Abstract
Epidermal growth factor receptor tyrosine kinase inhibitors (EGFR-TKIs) are effective against non-small cell lung cancer (NSCLC) with EGFR-activating mutations. The mechanisms underlying EGFR-TKI resistance are not fully understood. This study aimed to analyze the effects of seven EGFR ligands on EGFR-TKI sensitivity in NSCLC cells and patients. Cells with EGFR E746-A750del mutation were treated with recombinant EGFR ligands, and analyzed for cell viability, proliferation, and apoptosis. shRNA knockdown of endogenous Epiregulin (EREG) or overexpression of exogenous EREG and immunofluorescence experiments were carried out. Public gene expression datasets were used for tumor microenvironment and clinical assessment. Among the EGFR ligands, EREG significantly diminished cellular sensitivity to TKIs and was associated with decreased response to erlotinib in NSCLC patients. EREG induced AKT phosphorylation and attenuated TKI-induced cellular apoptosis in an ErbB2-dependent manner. EREG induced the formation of the EGFR/ErbB2 heterodimer regardless of gefitinib treatment. However, overexpression or knockdown of EREG in cancer cells had little impact on TKI sensitivity. Single-cell RNA sequencing data revealed that EREG was predominantly expressed in macrophages in the tumor microenvironment. In addition, EREG-enriched macrophage conditional medium induced EGFR-TKI resistance. These findings shed new light on the mechanism underlying EGFR-TKI resistance, and suggest macrophage-produced intratumoral EREG as a novel regulator and biomarker for EGFR-TKI therapy in NSCLC.
Similar content being viewed by others
Introduction
Mutations of the epidermal growth factor receptor (EGFR) are frequent genetic alterations in non-small cell lung cancer (NSCLC). They occur in approximately 11–16% of lung adenocarcinoma patients in Caucasians, and in up to 50% of East Asian populations [1]. The most prevalent EGFR mutations (>90%) are exon 19 deletions and the L858R mutation in exon 21 within the kinase domain, resulting in uncontrolled activation of EGFR [2]. First-generation EGFR tyrosine kinase inhibitors (TKIs), including gefitinib and erlotinib, are generally recommended for the treatment of advanced NSCLC harboring EGFR mutations. Furthermore, the third-generation TKI osimertinib has been recommended as a first-line drug, because of its better clinical efficacy and fewer adverse effects [3]. However, intrinsic resistance or acquired resistance after a period of initial response remains a problem in the field of EGFR-TKI therapy [2, 4, 5].
Several mechanisms have been elucidated for EGFR-TKI resistance. The most common mechanism of acquired resistance to first- and second-generation TKIs is EGFR T790M mutation alone or in combination with EGFR amplification, which is present in nearly 50–60% of cases [5]. However, tumors with the T790M mutation are sensitive to the third-generation TKI osimertinib. Other mechanisms of acquired resistance include small cell histology transformation (10%), HER2 amplification (12%), MET amplification (5%), and mutations in PI3K, BRAF, or KRAS genes (<5%) [2, 5]. Unlike acquired resistance, the mechanism of intrinsic resistance is not well understood. Amplification or rare mutations in EGFR and activation of the by-pass parallel receptor tyrosine kinase (e.g., HER2, MET, ALK, FGFR) may contribute to a poor response to TKIs [4]. Nevertheless, other novel mechanisms underlying EGFR-TKI resistance remain to be explored.
Ligand-dependent activation of the EGFR signaling pathway is a putative mechanism for EGFR-TKI resistance. There are seven EGFR ligands that activate EGFR by inducing the formation of EGFR homodimers, or heterodimers with other ErbB family members [6, 7]. Upon activation, EGFR stimulates the downstream MEK/ERK and PI3K/AKT pathways, thereby promoting cellular proliferation and survival [8]. Kakiuchi et al. [9] reported that amphiregulin (AREG) and TGFα were overexpressed in non-responders compared to responders to gefitinib. Further studies confirmed that elevated serum levels of TGFα were associated with lower therapeutic response and poor outcome in patients following gefitinib treatment [10, 11]. In addition, EGF expression was upregulated in EGFR T790M tumors that developed resistance to osimertinib [12]. However, the effect of other EGFR ligands on EGFR-TKI sensitivity, such as Epiregulin (EREG), Epigen (EPGN), Betacellulin (BTC), and Heparin-binding EGF (HB-EGF), remains unclear. Systematic analysis of the effects of seven EGFR ligands on TKI sensitivity would uncover the mechanism(s) of TKI resistance.
Although several EGFR ligands have been implicated in TKI resistance, the cellular origins of these ligands have not been investigated. Previous evidence suggests that EGFR and its ligands mediate vital processes within the tumor microenvironment via autocrine and paracrine circuits [13]. Several ligands of EGFR, such as EGF, TGFα, AREG, and EREG, can be secreted by lung cancer cells [9]. However, these ligands may also derive from other cells within the tumor microenvironment, such as fibroblasts and macrophages [14,15,16]. The cellular origin of EGFR ligands that influence TKI sensitivity has not been clearly characterized, which limits our understanding of the resistance mechanism.
In this study, we assessed the response of EGFR-mutant cells to EGFR-TKIs in the presence of different EGFR ligands. In addition to the ligands already reported, EREG was found to be dominantly involved in EGFR-TKI resistance. High expression of EREG was correlated with low clinical response and poor progression-free survival (PFS) in NSCLC patients receiving TKI treatment. Further results revealed that EREG induced TKI resistance by preventing apoptosis possibly through the EGFR/ErbB2 heterodimer. Finally, EREG expressed by macrophages rather than cancer cells may contribute to resistance. Our results suggest that EREG produced by non-tumor cells in the tumor microenvironment may be part of a novel mechanism for EGFR-TKI resistance.
Results
EREG confers EGFR-TKI resistance in NSCLC cells and patients
Two cell lines harboring the EGFR E746-A750del mutation (PC9 and HCC827) were chosen to analyze the impact of seven EGFR ligands on EGFR-TKI sensitivity. The cell counting kit-8 (CCK-8) assay revealed that PC9 and HCC827 cells became more resistant to TKIs when treated with recombinant EGF and TGFα. In particular, the effect on gefitinib sensitivity was much more pronounced (Fig. 1A, B). Surprisingly, we found that the sensitivities of the two cells to TKIs were significantly reduced in the presence of recombinant EREG (Fig. 1C). The IC50 of the inhibitory effect on the sensitivity to gefitinib and erlotinib increased by more than two times in the EREG group compared with the control group. The addition of Epigen to the medium also increased to a certain degree of the resistance to TKIs, but it did not affect the sensitivity of HCC827 cells to erlotinib (Fig. 1D). In contrast, treatment with the other ligands, including amphiregulin, betacellulin, and HB-EGF, resulted in no remarkable change in the sensitivity to TKIs (Supplementary Fig. S1). Furthermore, the effect of EREG on the sensitivity of the third-generation TKI osimertinib was also confirmed (Fig. 1E).
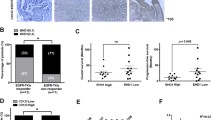
A–D HCC827 and PC9 cells were seeded in 96-well plates in medium containing 1% FBS. After overnight incubation, the cells were treated with EGFR ligands (50 ng/mL) and different doses of gefitinib or erlotinib for 48 h. The viability of cells was determined using the CCK-8 assay. The OD value at 450 nm was measured with an iMark microplate reader. Results are expressed as mean ± SEM from three independent experiments (n = 3). E The sensitivity of HCC827 (left) and PC9 (right) cells to Osimertinib was assayed by the above method. F Association of EREG expression with erlotinib response in NSCLC. EREG expression levels in tumors were compared between patients with different responses to erlotinib (left panel). p value was calculated by using Wilcoxon test. The 8-week disease control (DC) rate (middle panel) and PFS (right panel) of patients with high- and low-expression of EREG are presented. Statistical significance for differences in DC rate and PFS was calculated by Fisher’s exact test and log-rank test, respectively.
Next, we analyzed the correlation between EREG expression in tumor tissues and erlotinib response in NSCLC patients (Fig. 1F). The results showed that EREG expression was positively correlated with disease progression following erlotinib treatment. The 8-week disease control rate was 23.1% in the EREG-high subgroup and 69.2% in the EREG-low subgroup. Accordingly, higher expression of EREG indicated a significantly shorter PFS of patients. However, the expression levels of the other six EGFR ligands were not correlated with erlotinib response or PFS of patients (Supplementary Fig. S2).
EREG induces EGFR-TKI resistance by enhancing anti-apoptosis rather than proliferation
We next investigated whether EREG-induced TKI resistance was associated with cell proliferation. The CCK-8 assay was performed to examine the effects of EREG on the growth of PC9 and HCC827 cells. The results showed that EREG did not promote the growth of these cells, either in the presence or absence of TKI (Supplementary Fig. S3A, B). In addition, as shown by immunoblotting experiments, the phosphorylation levels of the extracellular signal-regulated kinase (ERK) in PC9 and HCC827 were comparable between the EREG treatment and control groups (Supplementary Fig. S3C, D).
We hypothesized that EREG might affect cellular apoptosis induced by EGFR-TKIs. Therefore, we analyzed apoptosis in PC9 cells treated with TKIs alone, EREG alone, or their combination. Flow cytometric analysis revealed that EREG prevented gefitinib-induced apoptosis in a dose-dependent manner (Fig. 2A, B). In particular, the percentage of apoptotic cells induced by 15 nM gefitinib was 44.3%, compared to 25.7% in the gefitinib plus EREG group. Similarly, the percentage of apoptosis induced by 15 nM erlotinib was reduced from 15.3 to 9.2% after treatment with EREG. Furthermore, we examined the effects of EREG on the levels of proteins associated with apoptosis. Western blot analysis showed that gefitinib and erlotinib enhanced cleaved caspase 9 levels compared with the control group, which was not observed after the addition of EREG (Fig. 2C). We observed similar results for the apoptotic markers cleaved caspase 8 and cleaved caspase 3 in HCC827 cells (Fig. 2D). When cells were pretreated with the pan-caspase inhibitor Z-VAD-FMK, the differences in apoptosis rates between the EREG and control groups were diminished (Supplementary Fig. S4A). The same was true for apoptotic protein levels (Supplementary Fig. S4B, C). Taken together, these results suggest that EREG-induced TKI resistance is more likely associated with prevention of apoptosis rather than fostering proliferation in NSCLC cells.
A PC9 cells were seeded in 6-well plates in medium containing 1% FBS. After overnight incubation, the cells were treated with 0, 5, 15 nM gefitinib/erlotinib and/or 50 ng/mL recombinant EREG for 24 h. Cells were stained with Annexin V-FITC and PI according to a standard protocol, and analyzed using flow cytometry. B The apoptotic rate is presented as a histogram by GraphPad Prism 7. Results are expressed as mean ± SEM from three independent experiments (n = 3). C, D PC9 and HCC827 cells were seeded in 6-well plates in medium containing 1% FBS. After overnight incubation, the cells were treated with gefitinib/erlotinib and/or 50 ng/mL recombinant EREG for 24 h, and then washed with PBS and lysed in loading buffer. The expressions of cleaved caspase 9 in PC9 cells and cleaved caspase 3 and cleaved caspase 8 in HCC827 cells were assessed by western blot analysis, GAPDH or β-actin was used as protein level control. *p < 0.05; **p < 0.01; NS not significant.
EREG-conferred EGFR-TKI resistance depends on ErbB2
EREG induces homodimerization of EGFR and ErbB4, and ErbB family members. Therefore, we sought to analyze the role of the ErbB family. PC9 cells were transfected with shRNAs targeting ErbB2, ErbB3, and ErbB4, or with scrambled control shRNA. CCK-8 assay revealed that knockdown of ErbB2 using two different shRNAs attenuated the EREG-induced resistance to TKIs, as compared with the scrambled control (Fig. 3A, B). However, knockdown of ErbB3 or ErbB4 had no effect on the EREG-induced resistance (Supplementary Fig. S5A–C). ErbB2 knockdown also attenuated the inhibitory effect of EREG on apoptosis, by increasing the percentage of apoptotic cells and cleaved caspase 9 expression (Fig. 3C, D). The same was true for HCC827 cells (Fig. 3E–G).
PC9 and HCC827 cells were transfected with shRNAs (scramble, HER2 shRNA sequences 1–2). A, E PC9 and HCC827 cells were harvested, and mRNA and proteins were extracted. Quantitative real-time RT-PCR was performed as described in the Methods (left panel), and the expression of ErbB2 was assayed by western blot analysis, GAPDH was used as protein level control (right panel). B, F PC9 and HCC827 cells were seeded in 96-well plates in medium containing 1% FBS. After overnight incubation, the cells were treated with different doses of gefitinib and/or 50 ng/mL recombinant EREG for 48 h. The viability of cells was determined using the CCK-8 assay. Results are expressed as mean ± SD (n = 3). C PC9 cells were harvested and incubated in 6-well plates in medium containing 1% FBS. After overnight incubation, the cells were treated with 50 ng/mL recombinant EREG and/or 0, 5, 15 nM gefitinib for 24 h. Cells were stained with Annexin V-FITC and PI according to a standard protocol, and analyzed using flow cytometry. The apoptotic rate is presented as a histogram by GraphPad Prism 7. Results are expressed as mean ± SEM from three independent experiments (n = 3). D, G The expressions of cleaved caspase 9 in PC9 cells and cleaved caspase 3 and cleaved caspase 8 in HCC827 cells were assayed by western blot analysis, GAPDH was used as protein level control. H, I Predictive value of EREG for erlotinib therapy in patient subgroups stratified by ErbB2 expression levels. Patients were grouped by the median value of ErbB2 expression. The association of EREG with 8-week disease control (DC) and PFS following erlotinib therapy was analyzed in the ErbB2-high (H) and ErbB2-low (I) subgroups, respectively. Wilcoxon test, Fisher’s exact test, and log-rank test were used to estimate the significance of differences in EREG expression, DC rate, and PFS, respectively. *p < 0.05; **p < 0.01; NS not significant.
Next we analyzed the requirement of ErbB2 for TKI resistance using patient’s data. Patients were divided into ErbB2-high and -low subgroups according to their gene expression levels. The results showed that the correlation between EREG expression and patient response to erlotinib was statistically significant in the ErbB2-high subgroup, but not in the ErbB2-low subgroup (Fig. 3H, I).
We further investigated the molecular mechanism underlying EREG-induced TKI resistance. Immunofluorescence assays showed a strong colocalization between EGFR and ErbB2 in the presence of EREG, which was maintained after gefitinib treatment (Fig. 4A). The downstream signaling pathways were detected following EREG treatment in the presence or absence of gefitinib. EREG recovered the phosphorylation level of ERK that were reduced by gefitinib, in cells expressing ErbB2 or being knockout for ErbB2. Although the pAKT levels were recovered by EREG in scrambled shRNA transfected cells, the recovery was attenuated when ErbB2 was knocked down (Fig. 4B, C). These results suggest that EREG-induced EGFR-TKI resistance may depend on the EGFR/ErbB2 heterodimer and the downstream PI3K/AKT pathway.
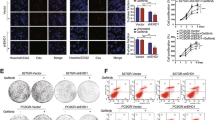
A Imaging analysis of EGFR-ErbB2 heterodimer: PC9 cells were seeded on 14-mm glass coverslips in 24-well plates, after serum starved overnight, 50 ng/mL EREG, 10 nM gefitinib, and EREG plus gefitinib were added to cells for 30 min. Cells were stained for immunofluorescence. Pictures were taken by Nikon A1 laser scanning confocal microscope with a 100× oil lens. The blue color represents DAPI, the green color represents ErbB2, and the red color represents EGFR. B PC9 cells were transfected with scramble or ErbB2 shRNA. Then the cells were harvested and incubated in 6-well plates. After serum starved overnight, the cells were treated with 2 nM gefitinib and/or different concentrations of recombinant EREG for 15 min. The expression of pEGFR, pAKT, and pERK was assayed by western blot analysis, GAPDH was used as protein level control. C Quantification of fold expression of ERK and AKT phosphorylation over GAPDH and normalization to DMSO control.
Cancer cell-derived EREG does not affect EGFR-TKI sensitivity
We have examined the role of extracellular addition of recombinant EREG on TKI resistance. We further assessed whether EREG expression in EGFR-mutant cell lines has an impact on EGFR-TKI resistance. First, we examined the expression of EREG in NSCLC cells that were sensitive (PC9 and HCC827) or resistant (H1650 and H1975) to EGFR-TKIs. Western blot results showed that HCC827 cells expressed the highest levels of EREG, while the other three cells expressed low levels of EREG. Determination of the EREG mRNA levels in the four cell lines by qPCR showed similar results (Fig. 5A). Next, we extracted mRNA and proteins from HCC827GR and PC9GR cells with acquired resistance to gefitinib. Although the expression of EREG in HCC827GR cells was significantly higher than that in parental HCC827 cells, there was no significant difference between PC9GR and PC9 cells (Fig. 5B).
A PC9, HCC827, H1650, and H1975 cells were seeded in 6-well plates overnight, then mRNA and proteins were extracted. Quantitative real-time RT-PCR was performed as described in the Methods (top panel), and the expression of EREG was assayed by western blot analysis, GAPDH was used as protein level control (bottom panel). B HCC827, HCC827GR and PC9, PC9GR cells were seeded in 6-well plates overnight, and mRNA and proteins were extracted. Quantitative real-time RT-PCR was performed as described in the Methods (left), and the expression of EREG was assayed by western blot analysis, GAPDH was used as protein level control (right). C HCC827 cells were transfected with shRNAs (scramble, EREG shRNA) and PC9 cells were transfected with EREG cDNAs using lentivirus vectors for 24 h, cell lysates were prepared, and the expression of EREG was assayed by western blot analysis, GAPDH was used as protein levels control (left), The viability of cells was determined using the CCK-8 assay. The OD value at 450 nm was measured with an iMark microplate reader. Results are expressed as mean ± SEM from three independent experiments (n = 3) (right). D PC9, PC9-expEREG, and HCC827 cell extracts were prepared. The expression of pAKT, pERK, and EREG was assayed by western blot analysis. GAPDH was used as protein level control.
Meanwhile, we knocked down EREG expression in HCC827 cells by shRNA lentivirus and introduced EREG cDNA into PC9 cells. The CCK-8 assay revealed that neither depletion nor overexpression of EREG had a significant effect on the sensitivity of gefitinib (Fig. 5C). EREG expression has been reported to be EGFR dependent and attenuated by specific inhibitors of EGFR. Consistently, western blot analysis showed that gefitinib could inhibit the expression of EREG in HCC827 and PC9-EREG-overexpression cells (Fig. 5D), indicating that EREG may originate from other cells in the tumor microenvironment.
Macrophages are potential source of EREG that confers EGFR-TKI resistance
By reanalyzing the single-cell RNA sequencing data of the tumor microenvironment of NSCLCs (GSE127465), we found that EREG was highly expressed in the macrophage population (Fig. 6A, B). We induced the differentiation of THP-1 cells to macrophages (M0) with PMA and then polarized the macrophages with LPS/IFN-γ and IL-4/IL-13, respectively. The expression of EREG protein in macrophages was significantly higher than that in control cells. Unlike in cancer cells, gefitinib did not inhibit the expression of EREG in macrophages (Fig. 6C). After treatment with conditioned medium (CM) from macrophages, NSCLC cells pronounced resistance to gefitinib (Fig. 6D). Similar results were obtained in the case of PBMCs differentiated into macrophages by M-CSF (Supplementary Fig. S6A, B). Collectively, these results indicate that paracrine secretion of EREG by macrophages in the tumor microenvironment may promote resistance to EGFR-TKIs in EGFR-mutant NSCLC cells (Fig. 6E).
A The single-cell landscape of tumor microenvironment components visualized by t-distributed stochastic neighbor embedding (t-SNE) plot. Each dot represents a single cell that is colored according to EREG expression level. B Percentage of EREG+ cells in cell populations in (A). Data of seven individual patients are presented. The patient-specific populations were excluded because there were only presented in one patient. p value was calculated by Kruskal–Wallis rank sum test. C THP-1 cells were treated with PMA to produce macrophages (M0) and then polarized with LPS/IFN-γ for 24 h to produce macrophages (M1) and IL-4/IL-13 for 72 h to produce macrophages (M2). The M1 and M2 macrophages were cultured in 10 mL serum-free RPMI 1640 medium for 48 h. Then, the conditioned medium (CM) were harvested, and cell lysates of M0, M1, and M2 macrophages were prepared. The expression of EREG was assayed by western blot analysis. GAPDH was used as protein levels control (left panel). The M2 macrophages were treated with 2 nM gefitinib for 24 h, M2 macrophage proteins were extracted. The expression of EREG was assayed by western blot analysis. GAPDH was used as protein level control (right panel). D PC9 cells were seeded in 96-well plates in the CM. The next day, the cells were treated with different doses of gefitinib for another 48 h. The viability of cells was determined using the CCK-8 assay. The OD value at 450 nm was measured with an iMark microplate reader. Results are expressed as mean ± SEM from three independent experiments (n = 3). E Schematic cartoon illustrates that EREG secreted by macrophages promote EGFR-TKI resistance in an EGFR/ErbB2-AKT-axis-dependent manner.
Discussion
This study systemically analyzed the seven ligands of EGFR (EGF, TGFα, AREG, EREG, EPGN, BTC, and HB-EGF) that affect the sensitivity of NSCLC cells to EGFR-TKIs in vitro. Consistent with previous studies, our results showed that EGF and TGFα were associated with reduced sensitivity to gefitinib or erlotinib in EGFR-mutant cells. The predictive value of serum amphiregulin for the response of patients with advanced NSCLC to EGFR-TKI treatment is controversial [10, 11]. We revealed that amphiregulin treatment of NSCLC cells did not influence the sensitivity to EGFR-TKIs, suggesting that the role of amphiregulin needs further investigation. HB-EGF and BTC, two ligands that promote lysosome degradation rather than membrane recycling of EGFR [17], were found to have little effect on TKI sensitivity according to our data. Surprisingly, we found that EREG and EPGN induced cellular resistance to EGFR-TKIs, with the exception of the effect of EPGN on erlotinib resistance in HCC827 cells. EREG and EPGN have been reported to interact with EGFR with relatively low affinity, but induced more sustained EGFR signaling than EGF and TGFα [6]. These results suggest that different EGFR ligands have distinct effects on EGFR-TKI sensitivity, which may depend on the mechanism of action of EGFR and subsequent cellular responses.
Dysregulation of EREG may contribute to the progression of various types of human malignancies, including lung cancer [18,19,20]. However, the effect of EREG on the sensitivity of NSCLC cells to EGFR-TKIs remains unclear. In this study, we revealed that soluble EREG induces resistance to EGFR-TKIs in EGFR-mutant cells by inhibiting cellular apoptosis. In contrast, we found that EREG stimulation did not directly promote proliferation of these cells. This might be explained by the fact that EREG induced sustained EGFR signaling and preferentially induced cellular differentiation but not proliferation [6]. In agreement with a previous report [6], we found that sustained downstream AKT activation after EREG stimulation may contribute to anti-apoptosis and cell survival.
EREG binds to EGFR and ErbB4 to stimulate homodimerization of the two receptors or their heterodimerization, thereby activating downstream pathways [19, 21]. It is necessary to determine whether EREG-induced EGFR-TKI resistance depends on other ErbB family members. By inhibiting the expression of ErbB-2, 3, and 4 using shRNAs targeting these genes, we found that only ErbB2 is required for EREG-induced cellular resistance to EGFR-TKIs. EREG stimulated subcellular colocalization of EGFR and ErbB2 in EGFR-mutant cells, suggesting the formation of heterodimers. Previous studies have demonstrated that ErbB2 genetic amplification [22] and mutation [23] contribute to EGFR-TKI resistance in NSCLC. Our data provided evidence that dysregulation of ErbB2 function by EREG may also affect TKI sensitivity. These results suggest that EREG-stimulated EGFR/ErbB2 signaling is a promising novel mechanism for TKI resistance.
The expression of EREG was elevated along with the activation of EGFR or downstream KRAS and BRAF, indicating a positive feedback loop of EREG and EGFR signaling [20, 21]. However, we found that the expression of EREG in cancer cells was not associated with either intrinsic or acquired resistance to EGFR-TKIs. This is partially explained by the fact that EGFR-TKIs could inhibit the endogenous expression of EREG, as seen in our results and previous reports [20]. Our results showed that expression of exogenous EREG in PC9 cells was also decreased in the presence of gefitinib. These results collectively suggest that EREG expression in cancer cells is inhibited by EGFR-TKIs and, therefore, is not associated with EGFR-TKI sensitivity.
Thus, it is necessary to explore the role of EREG in the tumor microenvironment which is composed of macrophages, fibroblasts, endothelial cells, and lymphocytes [24]. By reanalyzing previously published single-cell RNA sequencing data from NSCLC tumor tissues [25], we found that EREG was highly expressed macrophages/monocytes. These findings are supported by previous studies that have reported that monocytes and macrophages express EREG [16]. We also demonstrated that NSCLC cells became resistant to EGFR-TKIs when co-cultured with EREG-enriched macrophage conditional medium in vitro. Unlike in cancer cells, gefitinib did not inhibit EREG expression in macrophages, which may be regulated in an EGFR-independent manner. In fact, Massip-Copiz et al. [26] reported that EREG expression could be induced by the pro-inflammatory IL-1β/NFκB pathway. Recent evidence has shown that the peripheral inflammatory index, neutrophil-to-lymphocyte ratio, was inversely correlated with EGFR-TKI treatment response and clinical prognosis in NSCLC patients [27]. EREG has been implicated in inflammatory responses under both physiological and pathological conditions [19]. Taken together with our results, it is proposed that the macrophage-derived EREG may be involved in the mechanism of EGFR-TKI tolerance induced by inflammation.
In addition to EGFR mutations, there are few biomarkers that can effectively predict EGFR-TKI sensitivity in advanced NSCLC. Previous studies revealed that the objective response rates of the first-, second-, and third-generation TKIs were 62–83%, 56–75%, and 71–80%, respectively [1, 3]. These results indicate that a certain proportion of EGFR-mutant NSCLCs are intrinsically resistant to EGFR-TKIs. Meanwhile, a subgroup of patients with wild-type or unknown EGFR status may benefit from EGFR-TKI therapy [28, 29]. Therefore, it would be helpful to explore novel biomarkers for EGFR-TKI treatment. Our results suggested that the expression of EREG in tumors can predict the response and PFS of patients receiving EGFR-TKIs. The predictive value of EREG was dependent on ErbB2 expression, which was consistent with results from in vitro experiments showing that ErbB2 is required for EREG-induced EGFR-TKI resistance. One limitation of our study is the relatively small sample size of the clinical dataset. Therefore, validation of the results in a larger prospective cohort is warranted. Nevertheless, our findings suggest a trend toward the predictive value of EREG for EGFR-TKI therapy in advanced NSCLC.
In summary, we systemically assessed the effects of seven EGFR ligands on NSCLC cells in response to EGFR-TKIs. EREG induced cellular resistance to EGFR-TKIs by preventing apoptosis in an ErbB2-dependent manner. A paracrine effect of EREG secreted by macrophages in the tumor microenvironment has been proposed. High expression of EREG in tumor tissues is associated with EGFR-TKI resistance in patients with advanced NSCLC. These results suggest that high expression of intratumoral EREG may be a novel mechanism of resistance to EGFR-TKIs in NSCLC patients.
Materials and methods
Materials
EGF (cat. no. AF-100-15), TGFα (cat. no. 100-16A), epiregulin (cat. no. 100-04), Epigen (cat. no. 100-51), amphiregulin (cat. no. 200-55B), betacellulin (cat. no. 100-50), and HB-EGF (cat. no. 100-47) were purchased from PeproTech (Rocky Hill, NJ, USA). CCK-8 (cat. no. FXP132-1000) was purchased from 4A Biotech (Beijing, China). Gefitinib (cat. no. HY50895) and erlotinib (cat. no. HY50896) were purchased from MCE (Shanghai, China). Antibodies against epiregulin (cat. no. 12048), anti-phospho-AKT (cat. no. 4060), anti-phospho-ERK1/2 (cat. no. 9101), anti-ErbB2 (cat. no. 2165), anti-ErbB3 (cat. no. 12708), anti-ErbB4 (cat. no. 4795), anti-cleaved caspase 3 (cat. no. 9661), anti-caspase 8 (cat. no. 9746), anti-cleaved caspase 9 (cat. no. 9501) and anti-rabbit IgG (H+L), F(ab’) 2Fragment (Alexa Fluor 488) (cat. no. 4412) were purchased from Cell Signaling Technology (Danvers, MA, USA). Anti-EGFR antibodies (cat. no. ab52894) and anti-mouse IgG H&L conjugated to Alexa Fluor 647 (cat. no. ab150115) were purchased from Abcam (Cambridge, MA, USA). Antibodies against GAPDH (cat. no. EM1101) and the secondary antibody of anti-rabbit lgG (cat. no. HA1001) were purchased from HUA BIO (Hangzhou, China). 4’,6-Diamidino-2-phenylindole dihydrochloride (DAPI) (cat. no. 28718–90-3) and puromycin (cat. no. P8833) were purchased from Sigma-Aldrich (St. Louis, MO, USA). The anti-fluorescent quencher was purchased from Biosharp (cat. no. BL701A) (Hefei, China). Annexin V-FITC Apoptosis Detection Kit (cat.no. KGA108) was purchased from KeyGene BioTech (Nanjing, China).
Lentiviral-based shRNA constructs targeting human EREG and nonspecific control shRNAs were synthesized by GeneChem Biotech (Shanghai, China). The sequences of shRNAs targeting EREG were 5′-TCGGTTCCACATATTATTT-3′ and 5′-TGTGGCTCAAGTGTCAATA-3′, and the control shRNA sequence was 5′-TTCTCCGAACGTGTCACGT-3′. Lentiviral-based shRNA constructs targeting human ErbB2 and nonspecific control shRNAs were synthesized by Tsingke Biotechnology Co. (Beijing, China). The sequences of shRNAs targeting ErbB2 were 5′-GCCATCAAAGTGTTGAGGGAA-3′ and 5′-CCCTGTTCTCCGATGTGTA-3′, and control shRNA sequence was 5′-ccggUUCUCCGAACGUGUCACGUctcgagACGUGACACGUUCGGAGAAttttt-3′. RNeasy Mini Kit (cat. no. 74104), Gel Extraction Kit (cat. no. 28706), and Plasmid Maxi kit (cat. no. 12162) were purchased from Qiagen (Hilden, Germany). iTaq Universal SYBR Green One-Step Kit was purchased from Bio-Rad Laboratories (Hercules, CA, USA). OPTI-MEM (cat. no. 31985-062) was purchased from Gibco (Grand Island, NY, USA). The X-treme GENE HP DNA transfection reagent (cat. no. 06366236001) was purchased from Roche (Mannheim, Germany). The PCR Mycoplasma Test Kit (cat. no.K0103) was purchased from HUA BIO.
Cell culture
All cell lines were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA) and the cell bank of the Chinese Academy of Sciences (Shanghai, China), and were maintained in RPMI 1640 medium (Gibco) supplemented with 10% fetal bovine serum (BI, Kibbutz Beit-Haemek, Israel) at 37 °C in 5% CO2. All cell lines were tested for mycoplasma infection using the PCR Mycoplasma Test Kit (HUA BIO).
CCK-8 assay
HCC827 and PC9 cells were seeded in 96-well plates at a density of 5000 cells/well in medium containing 1% FBS or macrophage CM. After overnight incubation, the cells were treated with different reagents at various concentrations. Cell viability was determined using the CCK-8 assay. Ten microliters of CCK-8 solution was added to each well. The plates were incubated for 2 h at 37 °C, and the OD values at 450 nm were measured using the Bio-Rad microplate reader. Each CCK-8 assay was repeated three times.
Real-time PCR
Total RNA was extracted using RNeasy Mini Kit. RNA concentration was determined using a NanoDrop 2000 spectrophotometer. The iTaq Universal SYBR Green One-Step Kit was used for quantitative real-time reverse transcriptase-PCR with 50 ng RNA on the CFX-Connect real-time PCR detection system. Relative gene expression at the mRNA level was calculated using 2cq value of GAPDH – cq value of the target gene. GAPDH was used as an internal control. Each qPCR assay was repeated three times. The primer sequences used were as follows:
EREG forward primer 5′-TGGACATGAGTCAAAACTACT-3′,
EREG reverse primer 5′-GAAGTGTTCACATCGGACACC-3′,
ErbB2 forward primer 5′-TCAGTGACCTGTTTTGGACCG-3′,
ErbB2 reverse primer 5′-CGGGCCACGCAGAAGGGAGGG-3′,
GAPDH forward primer 5′-ACCCAGAAGACTGTGGATGG-3′,
GAPDH reverse primer 5′-TTCAGCTCAGGGATGACCTT-3′.
Apoptosis assays
PC9 and HCC827 cells were grown in 6-well plates at a density of 3 × 105 cells/well in a medium containing 1% FBS overnight. Cells were treated with 0, 5, or 15 nM gefitinib/erlotinib and/or 50 ng/mL recombinant EREG for 24 h, and DMSO was used as a control. Cells were harvested by exposure to trypsin without EDTA, washed with PBS, and centrifuged at 500 g for 5 min. Then, the cells were suspended in binding buffer at a concentration of 1 × 106 cells/mL. FITC-conjugated annexin V and PI (5 μL) were added to 500 μL of cell suspension. The samples were then analyzed using BD FACS Canto II flow cytometer. The experiments were repeated three times.
Western blot
Cells were treated with different reagents for various times, washed with PBS, and lysed in loading buffer. The samples were then subjected to SDS-PAGE. After blocking, the membranes were incubated with primary antibodies. If necessary, the membranes were stripped and re-probed with another antibody. The results were analyzed using Tanon 5200 chemiluminescence imager.
Lentiviral production and virus transduction
The lentiviruses were packaged by mixing 3 μg lentiviral vector with 2.7 μg of helper plasmid pCMV-dR8.91 and 0.3 μg envelope plasmid (vesicular stomatitis virus-G) for 5 min. Then, 582 μL of serum-free Opti-MEM and 18 μL XtremeGene HP were added. The tubes were incubated at room temperature for 15 min. The mixture was added dropwise to 10 cm dishes containing 293T cells. After 10 h, the medium was replaced with 10 mL growth medium containing 20% serum for virus production. The cells were incubated for another 24 h, and the lentivirus-containing medium was collected.
When the cells were approximately 35% confluent in a 10-cm dish, the growth medium was replaced with 10 mL fresh medium containing 2 mL virus and 8 μg/mL polybrene. After 24 h, the medium was replaced with 10 mL fresh growth medium containing 2 μg/mL puromycin. The cells were incubated for 48 h under puromycin selection. Cells were harvested and divided into three parts according to standard procedures for western blotting, cell proliferation, and apoptosis assays.
Immunofluorescence assay
PC9 cells were grown on 14-mm glass coverslips in 24-well plates. After overnight serum starvation, 50 ng/mL EREG and 10 nM gefitinib were added to the cells for 30 min. The cells were washed three times with ice-cold PBS and fixed with 4% paraformaldehyde at room temperature for 15 min. The cells were permeabilized with ice-cold PBS containing 0.25% Triton-X100 for 5 min after being washed three times with PBS. The cells were then incubated with 10% bovine serum albumin for 1 h at 37 °C. Anti-EGFR and anti-ErbB2 antibodies were incubated with the cells at 4 °C overnight. The cells were washed three times and incubated with Alexa488-conjugated anti-rabbit secondary antibody and Alexa Fluor 647-conjugated anti-mouse secondary antibody for 1 h at room temperature. The cells were then washed three times with PBS. After the nuclei were stained with 5 μg/mL DAPI for 5 min, the cells were washed three times with PBS, and the coverslips were removed and placed on slides with an anti-fluorescent quencher. Pictures were taken by Nikon A1 laser scanning confocal microscope with a 100× oil lens.
Polarization of M1 and M2 macrophages
In all, 1 × 107 THP-1 cells growing in a 10 cm dish were incubated in 10 mL complete medium containing 200 ng/mL PMA for 24 h and subsequently further incubated in 10 mL fresh complete medium for another 24 h to produce THP-1 macrophages (M0). For M1 polarization, the M0 macrophages were then cultured for another 24 h in 10 mL fresh complete medium containing 100 ng/mL LPS and 20 ng/mL IFN-γ. For M2 polarization, the M0 macrophages were cultured in 10 mL fresh complete medium with 20 ng/mL IL-4 and 20 ng/mL IL-13 for 72 h. After polarization, M1 and M2 macrophages were cultured in 10 mL of serum-free RPMI 1640 medium for 48 h. Then, the CM was harvested by centrifugation at 524 g for 5 min, and the supernatant was stored at ~80 °C for subsequent use.
Public datasets
The single-cell transcriptomics of tumor tissues from seven patients with NSCLC were obtained from a previous study [25]. Normalized single-cell RNA sequencing data and the cell population annotation file were downloaded from the Gene Expression Omnibus (GEO, GSE127465). There were four women and three men in this cohort, with a median age of 74 years (range, 61–83 years). Most of the patients (5/7, 71.4%) were histologically diagnosed with adenocarcinoma.
Gene expression profiles of NSCLC were previously obtained from biopsy tissues in the Biomarker-Integrated Approaches of Targeted Therapy for Lung Cancer Elimination (BATTLE) trial [29, 30]. Normalized gene expression data and corresponding clinical information were downloaded from GEO (GSE33072). A sub-cohort of 26 patients who received erlotinib therapy was enrolled for the analysis in this study.
Statistical analysis
The sigmoid dose-response curve and median inhibition concentration (IC50) for gefitinib or erlotinib were calculated by using GraphPad Prism 7 software. Unpaired Student’s t-test was used to estimate the statistical significance of the flow cytometry data and qPCR data. Gene expressions of EREG and other EGFR ligands were compared between patient subgroups using Student’s t-test or Wilcoxon test if the data followed a normal distribution. PFS of patients was analyzed using the Kaplan–Meier curve and log-rank test. The single-cell transcriptomic atlas was visualized using a t-distributed stochastic neighbor embedding plot, with EREG expression levels labeled for cells. The percentage of EREG+ cells in each cell population was calculated for each patient. The differences in EREG+ rates among different cell populations were compared by using the Kruskal–Wallis rank sum test. Except for specified, statistical analyses were performed using the R software (v3.5.1, https://www.r-project.org/). Statistical significance was defined as a two-sided p value < 0.05.
References
Recondo G, Facchinetti F, Olaussen KA, Besse B, Friboulet L. Making the first move in EGFR-driven or ALK-driven NSCLC: first-generation or next-generation TKI? Nat Rev Clin Oncol. 2018;15:694–708.
Westover D, Zugazagoitia J, Cho BC, Lovly CM, Paz-Ares L. Mechanisms of acquired resistance to first- and second-generation EGFR tyrosine kinase inhibitors. Ann Oncol. 2018;29:I10–9.
Soria JC, Ohe Y, Vansteenkiste J, Reungwetwattana T, Chewaskulyong B, Lee KH, et al. Osimertinib in untreated EGFR-mutated advanced non-small-cell lung cancer. N Engl J Med. 2018;378:113–25.
Santoni-Rugiu E, Melchior LC, Urbanska EM, Jakobsen JN, de Stricker K, Grauslund M, et al. Intrinsic resistance to EGFR-tyrosine kinase inhibitors in EGFR-mutant non-small cell lung cancer: differences and similarities with acquired resistance. Cancers. 2019;11:57.
Nagano T, Tachihara M, Nishimura Y. Mechanism of resistance to epidermal growth factor receptor-tyrosine kinase inhibitors and a potential treatment strategy. Cells. 2018;7:16.
Freed DM, Bessman NJ, Kiyatkin A, Salazar-Cavazos E, Byrne PO, Moore JO, et al. EGFR ligands differentially stabilize receptor dimers to specify signaling kinetics. Cell. 2017;171:683–95.e618.
Chen JC, Zeng FH, Forrester SJ, Eguchi S, Zhang MZ, Harris RC Expression and function of the epidermal growth factor receptor in physiology and disease. Physiol Rev. 2016;96:1025–69.
Du ZF, Lovly CM. Mechanisms of receptor tyrosine kinase activation in cancer. Mol Cancer. 2018;17:13.
Kakiuchi S, Daigo Y, Ishikawa N, Furukawa C, Tsunoda T, Yano S, et al. Prediction of sensitivity of advanced non-small cell lung cancers to gefitinib (Iressa, ZD1839). Hum Mol Genet. 2004;13:3029–43.
Ishikawa N, Daigo Y, Takano A, Taniwaki M, Kato T, Hayama S, et al. Increases of amphiregulin and transforming growth factor-alpha in serum as predictors of poor response to gefitinib among patients with advanced non-small cell lung cancers. Cancer Res. 2005;65:9176–84.
Vollebergh MA, Kappers I, Klomp HM, Buning-Kager JC, Korse CM, Hauptmann M, et al. Ligands of epidermal growth factor receptor and the insulin-like growth factor family as serum biomarkers for response to epidermal growth factor receptor inhibitors in patients with advanced non-small cell lung cancer. J Thorac Oncol. 2010;5:1939–48.
Kim TM, Song A, Kim DW, Kim S, Ahn YO, Keam B, et al. Mechanisms of acquired resistance to AZD9291: a mutation-selective, irreversible EGFR inhibitor. J Thorac Oncol. 2015;10:1736–44.
De Luca A, Carotenuto A, Rachiglio A, Gallo M, Maiello MR, Aldinucci D, et al. The role of the EGFR signaling in tumor microenvironment. J Cell Physiol. 2008;214:559–67.
Poole JA, Nordgren TM, Heires AJ, Nelson AJ, Katafiasz D, Bailey KL, et al. Amphiregulin modulates murine lung recovery and fibroblast function following exposure to agriculture organic dust. Am J Physiol Lung Cell Mol Physiol. 2020;318:L180–91.
Wang Y, Jing Y, Ding L, Zhang X, Song Y, Chen S, et al. Epiregulin reprograms cancer-associated fibroblasts and facilitates oral squamous cell carcinoma invasion via JAK2-STAT3 pathway. J Exp Clin Cancer Res. 2019;38:274.
Vlaicu P, Mertins P, Mayr T, Widschwendter P, Ataseven B, Högel B, et al. Monocytes/macrophages support mammary tumor invasivity by co-secreting lineage-specific EGFR ligands and a STAT3 activator. BMC Cancer. 2013;13:197.
Roepstorff K, Grandal MV, Henriksen L, Knudsen SL, Lerdrup M, Grovdal L, et al. Differential effects of EGFR ligands on endocytic sorting of the receptor. Traffic. 2009;10:1115–27.
Bauer AK, Velmurugan K, Xiong KN, Alexander CM, Xiong J, Brooks R. Epiregulin is required for lung tumor promotion in a murine two-stage carcinogenesis model. Mol Carcinog. 2017;56:94–105.
Riese DJ, Cullum RL. Epiregulin: roles in normal physiology and cancer. Semin Cell Dev Biol. 2014;28:49–56.
Zhang J, Iwanaga K, Choi KC, Wislez M, Raso MG, Wei W, et al. Intratumoral epiregulin is a marker of advanced disease in non-small cell lung cancer patients and confers invasive properties on EGFR-mutant cells. Cancer Prev Res. 2008;1:201–7.
Sunaga N, Kaira K. Epiregulin as a therapeutic target in non-small-cell lung cancer. Lung Cancer. 2015;6:91–98.
Takezawa K, Pirazzoli V, Arcila ME, Nebhan CA, Song XL, de Stanchina E, et al. HER2 amplification: a potential mechanism of acquired resistance to EGFR inhibition in EGFR-mutant lung cancers that lack the second-site EGFR(T790M) mutation. Cancer Disco. 2012;2:922–33.
Hsu CC, Liao BC, Liao WY, Markovets A, Stetson D, Thress K, et al. Exon 16-skipping HER2 as a novel mechanism of osimertinib resistance in EGFR L858R/T790M-positive non-small cell lung cancer. J Thorac Oncol. 2020;15:50–61.
Lim B, Lin Y, Navin N. Advancing cancer research and medicine with single-cell genomics. Cancer Cell. 2020;37:456–70.
Zilionis R, Engblom C, Pfirschke C, Savova V, Zemmour D, Saatcioglu HD, et al. Single-cell transcriptomics of human and mouse lung cancers reveals conserved myeloid populations across individuals and species. Immunity. 2019;50:1317–34.e1310.
Massip-Copiz M, Clauzure M, Valdivieso AG, Santa-Coloma TA. Epiregulin (EREG) is upregulated through an IL-1 autocrine loop in Caco-2 epithelial cells with reduced CFTR function. J Cell Biochem. 2018;119:2911–22.
Ono T, Igawa S, Kurahayashi S, Okuma Y, Sugimoto A, Kusuhara S, et al. Impact of neutrophil-to-lymphocyte ratio in patients with EGFR-mutant NSCLC treated with tyrosine kinase inhibitors. Invest. New Drugs. 2020;38:885–93.
Mok T, Ladrera G, Srimuninnimit V, Sriuranpong V, Yu CJ, Thongprasert S, et al. Tumor marker analyses from the phase III, placebo-controlled, FASTACT-2 study of intercalated erlotinib with gemcitabine/platinum in the first-line treatment of advanced non-small-cell lung cancer. Lung Cancer. 2016;98:1–8.
Byers LA, Diao L, Wang J, Saintigny P, Girard L, Peyton M, et al. An epithelial-mesenchymal transition gene signature predicts resistance to EGFR and PI3K inhibitors and identifies Axl as a therapeutic target for overcoming EGFR inhibitor resistance. Clin Cancer Res. 2013;19:279–90.
Kim ES, Herbst RS, Wistuba II, Lee JJ, Blumenschein GR, Jr, Tsao A, et al. The BATTLE trial: personalizing therapy for lung cancer. Cancer Disco. 2011;1:44–53.
Acknowledgements
We would like to thank Dr. Changmin Chen for his help with cell viability analysis.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 81602731), the Science and Technology Department of Sichuan Province (Grant Nos. 2019YJ0573 and 2020YJ0453), and the Innovation Program of Sichuan Medical Association Science for Youth (Grant No. Q18009).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Ma, S., Zhang, L., Ren, Y. et al. Epiregulin confers EGFR-TKI resistance via EGFR/ErbB2 heterodimer in non-small cell lung cancer. Oncogene 40, 2596–2609 (2021). https://doi.org/10.1038/s41388-021-01734-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-021-01734-4
- Springer Nature Limited
This article is cited by
-
TRPV1 inhibition overcomes cisplatin resistance by blocking autophagy-mediated hyperactivation of EGFR signaling pathway
Nature Communications (2023)
-
Epiregulin increases stemness-associated genes expression and promotes chemoresistance of non-small cell lung cancer via ERK signaling
Stem Cell Research & Therapy (2022)
-
Targeting epiregulin in the treatment-damaged tumor microenvironment restrains therapeutic resistance
Oncogene (2022)
-
Oncogenic KRAS promotes growth of lung cancer cells expressing SLC3A2-NRG1 fusion via ADAM17-mediated shedding of NRG1
Oncogene (2022)
-
Protein tyrosine kinase inhibitor resistance in malignant tumors: molecular mechanisms and future perspective
Signal Transduction and Targeted Therapy (2022)