and repress splicing (Fig. 2g). Our results suggested that alternative splicing changes of splicing factors might contribute to the abnormal splicing across cancer types.
Alternative splicing of splicing factors affects cancer-specific splicing patterns
To investigate whether the alternative splicing changes between tumor and adjacent normal samples were affected by splicing events of splicing factors, we analyzed the relationship between different alternative splicing events of splicing factors (SDASEs) and the second principal component (PC2) of the DASEs set (Supplementary Fig. S4). We found high correlations between PSI values of SDASEs and cancer-specific splicing patterns (Fig. 3a and Supplementary Table S4). In particular, the PSI value of MBNL1_ES_67324 was correlated with cancer-specific splicing patterns among most cancer types, including BLCA, BRCA, CHOL, COAD, KICH, KIRC, LUAD, LUSC, PRAD, READ, and STAD. Another event MBNL1_ES_67315 related to MBNL1 was also correlated with seven cancer types, including BLCA, COAD, KICH, PRAD, READ, STAD, and UCEC. We also used the same method to analyze the relationship between different alternative splicing events of splicing factors (SDASEs) and the second principal component (PC2) of random control set of DASEs (Supplementary Fig. S5). Among the 16 cancer types, the number of SDASEs that correlations with PC2 of the random control set were less than that associated with PC2 of the DASEs set (P < 0.05).
a Bubble plot showed the SDASEs that highly correlated with the second principal component (PC2) of the DASEs set are shown in the bubble plot (P < 0.05 and R > 0.5, Pearson). The size of each bubble corresponds to the correlation. The color of each bubble corresponds to the ΔPSI value. b The unspliced and spliced isoforms of MBNL1_ES_67324 and MBNL1_ES_67315. Blue rectangles were represented exons in the CDS region. c Protein structures of MBNL1 unspliced protein (red), MBNL1_ES_67315 spliced protein (yellow) and MBNL1_ES_67324 spliced protein (blue). d Root mean square deviation (RMSD) plot of unspliced and spliced proteins of MBNL1 over 50 ns of simulation. The yellow line represented MBNL1_ES_67315 spliced protein, the red line represented MBNL1 unspliced protein, and the blue line represented MBNL1_ES_67324 spliced protein. e Solvent-accessible surface area (SASA) plot of unspliced and spliced proteins of MBNL1 over 50 ns of simulation. The yellow line represented MBNL1_ES_67315 spliced protein, the red line represented MBNL1 unspliced protein, and the blue line represented MBNL1_ES_67324 spliced protein. f The radius of gyration (Rg) plot of unspliced and spliced proteins of MBNL1 over 50 ns of simulation. The yellow line represented MBNL1_ES_67315 spliced protein, the red line represented MBNL1 unspliced protein, and the blue line represented MBNL1_ES_67324 spliced protein.
Compared with the unspliced isoforms, the spliced isoforms, MBNL1_ES_67324 and MBNL1_ES_67315, skipped exon 8 and 10–11, respectively (Fig. 3b). We found that spliced proteins did not change the well-annotated protein domains, indicating that protein structure stability might altered. To illustrate the impact of the alternative splicing on the stability of the protein structure at the molecular level, we conducted molecular dynamics simulations (MDS) for 50 ns for spliced and unspliced proteins (Fig. 3c). We obtained root mean square deviation (RMSD), solvent-accessible surface area (SASA), and radius of gyration (Rg) from the resultant trajectory files of 50 ns simulation to analyze the protein stability, compactness, hydrophobic and hydrophilic nature of the proteins. RMSD plot illustrated that unspliced protein had better stability than spliced proteins (Fig. 3d). Figure 3e illustrated that the SASA of unspliced protein was larger than spliced proteins. Compared with unspliced protein, the Rg value of spliced proteins was lower, indicating that density was increased after splicing (Fig. 3f). The results indicated that the capability of MBNL1 unspliced proteins binding to other elements might stronger. Meanwhile, we validated the splicing isoforms of MBNL1 in BRCA and COAD. The results showed that the proportion of unspliced proteins in tumor samples was higher than in normal samples (Supplementary Fig. S6a–d).
Common splicing patterns across cancer types are mediated by alternative splicing of splicing factors
To further characterize whether common cancer splicing patterns are associated with alternative splicing of splicing factors. We identified common patterns of splicing changes between pairs of cancer types by selecting alternative splicing events correlated with the SDASEs in the two cancer types. Here, only the SDASEs with more than 50 correlation events in paired cancer types were shown. Figure 4a showed the ΔPSI of common events between cancer pairs. Here, we found a positive correlation in most pairs of cancer types. It indicated potential common splicing regulators in different cancer types. In addition, COAD and LUSC were highly correlated with 14 other cancer types, while LIHC was only correlated with THCA. Consistent with our t-SNE results, COAD and LUSC were unclearly separated from other cancer types. On the contrary, LIHC was distinguished well (Fig. 1a).
a Common events and SDASEs between pairs of cancer types. For each pair of cancer types and each SDASEs, we plotted the correlation of the ΔPSI value for events that have a correlation of |R | > 0.5 (Pearson) with these SDASEs in both cancer types. Each Point represented an alternative splicing event. b, c The common events were separated based on their potential SDASEs regulators, and the ΔPSI correlation for the pairs LUAD_LUSC and BRCA_KIRC (Pearson). Each Point represented an alternative splicing event.
The correlation between LUAD and LUSC tumors was 0.97 (Fig. 4b), and there were 1389 shared events associated with 13 SDASEs. In contrast, BRCA and KIRC shared 406 events associated with 4 SDASEs (Fig. 4c). These results suggested that abnormal splicing events of splicing factors could serve as potential common regulators for alternative splicing in different cancer types.
Putative neoepitopes derived from alternative splicing across cancer types
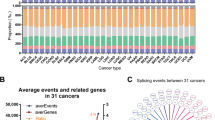
The direct oncogenic effect is a consequence of alternative splicing changes of splicing factors across cancer types. Cancer-specific isoform regulated by abnormal splicing of splicing factors was translated and could potentially lead to specific neoepitopes. We proposed a comprehensive workflow for the analysis of neoepitopes derived from alternative splicing. In our workflow, we first obtained TCGA alternative splicing data. Then, we got amino acid sequences from spliced and unspliced isoforms. After chopping up the two proteins into 9-mer peptides, we filtered the normal peptides set to obtain the novel peptides. Finally, we used NetMHCpan-4.1 [27] to predict the possibility for peptides to bind the 227 HLA types (Fig. 5a and Materials and methods). According to the result from our workflow, we defined putative neoepitopes generated by alternative splicing as ASPNs.
a The workflow of identification of alternative splicing neoepitopes. b Heatmaps showed the number of alternative splicing-derived neoepitopes produced by splicing factors across cancer types. c Bar plots showed the average number of neoepitopes and per gene related to DASEs across 16 cancer types. d Heatmaps showed the top 5 ASPNs that can bind the most HLA types in each cancer type.
We observed multiple ASPNs related to splicing factors (Fig. 5b), and found that genes related to DASEs would averagely produce multiple ASPNs (Fig. 5c). The result of statistical analysis of the top 5 ASPNs, which can be a binder with most of HLA types in each cancer, was shown that these ASPNs could be shared across multiple cancer types (Fig. 5d). ASPNs might use for expanding the repertoire of potential targets for immunotherapy. We constructed a database (http://jianglab.org.cn/ASPN/) which enabled the search of corresponding ASPNs through screening of cancer and HLA types.
Discussion
We reported alternative splicing changes of splicing factors in 6904 patients from 16 cancer types. Our comprehensive survey revealed that alternative splicing changes of splicing factors were pervasive in cancers and powerfully affected cancer-specific splicing patterns. This study suggested that the importance of alternative splicing changes of splicing factors in mediating common splicing patterns across cancer types. Focusing on neoantigens in tumor cells, possible neoepitopes derived from alternative splicing are usually overlooked [11]. We constructed a database (http://jianglab.org.cn/ASPN/) for the putative neoepitopes derived from alternative splicing across 16 cancer types.
Using TCGA RNA-seq data, we characterized the alternative splicing landscape across 16 cancer types. We found that alternative splicing can generate different isoforms which were different 5ʹUTRs/3ʹUTRs but encode identical amino acid sequences. These splicing events still played an important role in the survival of patients. The sequence characteristics within 5′UTRs and 3ʹUTRs played important roles in differential regulation of translation efficiency [28, 29]. Our result showed that alternative splicing affected cancers by changing the complexity of the protein. In addition, it can also play its potential function by changing the UTR sequence of isoforms.
Alternative splicing is regulated by multiple splicing factors and influences the expression of most eukaryotic genes [30]. Abnormal splicing is correlated with different aspects of cancer biology [31,32,33,34], such as cell proliferation and metastasis [35,36,37], apoptosis [7, 38, 39], cell differentiation [40, 41], angiogenesis[34, 42,43,44], and energy metabolism [45,46,47]. Although changes in the expression and mutations of genes encoding splicing factors have been increasingly recognized [19], alternative splicing of splicing factors over large patient sample cohorts have not been reported. Here, we performed a comprehensive characterization of 167 selected splicing factors across the 16 cancer types. This report constituted the first pan-cancer exploration of alternative splicing of splicing factors. Our results highlight that alternative splicing changes of splicing factors across cancer types were common. In normal tissue, abnormal spliced splicing factors formed a tightly interconnected network. Abnormal splicing of splicing factors could trigger a cascade of splicing alterations in cancer patients. We observed that the well-conservative binding motifs of abnormal spliced splicing factors were significantly over-represented in the transcript sequences of flanking the skipped/retained exons compared with non-differentially spliced exons. The difference in splicing causes the change in the ratio of splicing factor’s dominant functional protein. Especially, HNRNPA1 which as a splicing repressor would produce a nonfunctional protein through alternative splicing in Acute Myeloid Leukemia [21]. Consistent with the results, we further found that HNRNPA1, a gene in abnormal spliced splicing factors set, produced a dominant proportion of nonfunctional protein in normal adjacent samples within CHOL and LUSC.
In summary, this analysis, utilizing a large sample set size, revealed alternative splicing of splicing factors could contribute to the alternative splicing differences in cancer.
Our study of the alternative splicing landscape demonstrated that taking information of alternative splicing events of splicing factors into account was beneficial for characterizing cancer-specific splicing patterns. Systematic correlation analysis of splicing events in tumors demonstrated high correlations between PSI values of SDASEs and cancer-specific splicing patterns. These events, included MBNL1_ES_67324 and MBNL1_ES_67315, were strongly correlating with cancer-specific splicing patterns. A previous study has shown that MBNL1 regulates alternative splicing of NUMA1 through changes in expression [19]. Compared with adjacent normal samples, we found that MBNL1 expression was not significantly different in some cancer types, but the alternative splicing changes were significant. We found MBNL1, as a splicing factor, could modulate alternative splicing of pre-mRNAs more efficiently under unspliced conditions. Our results revealed that the proportion of MBNL1 unspliced isoform in the tumor samples was higher than in the normal samples. It may explain the previous study that there were more alternative splicing events in tumor samples compared with normal samples [48, 49]. In this context, we would like to note that alternative splicing of splicing factors could contribute to tumor development independently of expression alterations.
Immunotherapy has produced effective treatments for several previously incurable tumors [50]. Alternative processing of mRNA, a phenomenon that has been shown to alter the proteomic diversity of many tumors, may expand the number of suitable targets for immunotherapy [11]. In this work, we proposed a comprehensive workflow for identifying putative alternative splicing-derived neoepitopes. Taking together, we constructed a database (http://jianglab.org.cn/ASPN/), which provided the corresponding putative alternative splicing-derived neoepitopes of 16 cancer types. Overall, the putative neoepitopes derived from alternative splicing had the potential to contribute to immunotherapy.
In summary, our study provided further insight into the relationship between alternative splicing of splicing factors and cancer-specific splicing patterns that could be used to improve molecular understanding of the splicing transitions that take place during cancer development. We also revealed the potential mechanism of abnormal regulation in cancer. Furthermore, we emphasized the importance of alternative splicing in immunotherapy.
Materials and methods
Data acquisition
Alternative splicing data were downloaded in TCGA SpliceSeq [51]. Tumors with paired adjacent normal samples of at least nine were contained in this analysis (Supplementary Table S1). Alternative splicing events were quantified by percent spliced-in (PSI) values, which ranged from 0–1. PSI is the ratio of normalized read counts indicating inclusion of a transcript element over the total normalized reads for that event (both inclusion and exclusion reads) [51]. We filtered the results (percentage of samples with PSI values ≥80%, average PSI value ≥ 0.05) to generate a set of alternative splicing events. Level 3 RNA-seq and clinical data were obtained from the TCGA cohort by the UCSC Xena website (http://xena.ucsc.edu/). The 167 known and potential auxiliary splicing factors were obtained from Sebestyen, E. et al. [19]. Oncogenes and Tumor suppressor genes were obtained from NCG (http://ncg.kcl.ac.uk/index.php) and TSGens (https://bioinfo.uth.edu/TSGene/) [52, 53].
Differential expression and alternative splicing
A t-test was performed to identify different expressions with FPKM value and different alternative splicing with PSI value. P values were corrected for multiple testing using the Benjamini–Hochberg method. Different alternative splicing events (DASEs) were defined as mean PSI value varied more than 0.1 between tumor and the adjacent normal sample and FDR < 0.05. Moreover, if |log2(FC) | > 1 and the FDR value < 0.05, it was considered that the gene was differentially expressed [54, 55]. We used a standard dimensionality reduction technique t-distributed stochastic neighbor embedding (t-SNE) to visualize the splicing across 16 cancer types [56].
Survival analysis
Multivariate cox regression analysis was based on the PSI value of the alternative splicing events and the patient’s clinical information (overall survival time, survival status, stage, age, and gender) to identify the association between the PSI value of the alternative splicing events and patients’ overall survival used R packages ‘survival (3.1.8)’ and ‘survminer (0.4.7)’. Survival-related events were determined to be p value < 0.05. The samples were divided into two groups according to the average value of the PSI. The Kaplan–Meier curves were used to plot the overall survival rates of the two groups, and the log-rank test was used to analyze the differences between the two groups.
CDS sequence comparison and protein structure prediction
We obtained the unspliced isoforms according to the chromosome coordinates of DASEs. The spliced isoforms were created by deleting, adding, or changing the exons (e.g., the skipped exon) from the corresponding unspliced isoforms. We defined the high content isoforms in normal samples as the normal isoforms. The annotations were used from the UCSC (GRCh37/hg19). We used python to compare the spliced and unspliced isoforms’ CDS of each DASE. The gffread (v0.11.4) was used to retrieve each isoform’s nucleic acid sequence, and then they were translated into the amino acid sequence. Predicted motifs were retrieved from protein domain databases (i.e., Pfam, ProSite, and SMART) using InterProScan [57,58,59].
Molecular dynamic simulation
We used Groningen Machine for Chemical Structure (GROMACS) [60] to MDS analysis of MBNL1_unspliced, MBNL1_ES_67324 spliced, and MBNL1_ES_67315 spliced proteins. I-TASSER was used to predict the MBNL1_unspliced protein structure [61]. Swiss-model Server was used for homology modeling of MBNL1_ES_67324 and MBNL1_ES_67315 protein base on the amino acid sequence [62]. PyMOL was used to visualize the results [63].
Neopeptides prediction
We translated unspliced and spliced isoforms of each DASE into amino acid sequence from the translation start site to the stop codon and chopped up the amino acid sequences into 9-mer peptides. We defined the peptides produced by normal isoforms as normal peptides. We selected a total of 227 HLA types, of which labels are “common” in IDAWG (http://igdawg.org/). We used NetMHCpan-4.1 [27] to calculate the bind rank of peptides to HLA, and those rank <5% peptides were considered as putative neopeptides.
Binding motifs enrichment analyses
We used rMAPS to enrich the binding motifs of splicing factors across 16 cancer types [23]. rMAPS employs Wilcoxon’s rank sum test. P value < 0.05 was set as the cutoffs for the enriched binding motifs of splicing factors.
t-SNE
We used R package ‘Rtsne (0.15)’ for t-SNE analysis [64].
Protein-protein interaction networks
We used String (https://string-db.org/) and Cytoscape to generate biological networks for proteins [65, 66].
Validation of splicing isoforms
Five pairs of tumor and adjacent normal tissues were obtained from breast cancer patients treated at the Harbin Medical University (Harbin, China) after their written informed consent. HCT116 and NCM460 cells were purchased from The Global Bioresource Center, and no mycoplasma contamination was found after testing. RNAiso Plus (TAKARA, 9109) and PrimeScriptTM RT reagent Kit with gDNA Erase (TAKARA, RR047A) were used for total RNA extracted and reverse-transcribed. RT-PCR was performed in paired samples by splicing-specific primers (Supplementary Table S2) using GoTaq® Green Master Mix (Promega, M7128). Sanger sequencing (Comate, Jilin, China) to conform the isoform sequence.
Code availability
The code for the analyses described in this study is available at https://github.com/asd77088/Jiang-lab.
References
Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–5.
Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–63.
Blencowe BJ. The Relationship between Alternative Splicing and Proteomic Complexity. Trends Biochem Sci. 2017;42:407–8.
Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–6.
Dvinge H, Kim E, Abdel-Wahab O, Bradley RK. RNA splicing factors as oncoproteins and tumour suppressors. Nat Rev Cancer. 2016;16:413–30.
Bechara EG, Sebestyen E, Bernardis I, Eyras E, Valcarcel J. RBM5, 6, and 10 differentially regulate NUMB alternative splicing to control cancer cell proliferation. Mol Cell. 2013;52:720–33.
Coomer AO, Black F, Greystoke A, Munkley J, Elliott DJ. Alternative splicing in lung cancer. Biochim Biophys Acta Gene Regul Mech. 2019;1862:194388.
Zhang ZB, Zhou C, Tang LH, Gong YK, Wei ZT, Zhang GC, et al. ASNEO: identification of personalized alternative splicing based neoantigens with RNA-seq. Aging-Us. 2020;12:14633–48.
Kahles A, Lehmann KV, Toussaint NC, Huser M, Stark SG, Sachsenberg T, et al. Comprehensive Analysis of Alternative Splicing Across Tumors from 8,705 Patients. Cancer Cell. 2018;34:211–+.
Smart AC, Margolis CA, Pimentel H, He MX, Miao D, Adeegbe D, et al. Intron retention is a source of neoepitopes in cancer. Nat Biotechnol. 2018;36:1056–8.
Frankiw L, Baltimore D, Li GD. Alternative mRNA splicing in cancer immunotherapy. Nat Rev Immunol. 2019;19:675–87.
Black KL, Naqvi AS, Asnani M, Hayer KE, Yang SY, Gillespie E, et al. Aberrant splicing in B-cell acute lymphoblastic leukemia. Nucleic Acids Res. 2018;46:11357–69.
Kedzierska H, Piekielko-Witkowska A. Splicing factors of SR and hnRNP families as regulators of apoptosis in cancer. Cancer Lett. 2017;396:53–65.
Dvinge H, Guenthoer J, Porter PL, Bradley RK. RNA components of the spliceosome regulate tissue- and cancer-specific alternative splicing. Genome Res. 2019;29:1591–604.
Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–20.
Brooks AN, Choi PS, de Waal L, Sharifnia T, Imielinski M, Saksena G, et al. A pan-cancer analysis of transcriptome changes associated with somatic mutations in U2AF1 reveals commonly altered splicing events. Plos ONE. 2014;9:e87361.
Ellis MJ, Ding L, Shen D, Luo J, Suman VJ, Wallis JW, et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature. 2012;486:353–60.
Maguire SL, Leonidou A, Wai P, Marchio C, Ng CK, Sapino A, et al. SF3B1 mutations constitute a novel therapeutic target in breast cancer. J Pathol. 2015;235:571–80.
Sebestyen E, Singh B, Minana B, Pages A, Mateo F, Pujana MA, et al. Large-scale analysis of genome and transcriptome alterations in multiple tumors unveils novel cancer-relevant splicing networks. Genome Res. 2016;26:732–44.
Golan-Gerstl R, Cohen M, Shilo A, Suh SS, Bakacs A, Coppola L, et al. Splicing factor hnRNP A2/B1 regulates tumor suppressor gene splicing and is an oncogenic driver in glioblastoma. Cancer Res. 2011;71:4464–72.
Anande G, Deshpande NP, Mareschal S, Batcha AMN, Hampton HR, Herold T, et al. RNA Splicing Alterations Induce a Cellular Stress Response Associated with Poor Prognosis in Acute Myeloid Leukemia. Clin Cancer Res. 2020;26:3597–607.
Cieslak MC, Castelfranco AM, Roncalli V, Lenz PH, Hartline DK. t-Distributed Stochastic Neighbor Embedding (t-SNE): a tool for eco-physiological transcriptomic analysis. Mar Genom. 2020;51:100723.
Hwang JY, Jung S, Kook TL, Rouchka EC, Bok J, Park JW. rMAPS2: an update of the RNA map analysis and plotting server for alternative splicing regulation. Nucleic Acids Res. 2020;48:W300–W6. W1
Howard JM, Lin H, Wallace AJ, Kim G, Draper JM, Haeussler M, et al. HNRNPA1 promotes recognition of splice site decoys by U2AF2 in vivo. Genome Res. 2018;28:689–98.
Kim HJ, Kim NC, Wang YD, Scarborough EA, Moore J, Diaz Z, et al. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 2013;495:467–73.
Gui X, Luo F, Li Y, Zhou H, Qin Z, Liu Z, et al. Structural basis for reversible amyloids of hnRNPA1 elucidates their role in stress granule assembly. Nat Commun. 2019;10:2006.
Hollenbach JA, Holcomb C, Hurley CK, Mabdouly A, Maiers M, Noble JA, et al. 16(th) IHIW: immunogenomic data-management methods. report from the immunogenomic data analysis working group (IDAWG). Int J Immunogenet. 2013;40:46–53.
Davuluri RV, Suzuki Y, Sugano S, Plass C, Huang THM. The functional consequences of alternative promoter use in mammalian genomes. Trends Genet. 2008;24:167–77.
Mayr C. What Are 3’ UTRs Doing? Cold Spring Harb Perspect Biol. 2019;11:a034728, https://doi.org/10.1101/cshperspect.a034728.
Hahn CN, Venugopal P, Scott HS, Hiwase DK. Splice factor mutations and alternative splicing as drivers of hematopoietic malignancy. Immunol Rev. 2015;263:257–78.
Chabot B, Shkreta L. Defective control of pre-messenger RNA splicing in human disease. J Cell Biol. 2016;212:13–27.
Climente-Gonzalez H, Porta-Pardo E, Godzik A, Eyras E. The Functional Impact of Alternative Splicing in Cancer. Cell Rep. 2017;20:2215–26.
Martinez-Montiel N, Rosas-Murrieta NH, Anaya Ruiz M, Monjaraz-Guzman E, Martinez-Contreras R. Alternative Splicing as a Target for Cancer Treatment. Int J Mol Sci. 2018;19:545, https://doi.org/10.3390/ijms19020545.
Zhang Y, Qian J, Gu C, Yang Y. Alternative splicing and cancer: a systematic review. Signal Transduct Target Ther. 2021;6:78.
Hagen RM, Ladomery MR. Role of splice variants in the metastatic progression of prostate cancer. Biochem Soc Trans. 2012;40:870–4.
Xie R, Chen X, Chen Z, Huang M, Dong W, Gu P, et al. Polypyrimidine tract binding protein 1 promotes lymphatic metastasis and proliferation of bladder cancer via alternative splicing of MEIS2 and PKM. Cancer Lett. 2019;449:31–44.
Paschalis A, Sharp A, Welti JC, Neeb A, Raj GV, Luo J, et al. Alternative splicing in prostate cancer. Nat Rev Clin Oncol. 2018;15:663–75.
Lin JC, Tsao MF, Lin YJ. Differential Impacts of Alternative Splicing Networks on Apoptosis. Int J Mol Sci. 2016;17:2097, https://doi.org/10.3390/ijms17122097.
Schwerk C, Schulze-Osthoff K. Regulation of apoptosis by alternative pre-mRNA splicing. Mol Cell. 2005;19:1–13.
Norris AD, Calarco JA. Emerging Roles of Alternative Pre-mRNA Splicing Regulation in Neuronal Development and Function. Front Neurosci. 2012;6:122.
Pradella D, Naro C, Sette C, Ghigna C. EMT and stemness: flexible processes tuned by alternative splicing in development and cancer progression. Mol Cancer. 2017;16:8.
Wagner KD, El Mai M, Ladomery M, Belali T, Leccia N, Michiels JF. Altered VEGF Splicing Isoform Balance in Tumor Endothelium Involves Activation of Splicing Factors Srpk1 and Srsf1 by the Wilms’ Tumor Suppressor Wt1. Cells-Basel. 2019;8:41, https://doi.org/10.3390/cells8010041.
Hatcher JM, Wu G, Zeng C, Zhu J, Meng F, Patel S, et al. SRPKIN-1: a Covalent SRPK1/2 Inhibitor that Potently Converts VEGF from Pro-angiogenic to Anti-angiogenic Isoform. Cell Chem Biol. 2018;25:460–70. e6
Zhu H, Carpenter RL, Han W, Lo HW. The GLI1 splice variant TGLI1 promotes glioblastoma angiogenesis and growth. Cancer Lett. 2014;343:51–61.
Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–3.
Chen M, Zhang J, Manley JL. Turning on a fuel switch of cancer: hnRNP proteins regulate alternative splicing of pyruvate kinase mRNA. Cancer Res. 2010;70:8977–80.
Mazurek S. Pyruvate kinase type M2: a key regulator of the metabolic budget system in tumor cells. Int J Biochem Cell Biol. 2011;43:969–80.
Shah K, Gagliano T, Garland L, O’Hanlon T, Bortolotti D, Gentili V. Androgen receptor signaling regulates the transcriptome of prostate cancer cells by modulating global alternative splicing. Oncogene. 2020;39:6172–89. https://doi.org/10.1038/s41388-020-01429-2.
Venables JP. Aberrant and alternative splicing in cancer. Cancer Res. 2004;64:7647–54.
Riley RS, June CH, Langer R, Mitchell MJ. Delivery technologies for cancer immunotherapy. Nat Rev Drug Disco. 2019;18:175–96.
Ryan M, Wong WC, Brown R, Akbani R, Su XP, Broom B, et al. TCGASpliceSeq a compendium of alternative mRNA splicing in cancer. Nucleic Acids Res. 2016;44:D1018–D22. D1
Repana D, Nulsen J, Dressler L, Bortolomeazzi M, Venkata SK, Tourna A. The Network of Cancer Genes (NCG): a comprehensive catalogue of known and candidate cancer genes from cancer sequencing screens. Genom Biol. 2019;20. https://doi.org/10.1186/s13059-018-1612-0.
Zhao M, Sun JC, Zhao ZM. TSGene: a web resource for tumor suppressor genes. Nucleic Acids Res. 2013;41:D970–D6. D1
Kim TK. T test as a parametric statistic. Korean J Anesthesiol. 2015;68:540–6.
Ferreira JA, Zwinderman AH. On the Benjamini-Hochberg method. Ann Stat. 2006;34:1827–49.
Kobak D, Berens P. The art of using t-SNE for single-cell transcriptomics. Nat Commun. 2019;10:5416.
Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42(Database issue):D222–30.
Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30:1236–40.
Bairoch A. PROSITE: a dictionary of sites and patterns in proteins. Nucleic Acids Res. 1991;19:2241–5. Suppl
Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4: algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. J Chem Theory Comput. 2008;4:435–47.
Zhang Y. I-TASSER server for protein 3D structure prediction. Bmc Bioinforma. 2008;9:40.
Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42:W252–8. Web Server issue
Janson G, Zhang C, Prado MG, Paiardini A. PyMod 2.0: improvements in protein sequence-structure analysis and homology modeling within PyMOL. Bioinformatics. 2017;33:444–6.
Maaten LJTJoMLR. Accelerating t-SNE using tree-based algorithms. 2014;15:3221–45.
von Mering C, Huynen M, Jaeggi D, Schmidt S, Bork P, Snel B. STRING: a database of predicted functional associations between proteins. Nucleic Acids Res. 2003;31:258–61.
Otasek D, Morris JH, Boucas J, Pico AR, Demchak B. Cytoscape Automation: empowering workflow-based network analysis. Genom Biol. 2019;20:185.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Nos. 61822108 and 62032007 to QJ).
Author information
Authors and Affiliations
Contributions
Conception and design: QJ. Methodology: QJ, RC, LX, and WZ. Analysis and interpretation of data: QJ, RC, WZ, PW, KM, HC, FP, YH, XL, and YL. Writing-original draft: RC, LX, CX, ZX, XJ, ML, MW, and QJ.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Cheng, R., Xiao, L., Zhou, W. et al. A pan-cancer analysis of alternative splicing of splicing factors in 6904 patients. Oncogene 40, 5441–5450 (2021). https://doi.org/10.1038/s41388-021-01947-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-021-01947-7
- Springer Nature Limited
This article is cited by
-
AS-CMC: a pan-cancer database of alternative splicing for molecular classification of cancer
Scientific Reports (2022)
-
The aberrant upregulation of exon 10-inclusive SREK1 through SRSF10 acts as an oncogenic driver in human hepatocellular carcinoma
Nature Communications (2022)