Abstract
Epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (EGFR-TKIs) significantly prolong the survival time of non-small-cell lung cancer (NSCLC) patients with EGFR-activating mutations, but resistance develops universally. Activation of the phosphatidyl inositol-3 kinase (PI3K)/AKT signaling pathway and phenotypic alterations in epithelial–mesenchymal transition (EMT) are both mechanisms of acquired resistance to EGFR-TKIs. However, the mechanisms underlying this resistance remain unclear. In this study, EHD1 depletion significantly increased NSCLC cell sensitivity to EGFR-TKI, which was accompanied by EMT reversal. Microarray analysis showed that the PTEN/PI3K/AKT signaling pathway is a crucial pathway regulated by EHD1. Moreover, a PTEN inhibitor abolished EHD1 shRNA regulation of EGFR-TKI sensitivity, EMT, and cancer progression. Mass spectrometry showed that TUBB3 is a novel EHD1-interacting protein. EHD1 modulated microtubule stability by interacting with TUBB3. Furthermore, TUBB3 depletion significantly attenuated EHD1-induced EGFR-TKI resistance and EMT. Bioinformatics analysis revealed that EHD1 is significantly associated with the gene set, “Cellular Response to Interleukin-1β (IL-1β)”. As expected, treatment with IL-1β led to increased expression of EHD1, activation of PTEN/PI3K/AKT signaling, and induction of EMT in NSCLC cells. In patient specimens, EHD1 was highly expressed in EGFR-TKI-refractory specimens. EHD1 was positively associated with TUBB3 and IL-1R1 but negatively associated with PTEN. In addition, targeting the IL-1β/EHD1/TUBB3 axis mitigated cancer progression by inhibiting cell proliferation and metastasis and promoting apoptosis. Our study demonstrates the involvement of the IL-1β/EHD1/TUBB3 axis in EGFR-TKI resistance and provides a potential therapeutic approach for treating patients with NSCLC that has acquired EGFR-TKI resistance.
Similar content being viewed by others
Introduction
Lung cancer is the most common cause of cancer-related mortality worldwide [1]. The 5-year survival rate for patients with lung cancer is only 15%, and ~85% of these cases are non-small-cell lung cancer (NSCLC) [2]. EGFR-TKIs, including gefitinib and erlotinib, have demonstrated dramatic efficacy in NSCLC patients with EGFR-activating mutations [3]. Unfortunately, most patients do not achieve a complete response, and nearly all patients eventually acquire EGFR-TKI resistance, which leads to lethal disease progression [4]. The identified acquired resistance mechanisms have been mainly categorized as secondary mutations in the EGFR gene (T790M and other rare mutations), the activation of alternative signaling pathways (such as the phosphatidyl inositol-3 kinase (PI3K)/AKT pathway), and phenotypic changes such as epithelial–mesenchymal transition (EMT) [5, 6]. The unpredictability, diversity, and cooccurrence of EGFR-TKI resistance mechanisms present a challenge for efficiently developing innovative treatment strategies that can overcome EGFR-TKI resistance in patients.
C-terminal Eps15-homology (EH) domain-containing protein (EHD1) regulates multiple steps of endocytosis and vesicle trafficking [7, 8]. EHD1 has a substantial effect on the regulation of mitosis and pre-cytokinetic events such as microtubule dynamics [9, 10]. Furthermore, some studies have suggested that EHD1 plays a key role in tumor development and progression [11,12,13]. Recently, we reported that EHD1 is involved in promoting lung cancer progression and that EHD1 plays a role in erlotinib resistance using a lung cancer cell line and its EGFR-TKI-resistant daughter cell line, but the underlying mechanisms in EGFR-TKI resistance have not been fully elucidated [14, 15].
Microtubules are cytoskeletal proteins that comprise α- and β-tubulin heterodimers [16]. The dynamics and stability of microtubules play important roles in multiple cellular activities, including vesicle transport, cell movement, centrosome assembly, and chromosome segregation [17]. Notably, the dynamics and stability of microtubules are also reported to be related to EMT and cancer metastasis [18, 19]. Class III β-tubulin (βIII-tubulin, which is also referred to as TUBB3) is a microtubule protein that generates highly dynamic microtubules [20]. Previous studies have shown that TUBB3 promotes cancer development and induces EMT [21,22,23,24]. In addition, TUBB3 overexpression is associated with a reduced efficiency of tubulin-binding agents (TBAs) and DNA-damaging agents in cancers [25, 26]. However, the role of TUBB3 in tumor sensitivity to targeted therapies remains unknown.
Interleukin 1β (IL-1β) is an innate proinflammatory cytokine that has been reported to facilitate EMT and thus tumor metastasis in various cancer cells [27, 28]. In this mechanism, IL-1β induces β-catenin translocation, resulting in increased activation of transcription of target genes, such as SNAIL1 and CCDN1, which are involved in EMT [29]. Although previous research has confirmed that IL-1β is associated with the acquisition of an unresponsive phenotype to cetuximab, which is a monoclonal antibody that targets EGFR, the functional significance of IL-1β in EGFR-TKI resistance is unclear [30].
In this study, we aimed to establish a detailed mechanistic understanding of EHD1 as a significant factor in acquired EGFR-TKI resistance and tumor progression. We identified EHD1 through PTEN/PI3K/AKT signaling via TUBB3, which causes gefitinib resistance and promotes EMT. Moreover, we showed that IL-1β-induced EHD1 potentiates EGFR-TKI resistance and EMT. We demonstrated for the first time that TUBB3 and IL-1β play a crucial role in the resistance to gefitinib, suggesting novel therapeutic targets for treating this subset of patients with lung cancer. Our findings suggest that the IL-1β/EHD1/TUBB3 axis contributes to acquired gefitinib resistance in patients with NSCLC and an EGFR-activating mutation.
Results
EHD1 knockdown resensitizes gefitinib-resistant NSCLC cells to gefitinib
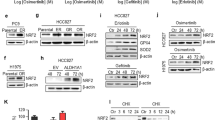
EGFR-TKI-resistant cell lines (HCC827GR and PC9GR cells) were generated by treating EGFR-TKI-sensitive HCC827 and PC9 cells, which harbor the EGFR exon 19 deletion, with increasing concentrations of gefitinib. The half-maximal inhibitory concentration (IC50) for gefitinib was dramatically increased in the EGFR-TKI-resistant cell lines compared with the EGFR-TKI-sensitive cell lines (Supplementary Fig. S1a, b). Expression vectors containing short hairpin RNAs (shRNAs) targeting EHD1 were transfected into EGFR-TKI-resistant cell lines. shRNA2 (shEHD1#2) and shRNA3 (shEHD1#3) most efficiently knocked down EHD1 expression and were therefore used in the subsequent experiments (Fig. 1a). EHD1 knockdown sensitized EGFR-TKI-resistant cells to different concentrations of gefitinib (Fig. 1b and Supplementary Fig. S1c). Next, we conducted an EdU incorporation assay and a CCK-8 assay to analyze cell viability. EHD1 depletion decreased the viability of HCC827GR and PC9GR cells, and gefitinib (IC2548h) treatment resulted in robust inhibition of EGFR-TKI-resistant cell viability (Fig. 1c, d and Supplementary Fig. S1d). In addition, we observed similar patterns in a long-term colony formation assay, suggesting that EHD1 depletion alleviates cell proliferation and sensitizes EGFR-TKI-resistant NSCLC cells (Fig. 1e). Flow cytometric analysis revealed that EHD1 knockdown significantly enhanced the apoptosis of HCC827GR and PC9GR cells following exposure to gefitinib (Fig. 1f). Overall, our results showed that EHD1 promotes cell proliferation and inhibits apoptosis in gefitinib-resistant lung cancer cells.
EHD1 knockdown overcomes TKI resistance in TKI-resistant lung cancer cells. a EHD1 protein expression levels were determined by Western blotting in HCC827GR (left panel) and PC9GR (right panel) cells stably expressing empty vector or shEHD1 (shEHD1#1, 2, 3). GAPDH served as a loading control. b HCC827GR-shEHD1 (left panel), PC9GR-shEHD1 (right panel) and corresponding vector control cells were treated with the indicated doses of gefitinib for 48 h, and cell viability was analyzed by a CCK-8 assay. Cell viability of the indicated cells cultured in the presence or absence of gefitinib (IC2548h) was analyzed by an EdU incorporation assay at 48 h (c) and by a CCK-8 assay at 24, 48, 72, and 96 h (d). e The colony-forming efficiency of HCC827GR-shEHD1, PC9GR-shEHD1 and corresponding vector control cells treated with or without gefitinib was determined. f Flow cytometric analysis of apoptosis in the indicated cells treated with or without gefitinib as assessed by Annexin V and 7-AAD staining. A representative flow profile is shown (upper panel), and a summary of the percentage of Annexin V-positive cells is shown (lower panel). *P < 0.05, **P < 0.01, and ***P < 0.001
EHD1 depletion reverses EMT in EGFR-TKI-resistant NSCLC cells
The Transwell assay showed that the gefitinib treatment alone (IC2548h) minimally affected the motility of EGFR-TKI-resistant cells. Interestingly, EHD1 depletion decreased the migration and invasion abilities of EGFR-TKI-resistant NSCLC cells; these effects were further enhanced when EHD1 depletion was combined with gefitinib (Fig. 2a and Supplementary Fig. S2a). The wound healing assay showed the same results in HCC827GR and PC9GR cells treated with gefitinib, transfected with shEHD1, and treated with a combination of gefitinib and shEHD1 transfection (Fig. 2b).
The effect of EHD1 knockdown on cell motility and EMT in TKI-resistant cells. a Transwell assays were conducted to assess TKI-resistant cell migration and invasion after EHD1 knockdown in cells cultured in the presence or absence of gefitinib compared with those of corresponding vector control cells (i.e., crystal violet staining of migratory and invasive cells). b A wound healing assay was performed in the indicated cells as described in a. c Representative phase contrast images of TKI-sensitive parental cells, TKI-resistant cells, and EHD1-knockdown cells (upper panel). Immunofluorescent staining confirmed the changes in the expression of EMT markers (lower panel). d Western blot analysis of E-cadherin, N-cadherin, Vimentin, and Snail expression in TKI-sensitive parental cells, TKI-resistant cells, and EHD1-knockdown cells. GAPDH served as a loading control. *P < 0.05, **P < 0.01 and ***P < 0.001
EMT is extensively correlated with therapeutic resistance to EGFR-TKIs [31]. As expected, we observed that long-term gefitinib treatment of TKI-sensitive cells changed the cellular phenotype to an elongated cell morphology and decreased cell–cell contacts, while the TKI-sensitive cells showed a cuboidal and cobblestone-like morphology (Fig. 2c). EHD1 knockdown induced a transition from a spindle-like to epithelial-like cell morphology, as indicated by the upregulation of E-cadherin expression and downregulation of Vimentin and N-cadherin expression (Fig. 2c). Consistent with the immunofluorescence staining results, Western blot analysis showed that the silencing of EHD1 effectively increased the E-cadherin expression level while suppressing the expression of mesenchymal markers in both HCC827GR and PC9GR cells (Fig. 2d and Supplementary Fig. S2b). Based on these findings, we concluded that EHD1 depletion mitigates cell motility and reverses EMT in EGFR-TKI-resistant lung cancer cells.
EHD1 knockdown inhibits PTEN/PI3K/AKT signaling activation
To show the potential mechanism by which EHD1 influences EGFR-TKI resistance and NSCLC progression, we performed microarray analysis to examine the global gene expression profile of NSCLC cells. Decreased activation of PTEN/PI3K/AKT signaling (notable upregulation of PTEN expression) was observed in EHD1-knockdown cells compared with the control (NC) cells (Fig. 3a and Supplementary Table S1). Consistently, Western blot analysis indicated that the level of PTEN expression was increased, whereas the phosphorylation of AKT (S473) and its downstream effectors Pras40 (Thr246), GSK3β (Ser9), and Foxo1 (Thr24)/3a (Thr32) was decreased in EHD1-knockdown EGFR-TKI-resistant cells compared with the control cells (Fig. 3b). Based on these findings, we concluded that EHD1 promotes PTEN/PI3K/AKT signaling activation.
PTEN/PI3K/AKT signaling is essential for EHD1-mediated TKI resistance and EMT. a Heatmaps showing the differential expression of PTEN/PI3K/AKT pathway gene signatures in control and EHD1-knockdown NSCLC cells. Red and green indicate high and low mRNA expression levels, respectively. b Western blot analysis of key signal transduction proteins in TKI-resistant cells stably expressing empty vector or shEHD1 and cultured in the presence of gefitinib. GAPDH served as a loading control. c The effect of a PTEN inhibitor (SF1670) on PTEN/PI3K/AKT signaling was analyzed by Western blotting with the indicated antibodies and samples. GAPDH served as a loading control. d Cell viability of HCC827GR-shEHD1 and PC9GR-shEHD1 cells after incubation with the indicated concentrations of gefitinib in the presence of the PTEN inhibitor (SF1670) for 48 h was analyzed by a CCK-8 assay. e A CCK-8 assay was used to examine changes in the proliferation rate of EHD1-knockdown cells cultured with gefitinib either alone or in combination with SF1670 at different time intervals (from 24 to 96 h). f HCC827GR-shEHD1 and PC9GR-shEHD1 cells seeded in Transwell culture chambers were treated with gefitinib either alone or in combination with SF1670. After 24–48 h, the migratory and invasive cells on the lower surface of the filter were stained with crystal violet and counted. g Protein expression levels of E-cadherin, N-cadherin, and Snail were analyzed by Western blotting. *P < 0.05, **P < 0.01 and ***P < 0.001
EHD1 induces EGFR-TKI resistance and EMT in a PTEN-dependent manner
Next, we used the PTEN inhibitor SF1670 to examine whether PTEN/PI3K/AKT signaling is responsible for EHD1-mediated gefitinib resistance and EMT. To further assess the potency of SF1670, we conducted dose-ranging experiments to estimate the level of SF1670 needed to achieve the maximal response (Supplementary Fig. S3a). SF1670 seemed to be a very potent PTEN inhibitor and could efficiently augment AKT phosphorylation at a concentration of 500 nM. Hence, we pretreated cells with SF1670 (500 nM) in the subsequent experiments. Western blot analysis indicated that SF1670 reversed the decrease in the PTEN/PI3K/AKT signaling activity observed in EGFR-TKI-resistant lung cancer cells subjected to EHD1 knockdown (Fig. 3c).
As demonstrated by the CCK-8 assay, the silencing of EHD1 increased the sensitivity to gefitinib, and SF1670 eliminated this effect (Fig. 3d). SF1670 caused a significant increase in the viability of EHD1 knockdown-resistant cells (Fig. 3e) and a higher colony-forming efficiency (Supplementary Fig. S3b). Similar results were also observed in the apoptosis analysis (Supplementary Fig. S3c).
Next, we found that SF1670 treatment led to increased migration and invasion abilities in EHD1-knockdown cells (Fig. 3f and Supplementary Fig. S3d). We also examined the expression levels of EMT markers treated with SF1670 by Western blot analysis. SF1670 downregulated E-cadherin expression and upregulated mesenchymal marker expression in EHD1-depleted cells (Fig. 3g). In addition, siRNAs targeting PTEN, among which siRNA-2 (siPTEN#2) and siRNA-3 (siPTEN#3) significantly repressed PTEN expression, were used to examine the effects of PTEN depletion on EHD1-induced activation of PTEN/PI3K/AKT signaling and EMT (Supplementary Fig. S3e–g). Overall, these data suggest that PTEN is required for EHD1-induced EGFR-TKI resistance and EMT.
EHD1 modulates microtubule stability through interacting with TUBB3
To elucidate the mechanism by which EHD1 promotes EGFR-TKI resistance and tumor progression, we analyzed proteins that interact with EHD1 using mass spectrometry. Notably, TUBB3 and the bait EHD1 were identified, and their peptide sequences are shown in Fig. 4a. We subsequently performed co-IP to validate whether EHD1 interacts with endogenous TUBB3. After IP of EHD1, we observed that TUBB3 interacted with EHD1, and vice versa, in HCC827GR and PC9GR cells (Fig. 4b, c). Confocal microscopy analysis demonstrated the subcellular localization and distribution of EHD1 and TUBB3 in the cytoplasm of the two cell lines (Fig. 4d). Furthermore, we found that the TUBB3 protein expression levels did not change when EHD1 was silenced, and TUBB3 knockdown did not affect the protein expression level of EHD1 (Supplementary Fig. S4).
EHD1 interacts with TUBB3 to enhance microtubule instability. a NSCLC cells transfected with the FLAG-tagged EHD1 construct or vector control plasmid were harvested after 48 h. IP and mass spectrometry analyses were performed on total proteins from the cells as described in the Supplementary Materials and Methods section. The domains of TUBB3 and the bait EHD1 that significantly interacted were identified, and the peptide sequences of TUBB3 and EHD1 are shown. b IP analysis of HCC827GR and PC9GR cells with an anti-EHD1 antibody and Western blot analysis of TUBB3 and EHD1 expression. c IP analysis of cells using an anti-TUBB3 antibody and Western blot analysis of TUBB3 and EHD1 expression. d Representative images of the colocalization of EHD1 (red) and TUBB3 (green) in HCC827GR and PC9GR cells. e Heatmaps showing that EHD1 was significantly correlated with cytoskeletal organization in control and EHD1-knockdown NSCLC cells. Red and green indicate high and low mRNA expression levels, respectively. f HCC827GR and PC9GR cells stably expressing empty vector or shEHD1 were treated with nocodazole for 0, 20, and 40 min. The polymeric and soluble dimeric fractions of tubulin were isolated and examined using Western blot analysis
How does EHD1 interact with TUBB3, which is composed of microtubules? A previous study reported that EHD1 is essential for proper central spindle formation and influences microtubule dynamics during mitosis [9]. Diverse cellular activities, including intracellular trafficking, cell migration, and cell division, depend on the dynamics and stability of microtubules [17]. Microarray analysis revealed a significantly positive correlation between EHD1 and cytoskeletal organization (Fig. 4e and Supplementary Table S2). Hence, we hypothesized that EHD1 modulates the stability of microtubules. To explore the role of EHD1 in microtubule stability, we treated the cells with a microtubule destabilizing agent (nocodazole) for 0, 20, and 40 min to make the microtubules depolymerize [32]. After cell extracts consisting of polymeric or soluble dimeric tubulin were obtained, Western blot analysis showed that nocodazole could depolymerize microtubules in a time-dependent manner. Moreover, a higher level of polymerized tubulin was detected in EHD1-knockdown cells compared with that in the control cells (Fig. 4f). Collectively, these results suggest that EHD1 regulates the stability of cytoskeletal microtubules through interacting with TUBB3 in vitro.
Suppression of TUBB3 expression increases gefitinib sensitivity
To examine the functional significance of TUBB3 in lung cancer, we showed that the TUBB3 mRNA expression level was significantly increased in NSCLC samples compared with normal lung samples from the cancer genome atlas (TCGA) datasets (Supplementary Fig. S5a). Next, Western blot analysis revealed higher protein expression levels of TUBB3 in the NSCLC samples than in adjacent noncancerous lung tissues (Supplementary Fig. S5b). Furthermore, the Kaplan–Meier plotter online tool was used to validate the effect of TUBB3 on lung cancer survival. The results consistently show that both LUAD and LUSC patients with elevated levels of TUBB3 expression had poorer OS (P = 0.016, P = 0.005) than did corresponding patients with low TUBB3 expression levels (Supplementary Fig. S5c). Using the web-based tools in The Human Protein Atlas (https://www.proteinatlas.org) based on the TCGA, low TUBB3 expression predicts a favorable prognosis in NSCLC (Supplementary Fig. S5d). As depicted in Supplementary Fig. S6a, the interference efficiency of shRNAs targeting TUBB3 was validated by Western blotting. shRNA-2 (shTUBB3#2) and shRNA-3 (shTUBB3#3) could significantly repress TUBB3 expression and were used for further studies (Supplementary Fig. S6a). TUBB3 depletion was shown to induce in vitro sensitization to gefitinib in EGFR-TKI-resistant lung cancer cells (Supplementary Fig. S6b). The Transwell and wound healing assays showed that silencing TUBB3 significantly attenuated the motility of resistant cells that were treated or untreated with gefitinib compared with that of the corresponding control cells (Supplementary Fig. S6c, d).
Next, the effect of TUBB3 on the activation of PTEN/PI3K/AKT signaling was determined. Compared with the controls, TUBB3 knockdown clearly decreased the expression levels of p-AKT, p-pras40, and p-GSK3β. Strikingly, TUBB3-knockdown cells showed markedly increased expression levels of PTEN (Supplementary Fig. S6e). These results suggest that TUBB3 promotes the resistance to gefitinib in EGFR-TKI-resistant cells via PTEN/PI3K/AKT signaling.
TUBB3 is essential for EHD1-induced gefitinib resistance and EMT
Next, we investigated whether TUBB3 is necessary for EHD1-mediated TKI resistance and EMT. First, we conducted an expression rescue experiment by transiently transfecting EHD1-knockdown EGFR-TKI-resistant cells with a vector encoding the human EHD1 gene (Fig. 5a). The CCK-8 assay showed that re-expression of EHD1 enhanced the resistance to gefitinib compared with the EHD1-knockdown cells (Fig. 5b). However, shTUBB3 abolished the EHD1 re-expression-induced EGFR-TKI resistance effect, cancer cell migration and invasion ability, EMT, and activation of PTEN/PI3K/AKT signaling (Fig. 5b–d). Thus, our data provide compelling evidence that TUBB3 is required in EHD1-mediated gefitinib resistance via EMT formation by triggering PTEN/PI3K/AKT signaling.
TUBB3 is required for EHD1-mediated TKI resistance and EMT. a Western blot analysis of EHD1 and TUBB3 in TKI-resistant cells transfected with the indicated constructs. GAPDH was used as a loading control. b Cell viability was analyzed by a CCK-8 assay in cells incubated with the indicated concentrations of gefitinib for 48 h. c Representative images (upper panel) and quantification (lower panel) of migratory and invasive cells as analyzed with a Transwell assay. d Proteins extracted from the indicated cells were subjected to Western blotting to analyze the expression of PTEN, phosphorylated AKT and EMT markers, such as E-cadherin and Vimentin. *P < 0.05, **P < 0.01, and ***P < 0.001
EHD1 promotes gefitinib resistance via PTEN/PI3K/AKT signaling in vivo
BALB/c-nu mice were distributed and assigned randomly to the following experimental groups: (1) PC9GR-vector (left) + PC9GR-shEHD1 (right) + water, (2) PC9GR-vector (left) + PC9GR-shEHD1 (right) + gefitinib, (3) PC9GR-shEHD1 (left) + gefitinib + SF1670. Representative tumor burdens in nude mice monitored by D-luciferin-based bioluminescence are shown in Fig. 6a, b. The tumor burden was reduced in the nude mice injected with PC9GR-shEHD1 cells compared with the nude mice injected with PC9GR-vector cells (Fig. 6a, c, d and Supplementary Fig. S7), suggesting that EHD1 promotes tumor growth. Intriguingly, a significant reduction in the tumor burden was observed in the shEHD1-transfected PC9GR cells treated with gefitinib compared with the control cells (Fig. 6b–d and Supplementary Fig. S7). These results strongly indicate that EHD1 plays an important role in tumor progression and EGFR-TKI resistance. Moreover, treatment with gefitinib and the PTEN inhibitor (SF1670) significantly restored tumor burden suppression in the nude mice injected with PC9GR-shEHD1 cells compared with the mice treated with gefitinib alone (Fig. 6b–d and Supplementary Fig. S7). Western blot analysis showed that the expression levels of PTEN and E-cadherin were significantly elevated, whereas the expression level of N-cadherin was decreased in the tumors formed by PC9GR-shEHD1 cells compared with the tumors formed by the control cells with or without gefitinib treatment (Fig. 6e). Moreover, the expression level of E-cadherin was lower in the tumor tissues treated with SF1670 compared with the tumor tissues treated with gefitinib alone, indicating that EHD1 induces EMT depending on PTEN/PI3K/AKT signaling in vivo (Fig. 6e). These results were also confirmed by IHC analysis (Fig. 6f). These results strongly support the hypothesis that EHD1 causes EGFR-TKI resistance by inducing EMT via the activation of PTEN/PI3K/AKT signaling in vivo.
Targeting EHD1 suppresses lung cancer growth and resistance to EGFR-TKI in a mouse xenograft model. PC9GR-vector cells or stable EHD1-knockdown (shEHD1) cells (each cell type expressed luciferase) were subcutaneously injected into the left and right dorsal regions of the posterior limbs, respectively, of nude mice. Mice receiving PC9GR-vector and PC9GR-shEHD1 xenografts were treated with water or gefitinib (100 mg/kg), and a subset of PC9GR-shEHD1 xenograft mice treated with gefitinib were also administered SF1670 (1 μmol/kg). a Tumor formation was monitored by bioluminescence imaging at the indicated time points to assess the therapeutic effect of EHD1 knockdown. b Bioluminescence imaging was used to analyze the therapeutic effect in PC9GR-shEHD1 mice treated with gefitinib either alone or in combination with SF1670. c Representative features of the tumors in the different treatment groups at 21 days. d The tumor volume in the nude mice from the five groups was measured at 4-day intervals from days 0 to 32 (n = 4 mice in each group). e Western blot analysis of the expression of the indicated markers in protein extracts obtained from harvested tumors. f Immunohistochemistry analysis to confirm the expression of EHD1, PTEN, E-cadherin, and N-cadherin in the indicated groups of tumor samples. Magnification, ×100 and ×400
IL-1β upregulates EHD1 and induces EGFR-TKI resistance
To identify the functional signatures of EHD1-bound genes that were enriched in the NSCLC cells, gene set enrichment analysis (GSEA) was conducted by using the TCGA database. We further ranked the gene sets in the GSEA from the TCGA data, and the most notable finding was that the functional gene set “Cellular Response to Interleukin-1β”, which is positively related to EHD1, was enriched in the NSCLC samples (Fig. 7a, b and Supplementary Fig. S8a). Accumulating evidence suggests that continuous exposure to inflammatory cytokines, including IL-1β, is known to promote both primary lung tumors and lung metastasis [33]. A positive correlation between EHD1 and IL-1β was observed in the lung cancer samples from the TCGA database (Fig. 7c). Western blot analysis showed that IL-1β stimulates the upregulation of EHD1 protein expression and increases the activity of PTEN/PI3K/AKT signaling (Fig. 7d). To investigate the effect of IL-1β on EHD1-induced EGFR-TKI resistance, we performed a CCK-8 assay, long-term colony formation assay, Transwell assay, wound healing assay and flow cytometry. The results revealed that IL-1β stimulation enhanced EHD1-mediated EGFR-TKI resistance (Fig. 7e–h and Supplementary Fig. S8b, c).
IL-1β upregulates EHD1 and enhances EGFR-TKI resistance. a GO analysis of genes that interact with EHD1 (indicated by the inverse log 10 of P values). b The heatmap shows the gene expression data for the cellular response to IL-1β gene signatures related to EHD1 based on the TCGA database. c The correlation between EHD1 and IL-1β mRNA expression was identified by the TCGA database. d EHD1-knockdown TKI-resistant cells were incubated with gefitinib alone or in combination with IL-1β (10 ng/mL) for 24 h. The cells were lysed, and the indicated proteins were analyzed by Western blotting. e HCC827GR-shEHD1 (left panel) and PC9GR-shEHD1 (right panel) cells that were nonstimulated or stimulated with IL-1β were treated with the indicated doses of gefitinib for 48 h, and cell viability was analyzed by a CCK-8 assay. f Cell viability was analyzed by a CCK-8 assay at 24, 48, 72, and 96 h. g Representative images (upper panel) and quantification (lower panel) of Annexin V and 7-AAD staining of HCC827GR-shEHD1 and PC9GR-shEHD1 cells incubated with gefitinib alone or in combination with IL-1β at 48 h. h Cell motility was assessed by Transwell assays via crystal violet staining of migratory and invasive cells. i The localization and expression of EMT markers in the indicated cells incubated in the presence or absence of IL-1β were analyzed by immunofluorescence microscopy and Western blotting (j). *P < 0.05, **P < 0.01, and ***P < 0.001
Considering that IL-1β plays a role in EMT progression, we analyzed EMT-related marker expression using immunofluorescence staining and Western blot analysis. We demonstrated that treatment with IL-1β leads to a lower E-cadherin expression level, whereas higher N-cadherin, Vimentin, and Snail expression levels were observed in the EHD1-knockdown EGFR-TKI-resistant cells compared with the control cells (Fig. 7i, j). These results revealed that IL-1β stimulation enhanced EGFR-TKI resistance and that EMT induction is mediated by the upregulation of EHD1. IL-1β triggers the canonical NF-κB pathway during which p-p65 expression is increased [34, 35]; therefore, we tested whether IL-1β induces EHD1 expression through the NF-κB pathway. BAY 11–7082, a specific inhibitor of NF-κB pathway that blocks IκB-α phosphorylation, was applied to inhibit IL-1β-induced NF-κB activation [36]. Interestingly, the results showed that IL-1β-mediated upregulation of EHD1 was abrogated when BAY 11–7082 was added (Supplementary Fig. S8d). These results indicate that IL-1β induces EHD1 through the NF-κB pathway.
EHD1 is correlated with TUBB3, IL-1R1, and PTEN in clinical NSCLC specimens
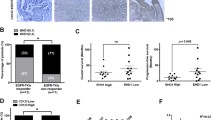
To determine the relationship between EHD1 expression and EGFR-TKI resistance, we examined NSCLC tissues from patients treated with EGFR-TKIs. In the IHC analysis, the protein expression level of EHD1 in these cases was classified as low (score = 0/1) or high (score = 2/3) based on the intensity and proportion of positively stained cells (Fig. 8a). As shown in Fig. 8b, the EHD1 expression level in the EGFR-TKI-insensitive group (PFS < 6 months) was higher than that in the EGFR-TKI-sensitive group (PFS ≥ 6 months). IHC analysis revealed that high EHD1 expression in clinical NSCLC specimens is significantly correlated with EGFR-TKI resistance.
Associations between EHD1, PTEN, TUBB3, and IL-1R1 expression in NSCLC tissues from patients. a Immunohistochemical staining yielded scores ranging from 0 to 3, which are representative of the intensity and proportion of EHD1-positive cells in lung cancer specimens. A score of 0–1 indicates low expression, and a score of 2–3 indicates high expression. Magnification, ×100 and ×400. b Representative immunostaining profiles of EHD1 in drug-sensitive (PFS ≥ 6 months, n = 23) and drug-insensitive (PFS < 6 months, n = 23) lung cancer tissues. c Representative images of immunohistochemical staining for EHD1, PTEN, TUBB3, and IL-1R1 in serial sections of lung cancer samples from patients. Case 1 is representative of a patient with EHD1-overexpressing lung cancer, whereas Case 2 is representative of a patient with non-EHD1-overexpressing lung cancer. d EHD1 expression determined by immunohistochemical staining was associated with PTEN, TUBB3 or IL-1R1 expression in 91 clinical NSCLC specimens. e A model of the IL-1β/EHD1/TUBB3 axis that contributes to EGFR-TKI resistance and tumor progression. The cytokine IL-1β upregulates EHD1 expression. Increased EHD1 expression modulates the microtubule polymerization dynamics by interacting with TUBB3, which in turn facilitates the activation of PTEN/PI3K/AKT signaling and EMT, thereby causing EGFR-TKI resistance
To further examine the relationship between EHD1 and tumor progression in human NSCLC, we performed IHC staining of EHD1, PTEN, TUBB3, and IL-1R1 in specimens from patients with NSCLC. Consistent with our observations in the tumor cell lines and xenograft models, the distribution and intensity of EHD1 were positively correlated with TUBB3 and IL-1R1 but negatively correlated with PTEN in NSCLC tissue specimens (Fig. 8c, d). Moreover, IHC analysis revealed that increased expression of EHD1 and overexpression of TUBB3 were correlated with the presence of positive lymph nodes (Supplementary Fig. S9).
Discussion
Although many patients harboring EGFR-activating mutations respond to EGFR-TKI treatment, the overall responses can vary, ranging from 5 to 90%, and remission could range from 3 months to longer than 5 years [37]. Consequently, understanding the unpredictability and diversity of EGFR-TKI resistance mechanisms and developing new treatment regimens that can combat resistance are urgently needed [38, 39]. In this study, we showed that the IL-1β/EHD1/TUBB3/PTEN signaling axis is crucial for EGFR-TKI resistance and lung cancer progression. In summary, IL-1β promotes the expression of EHD1, and EHD1 modulates microtubule stability through interacting with TUBB3, thereby activating PTEN/PI3K/AKT signaling. The activation of the PTEN/PI3K/AKT signaling axis potentiates lung cancer progression and EMT, leading to EGFR-TKI resistance (Fig. 8e).
Recent studies have emphasized the importance of upfront combination therapy to prevent the emergence of EMT rather than the strategy of resistance mechanism-based second-line therapies [40]. Our previous study revealed that EHD1 promotes lung cancer metastasis by inducing EMT [15]. Considering the impact of EMT on EGFR-TKI resistance, we confirmed EHD1-induced EGFR-TKI resistance through EMT in this study. Moreover, increasing evidence has demonstrated that acquired resistance to EGFR-TKIs in NSCLC is related to preexisting AKT activation, while treatment with EGFR-TKIs fails to suppress AKT signaling in intrinsically resistant tumor cells [41,42,43,44]. Notably, the activation of AKT signaling is a convergent feature in patients with NSCLC and an EGFR mutation with acquired resistance to EGFR-TKIs that may be due to multiple underlying mechanisms [45]. AKT signaling is activated in various cancers and induces enhanced cell proliferation, metastasis, EMT, and anti-apoptosis [46]. Consistent with these studies, our results indicated that the EHD1-activated AKT pathway leads to EMT and EGFR-TKI resistance. We demonstrated that EHD1 promotes tumor progression in an AKT pathway-dependent manner. The clinical trials demonstrated that patients with tumors benefit from treatment with AKT inhibitors. Progression-free survival (PFS) was longer in patients who received paclitaxel and the AKT inhibitor ipatasertib than in those who received placebo in triple-negative breast cancer [47]. Our results could help to improve strategies for the selection of patients with NSCLC who may particularly benefit from agents that selectively target the AKT pathway. In addition, PI3K inhibitors have demonstrated dramatic efficacy in patients with NSCLC. Edgar et al. [48] reported that patients with PTEN-negative tumors may preferentially benefit from treatment with a PI3K inhibitor. PI3K inhibitors exhibited satisfactory anticancer effects in preclinical and clinical studies in several types of cancer, including lung cancer [49, 50]. Our results show that EHD1/TUBB3 could significantly regulate PTEN expression; as a result, EHD1/TUBB3 could serve as biomarkers in guiding the selection of patients, who are most likely to respond to PI3K inhibitors.
For the first time, our results revealed that the microtubule component TUBB3 is an EHD1-interacting protein. TUBB3 is related to a poor prognosis and the metastasis of multiple malignancies, and 85% of small cell lung cancer cases exhibit TUBB3 overexpression [51,52,53,54]. In lung cancer, TUBB3 promotes tumorigenesis, EMT, and anoikis resistance through the AKT pathway [55]. In addition, TUBB3 depletion blocks AKT activation and delays the formation of the AKT signaling complex in response to glucose starvation [56]. Accordingly, we showed that TUBB3 regulates PTEN/PI3K/AKT signaling, which affects cell motility and EMT.
Alterations in the expression of microtubule proteins in cancer cells are recognized as important contributors to the resistance to chemotherapeutics, including microtubule-stabilizing and -destabilizing drugs [57, 58]. Previous studies have shown that microtubule destabilization facilitates focal adhesion assembly and thereby increases cell migration, and microtubule depolymerization contributes to the process of EMT [59,60,61]. We demonstrated that EHD1 modulates microtubule remodeling, possibly influencing cytoskeletal rearrangement, EMT, and cell motility in gefitinib-resistant cells. Overall, we proposed a working model that helps to explain the potential roles of EHD1 as an oncogene via TUBB3-mediated EMT. We aim to further investigate the precise mechanisms of the interactions between EHD1 and tubulins in a future study.
Chronic inflammation is recognized as one of the characteristics of carcinogenesis, tumor progression, and metastasis in various tumors [62]. Many inflammatory cytokines have been shown to be associated with therapeutic resistance to EGFR-TKIs and EMT [63]. A previous study has shown that a blockade of IL-8 signaling effectively reverses EMT in erlotinib-resistant cells and markedly increases their susceptibility to erlotinib [64]. EGFR-TKI treatment leads to STAT3 activation, which is caused by IL-6 in an autocrine-dependent manner [65]. It has been shown that IL-1 signaling activation induces the disassembly of adherens junction complexes and rearranges the membrane-tethered actin cytoskeleton via PI3K/Rac signaling [66]. In addition, the activation of IL-1 signaling is responsible for maintaining ERK and AKT signaling, and IL-1 is a therapeutic marker for patients who are resistant to cetuximab for anti-EGFR therapy [30]. Our results confirmed these previous observations regarding the role of the IL-1 pathway in cancer. In the context of EGFR-TKI resistance, we demonstrated that IL-1β upregulates EHD1, thereby induces gefitinib resistance and EMT. A large body of evidence from investigators supports the notion that the IL-1 pathway plays a carcinogenic role in EGFR-TKI resistance.
In summary, we uncovered a novel oncogenic signaling pathway involved in EHD1 that exclusively acts in acquired EGFR-TKI resistance and tumor progression by activating PTEN/PI3K/AKT signaling and promoting EMT in vitro and in vivo. These findings suggest that targeting IL-1β/EHD1/TUBB3 signaling could be a potential therapeutic strategy to enhance the efficacy of EGFR-TKIs in patients with acquired resistance.
Materials and methods
Cell culture, reagents, and lentivirus infection
The human NSCLC cell lines HCC827, HCC827GR, PC9, and PC9GR were obtained from Heilongjiang Cancer Institute (Harbin, China). These cells were cultured in DMEM or RPMI 1640 medium supplemented with 10% FBS at 37 °C. The cell lines were authenticated regularly by short tandem repeat DNA profiling and did not have any mycoplasma contamination [67]. The EGFR inhibitor gefitinib and the PTEN inhibitor SF1670 (Medchem Express, Monmouth Junction, NJ, USA) were used at concentrations indicated in the text. Lentiviral shRNA vectors targeting human EHD1 and TUBB3 and a nontargeting control vector were purchased from Genechem (www.genechem.com.cn; Shanghai, China). All the plasmid vectors were verified by sequencing.
Patients
Four pairs of fresh NSCLC tissue samples were collected from Harbin Medical University Cancer Hospital to determine the protein expression levels of TUBB3. We obtained paraffin-embedded samples from 91 NSCLC patients between May 2008 and August 2016 at the Harbin Medical University Cancer Hospital, including 46 patients with NSCLC harboring EGFR mutations. Patients were divided into the following two groups based on their response to TKIs: the EGFR-TKI insensitive group (n = 23) included patients with disease progression or stable disease without an extended (6 months) PFS, and the TKI-sensitive group (n = 23) included patients with a complete or partial response or stable disease with prolonged PFS (12 months) [68]. All identifying information relating to the samples was removed. This study was approved by the Institutional Research Ethics Committee of the Harbin Medical University Cancer Hospital for the use of these clinical materials for research purposes, and patient consent was obtained. The clinical implications of EHD1, TUBB3 and IL-1β were further analyzed with datasets from The Cancer Genome Atlas (TCGA) database.
Microarray processing and analysis
Detailed information on microarray processing and analysis has been described previously [15]. Pathway enrichment analysis was based on significant genes and was conducted using commercially available Ingenuity Pathway Analysis (IPA) software.
Animal experiments
BALB/c-nu mice (male, 4 weeks of age, Beijing Vital River Laboratory Animal Technology Co., Ltd.) were used to establish subcutaneous xenograft tumors. All animal studies were single blinded during group allocation and data analysis. Approximately 2 × 106 cells transduced with either vector or shEHD1 and a luciferase reporter in 100 μl of PBS were injected into the left and right dorsal regions, respectively, of the posterior limbs of the mice. When palpable tumors formed, the mice with two tumors were randomly allocated into groups to receive gefitinib (100 mg/kg) or sterile water by oral gavage every day. The mice with PC9GR-shEHD1 tumors were injected with SF1670 (1 μmol/kg) every other day. The tumor volume was calculated using the following equation: (length × width2)/2. Subsequently, 100 mg/kg D-luciferin was injected intraperitoneally into the mice, and the tumor size was monitored by measuring the bioluminescence signal every week until all the mice were sacrificed for collecting tumor tissues after three weeks of treatment. All animal studies were approved by the Institutional Animal Care and Use Committees of Harbin Medical University.
Statistical analysis
A detailed description of statistical analysis is provided in Supplementary Data section. Complete information about ‘Materials and methods’ is reported in the Electronic Supplementary Material.
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30.
Lee DH. Treatments for EGFR-mutant non-small cell lung cancer (NSCLC): the road to a success, paved with failures. Pharm Ther. 2017;174:1–21.
Kobayashi S, Boggon TJ, Dayaram T, Janne PA, Kocher O, Meyerson M, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. NEJM. 2005;352:786–92.
Soucheray M, Capelletti M, Pulido I, Kuang Y, Paweletz CP, Becker JH, et al. Intratumoral heterogeneity in EGFR-mutant NSCLC results in divergent resistance mechanisms in response to EGFR tyrosine kinase inhibition. Cancer Res. 2015;75:4372–83.
Sullivan I, Planchard D. Osimertinib in the treatment of patients with epidermal growth factor receptor T790M mutation-positive metastatic non-small cell lung cancer: clinical trial evidence and experience. Ther Adv Respir Dis. 2016;10:549–65.
George M, Ying G, Rainey MA, Solomon A, Parikh PT, Gao Q, et al. Shared as well as distinct roles of EHD proteins revealed by biochemical and functional comparisons in mammalian cells and C. elegans. BMC Cell Biol. 2007;8:3.
Daumke O, Lundmark R, Vallis Y, Martens S, Butler PJ, McMahon HT. Architectural and mechanistic insights into an EHD ATPase involved in membrane remodelling. Nature. 2007;449:923–7.
Reinecke JB, Katafiasz D, Naslavsky N, Caplan S. Novel functions for the endocytic regulatory proteins MICAL-L1 and EHD1 in mitosis. Traffic. 2015;16:48–67.
Hehnly H, Doxsey S. Rab11 endosomes contribute to mitotic spindle organization and orientation. Dev Cell. 2014;28:497–507.
Tong D, Liang YN, Stepanova AA, Liu Y, Li X, Wang L, et al. Increased Eps15 homology domain 1 and RAB11FIP3 expression regulate breast cancer progression via promoting epithelial growth factor receptor recycling. Tumour Biol. 2017;39:1010428317691010.
Liu Y, Liang Y, Li M, Liu D, Tang J, Yang W, et al. Eps15 homology domain 1 promotes the evolution of papillary thyroid cancer by regulating endocytotic recycling of epidermal growth factor receptor. Oncol Lett. 2018;16:4263–70.
Chan JK, Kiet TK, Blansit K, Ramasubbaiah R, Hilton JF, Kapp DS, et al. MiR-378 as a biomarker for response to anti-angiogenic treatment in ovarian cancer. Gynecol Oncol. 2014;133:568–74.
Wang X, Yin H, Zhang H, Hu J, Lu H, Li C, et al. NF-κB-driven improvement of EHD1 contributes to erlotinib resistance in EGFR-mutant lung cancers. Cell Death Dis. 2018;9:418.
Meng Q, Xing Y, Ren T, Lu H, Xi Y, Jiang Z, et al. Mammalian Eps15 homology domain 1 promotes metastasis in non-small cell lung cancer by inducing epithelial-mesenchymal transition. Oncotarget. 2017;8:22433–42.
Walczak CE, Cai S, Khodjakov A. Mechanisms of chromosome behaviour during mitosis. Nat Rev Mol Cell Biol. 2010;11:91–102.
Desai A, Mitchison TJ. Microtubule polymerization dynamics. Annu Rev Cell Dev Biol. 1997;13:83–117.
Satir P, Goltz JS, Wolkoff AW. Microtubule-based cell motility: the role of microtubules in cell motility and differentiation. Cancer Investig. 1990;8:685–90.
Etienne-Manneville S. Actin and microtubules in cell motility: which one is in control? Traffic. 2004;5:470–7.
Luduena RF. Are tubulin isotypes functionally significant. Mol Biol Cell. 1993;4:445–57.
Reiman T, Lai R, Veillard AS, Paris E, Soria JC, Rosell R, et al. Cross-validation study of class III beta-tubulin as a predictive marker for benefit from adjuvant chemotherapy in resected non-small-cell lung cancer: analysis of four randomized trials. Ann Oncol. 2012;23:86–93.
Lebok P, Ozturk M, Heilenkotter U, Jaenicke F, Muller V, Paluchowski P, et al. High levels of class III beta-tubulin expression are associated with aggressive tumor features in breast cancer. Oncol Lett. 2016;11:1987–94.
Hinsch A, Chaker A, Burdelski C, Koop C, Tsourlakis MC, Steurer S, et al. βIII-tubulin overexpression is linked to aggressive tumor features and genetic instability in urinary bladder cancer. Hum Pathol. 2017;61:210–20.
Sobierajska K, Wieczorek K, Ciszewski WM, Sacewicz-Hofman I, Wawro ME, Wiktorska M, et al. β-III tubulin modulates the behavior of Snail overexpressed during the epithelial-to-mesenchymal transition in colon cancer cells. Biochim Biophys Acta. 2016;1863:2221–33.
Du J, Li B, Fang Y, Liu Y, Wang Y, Li J, et al. Overexpression of class III beta-tubulin, Sox2, and nuclear survivin is predictive of taxane resistance in patients with stage III ovarian epithelial cancer. BMC Cancer. 2015;15:536.
Gan PP, Pasquier E, Kavallaris M. Class III beta-tubulin mediates sensitivity to chemotherapeutic drugs in non small cell lung cancer. Cancer Res. 2007;67:9356–63.
Mantovani A, Barajon I, Garlanda C. IL-1 and IL-1 regulatory pathways in cancer progression and therapy. Immunol Rev. 2018;281:57–61.
Steel JL, Terhorst L, Collins KP, Geller DA, Vodovotz Y, Kim J, et al. Prospective analyses of cytokine mediation of sleep and survival in the context of advanced cancer. Psychosom Med. 2018;80:483–91.
Li YJ, Wei ZM, Meng YX, Ji XR. Beta-catenin up-regulates the expression of cyclinD1, c-myc and MMP-7 in human pancreatic cancer: relationships with carcinogenesis and metastasis. World J Gastroenterol. 2005;11:2117–23.
Gelfo V, Mazzeschi M, Grilli G, Lindzen M, Santi S, D’Uva G, et al. A novel role for the interleukin-1 receptor axis in resistance to anti-EGFR therapy. Cancers. 2018;10:E355.
Weng CH, Chen LY, Lin YC, Shih JY, Lin YC, Tseng RY, et al. Epithelial-mesenchymal transition (EMT) beyond EGFR mutations per se is a common mechanism for acquired resistance to EGFR TKI. Oncogene. 2019;38:455–68.
Sun X, Shi X, Liu M, Li D, Zhang L, Liu X, et al. Mdp3 is a novel microtubule-binding protein that regulates microtubule assembly and stability. Cell Cycle. 2011;10:3929–37.
Apte RN, Krelin Y, Song X, Dotan S, Recih E, Elkabets M, et al. Effects of micro-environment- and malignant cell-derived interleukin-1 in carcinogenesis, tumour invasiveness and tumour-host interactions. Eur J Cancer. 2006;42:751–9.
Burke SJ, Stadler K, Lu D, Gleason E, Han A, Donohoe DR, et al. IL-1beta reciprocally regulates chemokine and insulin secretion in pancreatic beta-cells via NF-kappaB. Am J Physiol Endocrinol Metab. 2015;309:E715–726.
Wang D, Qiao J, Zhao X, Chen T, Guan D. Thymoquinone Inhibits IL-1beta-Induced Inflammation in human osteoarthritis chondrocytes by suppressing NF-kappaB and MAPKs signaling pathway. Inflammation. 2015;38:2235–41.
Pierce JW, Schoenleber R, Jesmok G, Best J, Moore SA, Collins T, et al. Novel inhibitors of cytokine-induced IkappaBalpha phosphorylation and endothelial cell adhesion molecule expression show anti-inflammatory effects in vivo. J Biol Chem. 1997;272:21096–103.
Yao Z, Fenoglio S, Gao DC, Camiolo M, Stiles B, Lindsted T, et al. TGF-beta IL-6 axis mediates selective and adaptive mechanisms of resistance to molecular targeted therapy in lung cancer. Proc Natl Acad Sci USA. 2010;107:15535–40.
Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci USA. 2004;101:13306–11.
Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. NEJM. 2004;350:2129–39.
Suda K, Murakami I, Yu H, Kim J, Tan AC, Mizuuchi H, et al. CD44 facilitates epithelial-to-mesenchymal transition phenotypic change at acquisition of resistance to EGFR kinase inhibitors in lung cancer. Mol Cancer Ther. 2018;17:2257–65.
Yuan TL, Cantley LC. PI3K pathway alterations in cancer: variations on a theme. Oncogene. 2008;27:5497–510.
Amann J, Kalyankrishna S, Massion PP, Ohm JE, Girard L, Shigematsu H, et al. Aberrant epidermal growth factor receptor signaling and enhanced sensitivity to EGFR inhibitors in lung cancer. Cancer Res. 2005;65:226–35.
Engelman JA, Janne PA, Mermel C, Pearlberg J, Mukohara T, Fleet C, et al. ErbB-3 mediates phosphoinositide 3-kinase activity in gefitinib-sensitive non-small cell lung cancer cell lines. Proc Natl Acad Sci USA. 2005;102:3788–93.
Ono M, Hirata A, Kometani T, Miyagawa M, Ueda S, Kinoshita H, et al. Sensitivity to gefitinib (Iressa, ZD1839) in non-small cell lung cancer cell lines correlates with dependence on the epidermal growth factor (EGF) receptor/extracellular signal-regulated kinase 1/2 and EGF receptor/Akt pathway for proliferation. Mol Cancer Ther. 2004;3:465–72.
Jacobsen K, Bertran-Alamillo J, Molina MA, Teixido C, Karachaliou N, Pedersen MH, et al. Convergent Akt activation drives acquired EGFR inhibitor resistance in lung cancer. Nat Commun. 2017;8:410.
Manning BD, Toker A. AKT/PKB signaling: navigating the network. Cell. 2017;169:381–405.
Kim SB, Dent R, Im SA, Espie M, Blau S, Tan AR, et al. Ipatasertib plus paclitaxel versus placebo plus paclitaxel as first-line therapy for metastatic triple-negative breast cancer (LOTUS): a multicentre, randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Oncol. 2017;18:1360–72.
Edgar KA, Wallin JJ, Berry M, Lee LB, Prior WW, Sampath D, et al. Isoform-specific phosphoinositide 3-kinase inhibitors exert distinct effects in solid tumors. Cancer Res. 2010;70:1164–72.
Hyman DM, Snyder AE, Carvajal RD, Gerecitano JF, Voss MH, Ho AL, et al. Parallel phase Ib studies of two schedules of buparlisib (BKM120) plus carboplatin and paclitaxel (q21 days or q28 days) for patients with advanced solid tumors. Cancer Chemother Pharm. 2015;75:747–55.
Juric D, Castel P, Griffith M, Griffith OL, Won HH, Ellis H, et al. Convergent loss of PTEN leads to clinical resistance to a PI(3)Kalpha inhibitor. Nature. 2015;518:240–4.
Mariani M, Zannoni GF, Sioletic S, Sieber S, Martino C, Martinelli E, et al. Gender influences the class III and V beta-tubulin ability to predict poor outcome in colorectal cancer. Clin Cancer Res. 2012;18:2964–75.
Duran GE, Wang YC, Moisan F, Francisco EB, Sikic BI. Decreased levels of baseline and drug-induced tubulin polymerisation are hallmarks of resistance to taxanes in ovarian cancer cells and are associated with epithelial-to-mesenchymal transition. Br J Cancer. 2017;116:1318–28.
Powell S, Kaizer A, Koopmeiners JS, Iwamoto C, Klein M. High expression of class III beta-tubulin in small cell lung carcinoma. Oncol Lett. 2014;7:405–10.
Riihimaki M, Hemminki A, Fallah M, Thomsen H, Sundquist K, Sundquist J, et al. Metastatic sites and survival in lung cancer. Lung Cancer. 2014;86:78–84.
McCarroll JA, Gan PP, Erlich RB, Liu M, Dwarte T, Sagnella SS, et al. TUBB3/betaIII-tubulin acts through the PTEN/AKT signaling axis to promote tumorigenesis and anoikis resistance in non-small cell lung cancer. Cancer Res. 2015;75:415–25.
Parker AL, Turner N, McCarroll JA, Kavallaris M. betaIII-Tubulin alters glucose metabolism and stress response signaling to promote cell survival and proliferation in glucose-starved non-small cell lung cancer cells. Carcinogenesis. 2016;37:787–98.
Jordan MA, Wilson L. Microtubules as a target for anticancer drugs. Nat Rev Cancer. 2004;4:253–65.
Kavallaris M. Microtubules and resistance to tubulin-binding agents. Nat Rev Cancer. 2010;10:194–204.
Liu BP, Chrzanowska-Wodnicka M, Burridge K. Microtubule depolymerization induces stress fibers, focal adhesions, and DNA synthesis via the GTP-binding protein Rho. Cell Adhes Commun. 1998;5:249–55.
Enomoto T. Microtubule disruption induces the formation of actin stress fibers and focal adhesions in cultured cells: possible involvement of the rho signal cascade. Cell Struct Funct. 1996;21:317–26.
Nakaya Y, Sukowati EW, Wu Y, Sheng G. RhoA and microtubule dynamics control cell-basement membrane interaction in EMT during gastrulation. Nat Cell Biol. 2008;10:765–75.
Solinas G, Marchesi F, Garlanda C, Mantovani A, Allavena P. Inflammation-mediated promotion of invasion and metastasis. Cancer Metastasis Rev. 2010;29:243–8.
Nickoloff BJ, Ben-Neriah Y, Pikarsky E. Inflammation and cancer: is the link as simple as we think? J Investig Dermatol. 2005;124:x–xiv.
Fernando RI, Hamilton DH, Dominguez C, David JM, McCampbell KK, Palena C. IL-8 signaling is involved in resistance of lung carcinoma cells to erlotinib. Oncotarget. 2016;7:42031–44.
Kim SM, Kwon OJ, Hong YK, Kim JH, Solca F, Ha SJ, et al. Activation of IL-6R/JAK1/STAT3 signaling induces de novo resistance to irreversible EGFR inhibitors in non-small cell lung cancer with T790M resistance mutation. Mol Cancer Ther. 2012;11:2254–64.
Franco-Barraza J, Valdivia-Silva JE, Zamudio-Meza H, Castillo A, Garcia-Zepeda EA, Benitez-Bribiesca L, et al. Actin cytoskeleton participation in the onset of IL-1beta induction of an invasive mesenchymal-like phenotype in epithelial MCF-7 cells. Arch Med Res. 2010;41:170–81.
Chalmers J, Jean A, Tardy F, Allatif O, Grosjean I, Blanquier B et al. Assessing mycoplasma contamination of cell cultures by qPCR using a set of universal primer pairs targeting a 1.5 kb fragment of 16S rRNA genes. Plos One 2017; 12: e0172358.
Jackman D, Pao W, Riely GJ, Engelman JA, Kris MG, Janne PA, et al. Clinical definition of acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancer. J Clin Oncol. 2010;28:357–60.
Funding
This study was supported in part by grants from the Certificate of China Postdoctoral Science Foundation Grant (2015M581477, 2017M621307), the Hei Long Jiang Postdoctoral Foundation (LBH-Z15145, LBH-Z17182), the Natural Science and Technology Foundation of Heilongjiang Province (LC2016037), the Fundamental Research Funds for the Provincial Universities, the Hai Yan Youth Fund and Top-Notch Youth Fund from Harbin Medical University Cancer Hospital (JJQN2018-11, JJQN2018-02 and BJQN2019-07), and the NSFC (Grant Nos. 81772474, 81572276, 81803023 and 81602717).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Huang, J., Lan, X., Wang, T. et al. Targeting the IL-1β/EHD1/TUBB3 axis overcomes resistance to EGFR-TKI in NSCLC. Oncogene 39, 1739–1755 (2020). https://doi.org/10.1038/s41388-019-1099-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-019-1099-5
- Springer Nature Limited
This article is cited by
-
Wnt/β-catenin signaling in the development and therapeutic resistance of non-small cell lung cancer
Journal of Translational Medicine (2024)
-
Targeting cytokine and chemokine signaling pathways for cancer therapy
Signal Transduction and Targeted Therapy (2024)
-
Deciphering the molecular mechanistic paths describing the chemotherapeutic potential and epigenetic regulation of curcumin in lung cancer: a mini review
Naunyn-Schmiedeberg's Archives of Pharmacology (2024)
-
CircZNF215 promotes tumor growth and metastasis through inactivation of the PTEN/AKT pathway in intrahepatic cholangiocarcinoma
Journal of Experimental & Clinical Cancer Research (2023)
-
EHD1-dependent traffic of IGF-1 receptor to the cell surface is essential for Ewing sarcoma tumorigenesis and metastasis
Communications Biology (2023)