Abstract
Chronic myeloid leukemia (CML) originates from normal hematopoietic stem cells acquiring BCR-ABL fusion gene, specific BCR-ABL inhibitors (e.g., imatinib mesylate, IM) have greatly improved patient management. However, some patients are still suffering from relapse and drug resistance, which urges better understanding of the growth/survival mechanisms of CML stem/progenitor cells. In the present study, the role and its underlying mechanism of heterogeneous nuclear ribonucleoprotein D-like (HNRPDL) in CML cells were investigated. Firstly, overexpression of HNRPDL promoted the growth of murine BaF3 cells in vitro and induced leukemia in vivo, which was enhanced by co-expression of BCR-ABL. Conversely, HNRPDL silencing inhibited colony-forming cell (CFC) production of CML CD34+ cells and attenuated BCR-ABL induced leukemia. In addition, HNRPDL modulated imatinib response of K562 cells and HNRPDL silencing sensitized CML CD34+ cells to imatinib treatment. Mechanistically, we found the stability of pre-B-cell leukemia homeobox 1 (PBX1) mRNA was sustained by HNRPDL through its binding to a specific motif (ACUAGC) in 3′-untranslated region (3′-UTR) of PBX1. The expression of PBX1 was significantly higher in CML CD34+ cells than that in control cells and PBX silencing inhibited the growth of CML cells and sensitized them to imatinib treatment. In contrast, overexpression of PBX1 elevated the CFC production of normal hematopoietic CD34+ cells and “rescued” HNRPDL silencing induced growth inhibition and imatinib sensitization. Taken together, our data have demonstrated that HNRPDL transforms hematopoietic cells and a novel HNRPDL/PBX1 axis plays an important role in human CML CD34+ cells.
Similar content being viewed by others
Introduction
Heterogeneous nuclear ribonucleoproteins (hnRNPs) represent a large RNA-binding protein family regulating many aspects of mRNA metabolism, including translational control, stability/decay of mRNA and alternative splicing [1,2,3]. Heterogeneous nuclear ribonucleoprotein D-like (HNRPDL) is a poorly studied member in this family. The cDNA of HNRPDL was isolated by two independent groups in 1998 [4, 5]. HNRPDL was present in both nucleus and cytoplasm and the C-terminus was responsible for its shuttling between nucleus and cytoplasm [6, 7]. HNRPDL bound to poly(A)+ RNAs rather than poly(A)− RNAs through a specific motif (ACUAGC) [8], and it bound to a single-stranded C-rich telomeric motif with limited specificity as well [9]. HNRPDL increased the internal translation initiation and maintained the stability of NF-κB-repressing factor (NRF) mRNA through its binding to 5′- and 3′-untranslated terminal region (UTR) of NRF [10, 11]. In addition, HNRPDL played a role in transcription regulation through its interaction with enhancers or other transcriptional factors [5, 12].
Heterozygous mutations of HNRPDL were identified in patients with limb girdle muscular dystrophy type 1G, and hnrpdl silencing in zebrafish caused the impairment of muscle development [13]. 4q21 microdeletion syndrome emerged as a newly identified disease with marked growth restriction, mental retardation, speech absence or delay; one of the affected genes was HNRPDL [14, 15]. The aberrant expression of HNRPDL was reported in various cancers as well, including hepatocellular carcinoma, prostate cancer, colorectal cancer, chronic myeloid leukemia (CML), and melanoma [16,17,18,19,20,21]. Overexpression of HNRPDL resulted in androgen independent growth of LNCaP cells (prostate cancer) and malignant transformation of NIH-3T3 cells [17, 19]; while silence of HNRPDL led to growth inhibition of SW620 cells (colorectal cancer) and K562 cells (CML) [19, 20]. HNRPDL was possibly involved in drug response of cancer cells as well [22,23,24]. For instance, quercetin inhibited the growth of Caco-2 cells (colon cancer) associated with the reduced HNRPDL expression [22].
CML is a hematological malignancy originated from normal hematopoietic stem cell acquiring BCR-ABL fusion gene. Despite the great improvement of patient management with tyrosine kinase inhibitors (e.g., IM), some patients are still suffering from drug resistance and relapse [25, 26]. The fact that the survival of CML stem/progenitor cells are not totally dependent on BCR-ABL signaling urges better understanding of the growth/survival mechanisms of these cells [27, 28]. Although we have reported the aberrant expression of HNRPDL in CML CD34+ cells [20], the role of HNRPDL in CML cells and its underlying mechanism remain unclear.
In the present study, we investigated whether HNRPDL was able to transform hematopoietic cells and how HNRPDL regulated the growth and IM response of human CML cells. The results have demonstrated that HNRPDL is an oncoprotein in hematological malignancies and identified a novel HNRPDL/pre-B-cell leukemia homeobox 1 (PBX1) axis playing an important role in modulation of the growth and IM response of CML cells.
Results
HNRPDL transforms murine BaF3 cells in vivo
To study the effect of HNRPDL overexpression on hematopoietic cells, the full-length HNRPDL was cloned into a retroviral vector [19]. The overexpression of HNRPDL in BaF3 cells was validated with western blot at first (Fig. 1a). In BaF3 cell, the shorter isoform of HNRPDL has much stronger expression than the larger one, while previous study showed that these two isoforms were similarly expressed in K562 cells [20]. The overexpression of HNRPDL enhanced the growth of these cells significantly in vitro (Fig. 1a). Similar results were obtained with K562 cells as well (Fig. S1). HNRPDL overexpressed BaF3 cells were more sensitive to mIL-3 treatment than control cells (Fig. 1b). In addition, these cells induced lethal leukemia in mice but not the control cells (Fig. 1c). The leukemic mice had significantly enlarged spleen and liver (Fig. S2b), and apparent impairment of spleen tissues was observed with hematoxylin-eosin (H&E) staining (Fig. S2c). In addition, FACS analysis showed that leukemic cells were present in bone marrow, spleen, and liver (Fig. S2d).
HNRPDL enables malignant transformation of BaF3 cells. a The growth of control and BaF3/HNRPDL cells was measured (n = 3), and the western blot to analyze these cells was shown. b Colony-forming cell (CFC) production of control and BaF3/HNRPDL cells were assessed in the presence of various concentrations of m-IL3 (n = 3). c Control and BaF3/HNRPDL cells were injected into lethally irradiated mice through tail vein (1.5 × 106 cells/mouse), each group had eight mice. The survival of these mice was analyzed with Kaplan–Meier method. d, e The growth and CFC production of control, BaF3/HNRPDL, BaF3/BCR-ABL, and BaF3/BCR-ABL + HNRPDL cells were measured in the presence of mIL-3 (n = 3). f BaF3/BCR-ABL and BaF3/BCR-ABL + HNRPDL cells were injected into lethally irradiated mice through tail vein (3.0 × 104 cells/mouse), there were eight mice in each group. Kaplan–Meier method was used to analyze the survival of these mice. *P < 0.05, **P < 0.01, and ***P < 0.001
Next, we asked whether HNRPDL and BCR-ABL collaboratively promoted the in vitro growth and malignant transformation in vivo. HNRPDL and BCR-ABL co-expressed BaF3 cells grew significantly faster than those expressed a single transgene (Fig. 1d), accordingly co-expressed cells generated more colony-forming cells (CFCs) than those expressed either HNRPDL or BCR-ABL alone (Fig. 1e). Lastly, BaF3/BCR-ABL cells and BaF3/BCR-ABL+HNRPDL cells were injected into mice, and the co-expressed cells induced a more aggressive disease than those expressing BCR-ABL alone (Figs. 1f and S3). These data suggested that the aberrant expression of HNRPDL in chronic phase (CP) might facilitate the malignant transformation of BCR-ABL. In addition, HNRPDL possibly played a role in the disease progression. Thus, the gene expression of HNRPDL was analyzed in CD34+ cells from 15 patients in CP and 10 patients in blast crisis (BC), and the data showed that HNRPDL expression was significantly higher in BC than that in CP (Fig. S4), which was also agreeable with previous in silico analysis of microarray data in Radich’s study [20, 29].
HNRPDL silencing inhibits in vitro and in vivo growth of CML cells
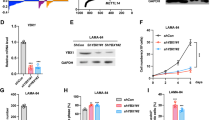
Transcript and protein analyses showed clearly that HNRPDL had higher expression in K562 cells than normal bone marrow (NBM) cells (Fig. S5), and then two independent shRNA sequences (shRNA#1 and shRNA#2) were validated to suppress HNRPDL expression in K562 cells (Fig. S6). The silence of HNRPDL inhibited the growth and CFC production of K562 cells significantly (Fig. 2a, b). A “rescue” experiment was also conducted to show that HNRPDL overexpression was able to reverse HNRPDL silencing induced growth inhibition (Fig. S7). Importantly, HNRPDL silencing inhibited CFC production of CD34+ cells from CML patients significantly (n = 4, Fig. 2c). In addition, HNRPDL silencing promoted erythroid differentiation of K562 cells; upon the treatment of hemin, more differentiated cells were observed in HNRPDL silenced group compared to control group (Fig. S8).
HNRPDL silencing inhibits the growth of BCR-ABL+ CML cells. a, b Two independent shRNA sequences were delivered into K562 cells, and then the growth and colony-forming cell (CFC) capacities of HNRPDL silenced and control cells were compared (n = 4). c One shRNA sequence (shRNA#2) was also delivered into CD34+ cells from CML patients (n = 4), and then the CFC productions of HNRPDL silenced and control cells were compared. d The control and HNRPDL silenced (shRNA#1) BaF3/BCR-ABL cells (1 × 105/mouse) were injected into lethally irradiated mice, each group had eight mice. Then the survival of these mice was analyzed with Kaplan–Meier method. e The coefficient of spleen of control and HNRPDL silenced group was compared (n = 4). f FACS analysis was perform to detect the leukemic cells (YFP+GFP+) in bone marrow and spleen of the diseased mice from each group and then compared (n = 4). shRNA#1 was suitable for HNRPDL silencing in both K562 and BaF3/BCR-ABL cells. *P < 0.05, **P < 0.01, and ***P < 0.001
Using BaF3/BCR-ABL cells as model, the inhibitory effect of HNRPDL silencing in vitro was confirmed with two independent shRNA sequences (shRNA#1 and shRNA#3) at first (Fig. S9). It was of note that shRNA#1 was suitable for HNRPDL silencing in both human and murine cells. HNRPDL silencing significantly delayed BCR-ABL induced leukemia (Fig. 2d). The coefficient of spleen in HNRPDL silenced group was significantly less than that in BCR-ABL group (Fig. 2e), the coefficient of liver was also decreased though not reaching statistical significance (Fig. S10). In addition, there were significantly less leukemic cells (GFP+YFP+) from both bone marrow and spleen in HNRPDL silenced group than those in BCR-ABL group (Fig. 2f). The representative FACS profiles and western blot analysis of leukemic cells from HNRPDL silenced and control groups were shown (Fig. S10). There was one survivor in HNRPDL silenced group without evident symptoms till 60 days post injection, which was killed then. FACS analysis showed that there were no detectable leukemic cells in bone marrow, spleen, and liver (Fig. S11).
HNRPDL modulates imatinib response of CML cells
The responses of control (Scramble) and HNRPDL silenced K562 cells upon imatinib treatment were studied. The data showed that HNRPDL silencing significantly increased imatinib sensitivity. In addition, more apoptotic cells were induced upon imatinib treatment in HNRPDL silencing group than those in control group (Fig. 3a), the representative FACS profiles to analyze apoptosis were shown (Fig. S12). In contrast, HNRPDL overexpression conferred imatinib resistance to K562 cells; at the same time, less apoptotic cells were induced upon imatinib treatment in HNRPDL overexpression group than those in control group (Figs. 3b and S13).
HNRPDL modulates imatinib mesylate response of CML cells. a Scramble and HNRPDL silenced K562 cells were plated for colony-forming cell (CFC) assay in the presence of imatinib mesylate (IM) (n = 3, left panel), and the apoptosis of these cells induced by IM treatment were analyzed with Annexin V/7-AAD method (n = 3, right panel). b Control and HNRPDL overexpressed K562 cells were analyzed with CFC assay in the presence of IM (n = 3, left panel), and apoptosis of these cells induced by IM treatment were shown (n = 4, right panel). c CD34+ cells from three CML patients were used for CFC analysis upon the treatment of IM or HNRPDL silencing alone or in combination. *P < 0.05 and **P < 0.01
Next, the effect of HNRPDL silencing on the imatinib response of CD34+ cells from patients was studied. The HNRPDL silencing and control CD34+ cells from three CML patients were subjected to CFC assay with or without imatinib. The data showed that the CFC production was decreased about 30% with imatinib treatment (5 μM) alone, suggesting these cells were imatinib insensitive according to Jiang’s study [30]. Imatinib treatment plus HNRPDL silencing produced significantly less CFC than imatinib treatment or HNRPDL silencing alone (Fig. 3c), indicating HNRPDL silencing enhanced chemo-response of imatinib insensitive CD34+ cells in vitro.
HNRPDL regulates the expression of PBX1
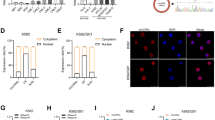
To obtain molecular insights of how HNRPDL modulates the growth and imatinib response of human CML cells, we generated microarray data comparing HNRPDL silenced K562 cells with control (Scramble) cells (GSE132975). Totally, 335 differentially expressed transcripts were yielded (Table S1) and they were displayed in a heatmap as well (Fig. 4a). Several known oncogenes were among the downregulated transcripts, including Myeloid ecotropic viral integration 2, PBX1 and Muscle RAS oncogene homolog, whose involvements of leukemic cell growth have been reported [31,32,33,34,35]. Using Q-RT-PCR, we were able to validate that the expression of these transcripts was significantly lower in HNRPDL silenced K562 cells than that in control cells (n = 4, Fig. 4b, left panel). The expression of PBX1 was also significantly decreased in HNRPDL silenced CML CD34+ cells than that in control cells (n = 5, Fig. 4b, right panel). Western blot showed that the protein expression of PBX1 was decreased upon HNRPDL silencing in K562 cells as well (Fig. 4c). Importantly, the expression of PBX1 in CML CD34+ cells (n = 8) was significantly higher (threefold) than that in NBM CD34+ cells (n = 7), suggesting a possible role of PBX1 in CML cells (Fig. 4d).
Microarray data reveal that HNRPDL regulates the expression of PBX1. a three independent HNRPDL silenced (shRNA#2) and control (Scramble) K562 cells were yielded for RNA preparation and microarray analysis. Totally, 335 differentially expressed transcripts were obtained, and they were displayed in a heatmap. b The expression levels of Myeloid ecotropic viral integration 2 (MEIS2), Pre-B-cell leukemia homeobox 1 (PBX1) and Muscle RAS oncogene homolog (MRAS) were measured in both HNRPDL silenced and control K562 cells (n = 4, left panel), and the expression of PBX1 was also measured with HNRPDL silenced and control CML CD34+ cells (n = 5, right panel). c The protein expression of PBX1 in HNRPDL silenced (shRNA#2) and control (Scramble) K562 cells was analyzed. d The PBX1 transcript was assessed with purified CD34+ cells from healthy donors (n = 7) and CML patients (n = 8) as well. *P < 0.05 and **P < 0.01
PBX1 is a novel regulator of CML cells
Two independent shRNA sequences were delivered with lentiviral vectors into K562 cells. Both of them suppressed PBX1 transcript expression significantly and PBX1 protein expression was inhibited as well (Fig. 5a). PBX1 silencing inhibited the growth and the CFC production of K562 cells significantly (Fig. 5b, c). In addition, the silence of PBX1 sensitized K562 cells to imatinib treatment (Fig. 5d). Importantly, the CFC production of CML CD34+ cells were inhibited by PBX1 silencing significantly as well (Fig. 5e). Conversely, PBX1 overexpression significantly promoted CFC production of normal hematopoietic CD34+ cells than control, and the significant promotion was also observed in secondary plating (Fig. 5f).
PBX1 plays an important role in the growth and imatinib response of CML cells. a Two independent shRNA sequences against PBX1 were delivered into K562 cells. The transcript of PBX1 were quantified (n = 4), and the protein expression was analyzed with western blot. b, c The growth and colony-forming cell (CFC) capacities of PBX1 silenced and control (Scramble) cells were compared (n = 4). d The percentages of survival CFC of PBX1 silenced and control K562 cells upon imatinib mesylate (IM) treatment were compared (n = 3). e One shRNA sequence (shPBX1#1) was also delivered into CD34+ cells from CML patients (n = 7), and then the CFC productions of PBX1 silenced and control cells were compared. f Lentiviral vector was used to deliver PBX1 into normal hematopoietic CD34+ cells. Then CFC assay was performed (first CFC), after counting the progeny cells were collected and plated for secondary CFC assay (second CFC). The total CFC yields normalized to 1000 initial input cells were calculated and compared (n = 3). *P < 0.05 and **P < 0.01
PBX1 is a functional target of HNRPDL
As HNRPDL regulates gene expression in both transcriptional and posttranscriptional fashions [5, 10,11,12], the expression of pre-mRNA of PBX1 was measured. The data showed that pre-mRNA of PBX1 did not change upon HNRPDL silencing (Fig. 6a). Next, HNRPDL silenced and control K562 cells were treated with actinomycin D (Act D), the relative expression of PBX1 compared with untreated cells was measured, which indicated that HNRPDL silencing increased the degradation rate of PBX1 mRNA (Fig. 6b).
HNRPDL/PBX1 axis plays a crucial role in the growth and imatinib response of human CML cells. a The expression of pre-mRNA of PBX1 in HNRPDL silenced and control K562 cells was assessed (n = 5). b The expression of mRNA of PBX1 in HNRPDL silenced and control K562 cells upon actinomycin D (Act D) treatment was measured (n = 5). c The schematic structure of 3′-untranslated region (3′-UTR) of PBX1 was plotted. Only one HNRPDL binding motif (ACUAGC) was identified, and a piece of DNA containing this motif and its mutant was subcloned and subjected to luciferase activity measurement. A pair of primers was designed to detect 3′-UTR of PBX1 in RNA immunoprecipitation (RIP) assay (upper panel). A control reporter vector (Ctrl), a reporter vector containing HNRPDL consensus binding motif (wide-type) and another reporter vector bearing mutated motif (mutant) were delivered into HNRPDL silenced (shRNA#2) and control (Scramble) K562 cells, and the reporter activities were quantified (n = 4, middle panel). The RIP assay was performed with K562 cells and a piece of PBX1 3′-UTR was detected with conventional RT-PCR (lower panel). d–f The growth (n = 3), CFC capacities (n = 6) and IM sensitivity (n = 4) of various transduced K562 cells were measured and compared. n.s. not significant; *P < 0.05 and **P < 0.01
A consensus RNA-binding motif (ACUAGC) recognized by HNRPDL was found in the 3′-UTR of PBX1. A piece of DNA sequence containing this motif and its mutant were subcloned into a luciferase reporter vector (Fig. 6c, upper panel). Firstly, the reporter assay showed that the wide-type motif conferred greater reporter activity (fourfold) compared with the empty vector, however this enhanced activity was almost totally abrogated when the mutant motif was introduced into the reporter vector. Next, the activity of the wide-type motif was significantly decreased upon HNRPDL silencing (Fig. 6c, middle panel). Lastly, RNA immunoprecipitation (RIP) experiment was performed with K562 cells, which showed that antibody against HNRPDL was able to enrich PBX1 mRNA compared with the isotype control antibody (Fig. 6c, lower panel).
Functionally, the overexpression of PBX1 was able to “rescue” HNRPDL silencing introduced growth inhibition and CFC reduction (Figs. 6d, e and S14). In addition, the sensitized imatinib response caused by HNRPDL silencing was restored by PBX1 overexpression as well (Fig. 6f).
Discussion
The aberrant expression of multiple RNA-binding proteins, including several hnRNP family members in CML cells have been reported [36,37,38,39]. Thus, it is reasonable to speculate whether HNRPDL has redundant function with some of them. In the present study, gene silencing experiments with various cellular models both in vitro and in vivo clearly show that HNRPDL plays a unique role in CML cells. Indeed the sequence similarity analysis by Akindahunsi et al. has shown that HNRPDL is a remote member isolated from hnRNP A1, K, E2, and Musashi 2 [40], supporting the experimental data in this study. Despite numerous reports pertaining hnRNP members in various cancer cells, few of them have clearly demonstrated that hnRNP member is able to transform normal cells [19, 41]. In the present study, we have provided the first piece of evidence that HNRPDL confers cytokine hypersensitivity of BaF3 cells in vitro and induces leukemia in vivo. In addition, HNRPDL and BCR-ABL cooperatively promote cell growth and induce leukemia. As the expression of HNRPDL is elevated in CD34+ cells from patients in BC than that in CP, our data suggest a possible role of HNRPDL in disease progression. Interestingly, Deneault et al. have reported that HNRPDL elevates the reconstitution of primitive hematopoietic cells, supporting the notion that HNRPDL promotes the growth of hematopoietic cells [42].
The fusion protein of E2A-PBX1 plays a pivotal role in a subset of human B-cell acute lymphoblastic leukemia patients [43,44,45]. However, the expression and function of PBX1 in myeloid leukemia cells has not been investigated directly [33, 34]. Our study has shown that PBX1 expression in CML CD34+ cells is significantly higher than that in NBM CD34+ cells, and PBX1 is required for the in vitro growth of CML cells. In addition, PBX1 overexpression promotes the CFC production of normal CD34+ cells (Fig. 5). These data are in line with the fact that Pbx1 deficiency impairs mice hematopoiesis including myelopoiesis in vivo and in vitro [46]. Thus our data have revealed a previously neglected role of PBX1 in myeloid leukemia cells.
HNRPDL is also known as an AU-rich element (ARE)-binding protein. Through the binding with ARE, HNRPDL has been reported to regulate the stability of NRF [10, 11]. At the same time, a motif (ACUAGC) recognized by HNRPDL has been identified through in vitro screening [8]; however, whether this motif has in vivo function has not been elucidated yet. In this study, we found that HNRPDL silencing inhibited PBX1 expression and Act D treatment caused a more rapid decay of PBX1 mRNA in HNRPDL silenced cells than control cells, suggesting HNRPDL possibly modulated the stability of PBX1. Sequence analysis showed there was no typical ARE in the 3′-UTR of PBX1 mRNA, while a consensus motif of “ACUAGC” was present. Thus luciferase assay and RIP experiment were performed, which showed that HNRPDL controlled the mRNA lifespan through the binding with “ACUAGC” motifs for the first time. Furthermore, HNRPDL silencing induced growth inhibition and IM hypersensitivity was “rescued” by PBX1 overexpression, suggesting a pivotal role of HNRPDL/PBX1 axis in human CML cells. Nevertheless, it is worth to note that “ACUAGC” motif is not present in murine Pbx1, thus how HNRPDL regulates murine hematopoietic cells needs further investigation.
Overall, the present study will stimulate more investigations on this newly defined oncoprotein. For instance, both ARE and “ACUAGC” motifs are recognized by HNRPDL, thus a globe sequencing of RIP enriched RNA is likely to provide more clues to delineate the function of HNRPDL. Previous studies have shown that HNRPDL may regulate gene expression through its interaction with other transcriptional factors or enhancers [5, 12]; thus it is interesting to study whether and how HNRPDL regulate gene expression at transcription level using the microarray data generated in current study.
Taken together, we have demonstrated that HNRPDL promotes the growth of CML cells and modulates their drug response partially through its regulation of PBX1 mRNA stability. The present study sheds new light on the molecular pathogenesis of CML and possibly provides new clues to improve disease management.
Materials and methods
Patients and cells
K562 and 293T cells were purchased from the Cell Bank of Shanghai Institutes for Biological Sciences of the Chinese Academy of Sciences (Shanghai, China). Murine BaF3 cell line was a gift from Dr Connie J. Eaves (Terry Fox Laboratory, British Columbia Cancer Research Center). An authentication test confirmed the identity of K562 cells and all these cell lines were free of mycoplasma contamination. The bone marrow cells from CML patients and healthy donors were collected, after the informed consent forms approved by the Ethics Committee of Soochow University (ECSU-201800062, Suzhou, China) were obtained. The clinical characteristics of CML patients were summarized (Table S2). Following gradient centrifuge with Lympholyte-H cell separation media (Cedarlane Laboratories, Burlington, NC, USA), the nucleated cells were yielded and then purified with the CD34 EasySep kit (Stem Cell Technologies, Vancouver, Canada) following the manufacturer’s instruction.
RNA extraction and Q-RT-PCR
RNA was obtained with the RNAprep Pure Micro kit (Tiangen, Beijing, China) and reversely transcribed with SuperScriptIII (Thermo Fisher Scientific, Waltham, MA, USA). Gene expression was assessed with SYBR Green PCR MasterMix with 7500 real time PCR system (Applied Biosystems, Foster City, CA, USA). The sequences of gene-specific primers were summarized (Table S3). In all Q-RT-PCR analyses, the gene expression was normalized against β-ACTIN.
Western blot
Protein samples were prepared with lysis buffer (Beyotime, Shanghai, China) supplemented with PMSF, and then transferred to the ImmobilonTM PVDF membrane (MilliporeSigma, Burlington, MA, USA) after SDS-PAGE separation (Bio-Rad, Hercules, CA, USA). The blots were performed with various antibodies, including anti-HNRPDL (ab83215, Abcam, Cambridge, MA, USA), anti-PBX1 (ab104247, Abcam) and anti-Tubulin (ab009-100, Multi Science, Hongzhou, China). The blot was developed using an enhanced chemiluminescence kit (GE Healthcare Life Sciences, Piscataway, NJ, USA) automatically.
Retroviral and lentiviral vectors, viral production, and infection
A retroviral vector (pMSCV-IRES-YFP, pMIY) was a gift from Dr Keith Humphries (Terry Fox Laboratory, British Columbia Cancer Research Center) and the packaging plasmid to produce retrovirus was from Dr Hudan Liu (Wuhan University, Wuhan, China). HNRPDL cDNA (full-length, NM_031372.3) was purchased (Origene, Rockville, MD, USA), and then subcloned into pMIY. The lentiviral vector to overexpress BCR-ABL was a gift from Dr Connie J. Eaves (Terry Fox Laboratory). PBX1 cDNA was a gift from Dr Xinliang Mao (Soochow University, Suzhou, China), and which was subcloned into a lentiviral vector containing yellow fluorescent protein (YFP). Lentiviral vectors to silence HNRPDL or PBX1 and the scramble control were from GenePharma Co, Ltd (Shanghai, China), the sequences were summarized (Table S4). The retroviral and lentiviral productions were performed as previously described [19, 20].
Colony-forming cell assay
The FACS purified primary cells were plated into methylcellulose media (MethoCult H4230, Stem Cell Technologies) supplemented with a cocktail of cytokines (SCF (50 ng/ml), IL3 (20 ng/ml), IL-6 (20 ng/ml), GM-CSF (20 ng/ml), G-CSF (20 ng/ml), and EPO (3 IU/ml)), and the colonies were numerated 14–16 days later.
Animals
Six- to eight-week-old female BalB/C mice were maintained in specific pathogen free animal facility of Soochow University. They were randomly allocated to each experimental group. The test and control cells were mixed with 7.5 × 105 mice bone marrow cells, and then injected intravenously into mice a few hours after lethal irradiation. Mice were monitored closely for signs of weight loss or lethargy. The diseased mice were dissected and the weights of spleens and livers were measured. The spleens were also analyzed with H&E staining. The cells from spleen, liver, and bone marrow were analyzed with flow cytometry. There was no exclusion of experimental mouse. All animal studies were blinded and conducted following an experimental protocol approved by the Ethics Committee of Soochow University (ECSU-201800061, Suzhou, China).
Apoptosis assay
Cell apoptosis was assessed with a kit (Cat#559763, BD Biosciences, Franklin Lakes, NJ, USA) following the manufacturer’s instruction. Briefly, aliquots of 1 × 105 cells were washed twice with PBS, and then resuspended with 100 μl 1 × binding buffer. Cells were incubated with Annexin V-PE (5 μl) and 7-AAD (5 μl) at room temperature in the dark. After 15 min, 400 μl 1 × binding buffer were added and then the cells were analyzed with flow cytometry.
RNA immunoprecipitation
The RIP experiment was performed with the EZ-mgana RIP kit (MilliporeSigma). In brief, 2 × 107 K562 cells were resuspended with 100 μl complete RIP lysis buffer. Anti-HNRPDL antibody and the IgG control antibody were mixed with magnetic beads, respectively. Later the RIP lysate and the antibody coated magnetic beads were incubated with RIP immunoprecipitation buffer at 4 °C overnight. RNA was purified with protease K treatment and phenol/chloroform extraction. The purified RNA samples were analyzed with conventional reverse transcription-polymerase chain reaction (RT-PCR) using specific primers to detect 3′-UTR of PBX1. The sequences of these primers were summarized (Table S3).
Luciferase assay
A piece of 3′-UTR of PBX1 containing “ACUAGC” motif was subcloned into psiCHECK-2 (Promega, Madison, WI, USA). A mutant of this piece of 3′-UTR was generated with point mutation technique. Similar to previous description [47], the control, wide-type 3′-UTR and its mutant were transferred into control and HNRPDL silenced K562 cells with Nucleofector device (Lonza, Basel, Switzerland). After 48 h, cells were collected and the reporter activities were assessed using Dual-Luciferase Reporter Assay System (Promega) with Luminoskan Ascent reader (Thermo Scientific, Waltham, MA, USA). After normalizing the activity of Renilla luciferase reporter to that of Firefly luciferase reporter, the relative activities of 3′-UTR and its mutant were calculated and compared.
Microarray analysis
Three biological replicates of control and HNRPDL silenced K562 cells were harvested for microarray analysis using Agilent whole human genome oligo-chips (4 × 180K) in Shanghai Biotechnology Corporation. The gene expression data have been assigned an accession ID as GSE132975. The differentially expressed transcripts were determined based on Student’s t-test (P < 0.05) and fold change (>2). All differentially expressed transcripts were clustered by Hierarchical Clustering method.
Statistical analysis
All values were represented as the mean ± SD from more than three biological independent experiments, and the statistical analysis was performed with Student’s t-test, in which a P value < 0.05 was considered significantly different. Kaplan–Meier curves were plotted to study survival tendency, and the P value was estimated using the log-rank test. The statistical tests in this study are justified as appropriate and the data meet the assumptions of the tests.
References
Dreyfuss G, Matunis MJ, Piñol-Roma S, Burd CG. hnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem. 1993;62:289–321.
Geuens T, Bouhy D, Timmerman V. The hnRNP family: insights into their role in health and disease. Hum Genet. 2016;135:851–67.
Wang Y, Xiao X, Zhang J, Choudhury R, Robertson A, Li K, et al. A complex network of factors with overlapping affinities represses splicing through intronic elements. Nat Struct Mol Biol. 2013;20:36–45.
Doi A, Shiosaka T, Takaoka Y, Yanagisawa K, Fujita S. Molecular cloning of the cDNA encoding A+ U-rich element RNA binding factor. Biochim Biophys Acta. 1998;1396:51–6.
Tsuchiya N, Kamei D, Takano A, Matsui T, Yamada M. Cloning and characterization of a cDNA encoding a novel heterogeneous nuclear ribonucleoprotein-like protein and its expression in myeloid leukemia cells. J Biochem. 1998;123:499–507.
Kawamura H, Tomozoe Y, Akagi T, Kamei D, Ochiai M, Yamada M. Identification of the nucleocytoplasmic shuttling sequence of heterogeneous nuclear ribonucleoprotein D-like protein JKTBP and its interaction with mRNA. J Biol Chem. 2002;277:2732–9.
Imasaki T, Shimizu T, Hashimoto H, Hidaka Y, Kose S, Imamoto N, et al. Structural basis for substrate recognition and dissociation by human transportin 1. Mol Cell. 2007;28:57–67.
Kamei D, Yamada M. Interactions of heterogeneous nuclear ribonucleoprotein D-like protein JKTBP and its domains with high-affinity binding sites. Gene. 2002;298:49–57.
Bandiera A, Tell G, Marsich E, Scaloni A, Pocsfalvi G, Akintunde Akindahunsi A, et al. Cytosine-block telomeric type DNA-binding activity of hnRNP proteins from human cell lines. Arch Biochem Biophys. 2003;409:305–14.
Reboll MR, Oumard A, Gazdag AC, Renger I, Ritter B, Schwarzer M, et al. NRF IRES activity is mediated by RNA binding protein JKTBP1 and a 14-nt RNA element. RNA. 2007;13:1328–40.
Omnus DJ, Mehrtens S, Ritter B, Resch K, Yamada M, Frank R, et al. JKTBP1 is involved in stabilization and IRES-dependent translation of NRF mRNAs by binding to 5′ and 3′ untranslated regions. J Mol Biol. 2011;407:492–504.
Boopathi E, Lenka N, Prabu SK, Fang JK, Wilkinson F, Atchison M, et al. Regulation of murine cytochrome c oxidase Vb gene expression during myogenesis: YY-1 and heterogeneous nuclear ribonucleoprotein D-like protein (JKTBP1) reciprocally regulate transcription activity by physical interaction with the BERF-1/ZBP-89 factor. J Biol Chem. 2004;279:35242–54.
Vieira NM, Naslavsky MS, Licinio L, Kok F, Schlesinger D, Vainzof M, et al. A defect in the RNA-processing protein HNRPDL causes limb-girdle muscular dystrophy 1G (LGMD1G). Hum Mol Genet. 2014;23:4103–10.
Bonnet C, Andrieux J, Béri-Dexheimer M, Leheup B, Boute O, Manouvrier S, et al. Microdeletion at chromosome 4q21 defines a new emerging syndrome with marked growth restriction, mental retardation and absent or severely delayed speech. J Med Genet. 2010;47:377–84.
Hu X, Chen X, Wu B, Soler IM, Chen S, Shen Y. Further defining the critical genes for the 4q21 microdeletion disorder. Am J Med Genet A. 2017;173:120–5.
Kurokawa Y, Matoba R, Takemasa I, Nakamori S, Tsujie M, Nagano H, et al. Molecular features of non-B, non-C hepatocellular carcinoma: a PCR-array gene expression profiling study. J Hepatol. 2003;39:1004–12.
Wu YY, Li H, Lv XY, Wei Q, Li X, Liu XY, et al. Overexpression of JKTBP1 induces androgen-independent LNCaP cell proliferation through activation of epidermal growth factor-receptor (EGF-R). Cell Biochem Funct. 2008;26:467–77.
de Wit M, Kant H, Piersma SR, Pham TV, Mongera S, van Berkel MP, et al. Colorectal cancer candidate biomarkers identified by tissue secretome proteome profiling. J Proteom. 2014;99:26–39.
Zhang P, Ji D, Hu X, Ni H, Ma W, Zhang X, et al. Oncogenic heterogeneous nuclear ribonucleoprotein D-like promotes the growth of human colon cancer SW620 cells via its regulation of cell-cycle. Acta Biochim Biophys Sin. 2018;50:880–7.
Zhou H, Ge Y, Sun L, Ma W, Wu J, Zhang X, et al. Growth arrest specific 2 is up-regulated in chronic myeloid leukemia cells and required for their growth. PLoS ONE. 2014;9:e86195.
Al-Ghoul M, Brück TB, Lauer-Fields JL, Asirvatham VS, Zapata C, Kerr RG, et al. Comparative proteomic analysis of matched primary and metastatic melanoma cell lines. J Proteome Res. 2008;7:4107–18.
van Erk MJ, Roepman P, van der Lende TR, Stierum RH, Aarts JM, van Bladeren PJ, et al. Integrated assessment by multiple gene expression analysis of quercetin bioactivity on anticancer-related mechanisms in colon cancer cells in vitro. Eur J Nutr. 2005;44:143–56.
Peng X, Gong F, Xie G, Zhao Y, Tang M, Yu L, et al. A proteomic investigation into adriamycin chemo-resistance of human leukemia K562 cells. Mol Cell Biochem. 2011;351:233–41.
Bertagnolo V, Grassilli S, Petretto A, Lambertini E, Astati L, Bruschi M, et al. Nuclear proteome analysis reveals a role of Vav1 in modulating RNA processing during maturation of tumoral promyelocytes. J Proteom. 2011;75:398–409.
Sherbenou DW, Druker BJ. Applying the discovery of the Philadelphia chromosome. J Clin Investig. 2007;117:2067–74.
Sloma I, Jiang X, Eaves AC, Eaves CJ. Insights into the stem cells of chronic myeloid leukemia. Leukemia. 2010;24:1823–33.
Corbin AS, Agarwal A, Loriaux M, Cortes J, Deininger MW, Druker BJ. Human chronic myeloid leukemia stem cells are insensitive to imatinib despite inhibition of BCR-ABL activity. J Clin Investig. 2011;121:396–409.
Hamilton A, Helgason GV, Schemionek M, Zhang B, Myssina S, Allan EK, et al. Chronic myeloid leukemia stem cells are not dependent on Bcr-Abl kinase activity for their survival. Blood. 2012;119:1501–10.
Radich JP, Dai H, Mao M, Oehler V, Schelter J, Druker B, et al. Gene expression changes associated with progression and response in chronic myeloid leukemia. Proc Natl Acad Sci USA. 2006;103:2794–9.
Jiang X, Forrest D, Nicolini F, Turhan A, Guilhot J, Yip C, et al. Properties of CD34+ CML stem/progenitor cells that correlate with different clinical responses to imatinib mesylate. Blood. 2010;116:2112–21.
Vegi NM, Klappacher J, Oswald F, Mulaw MA, Mandoli A, Thiel VN, et al. MEIS2 is an oncogenic partner in AML1-ETO-positive AML. Cell Rep. 2016;16:498–507.
Lai CK, Norddahl GL, Maetzig T, Rosten P, Lohr T, Sanchez Milde L, et al. Meis2 as a critical player in MN1-induced leukemia. Blood. Cancer J. 2017;7:e613.
Shimabe M, Goyama S, Watanabe-Okochi N, Yoshimi A, Ichikawa M, Imai Y, et al. Pbx1 is a downstream target of Evi-1 in hematopoietic stem/progenitors and leukemic cells. Oncogene. 2009;28:4364–74.
Xu X, Han K, Tang X, Zeng Y, Lin X, Zhao Y, et al. The ring finger protein RNF6 induces leukemia cell proliferation as a direct target of pre-B-cell leukemia homeobox 1. J Biol Chem. 2016;291:9617–28.
Guo X, Stratton L, Schrader JW. Expression of activated M-Ras in hemopoietic stem cells initiates leukemogenic transformation, immortalization and preferential generation of mast cells. Oncogene. 2006;25:4241–4.
Perrotti D, Bonatti S, Trotta R, Martinez R, Skorski T, Salomoni P, et al. TLS/FUS, a pro-oncogene involved in multiple chromosomal translocations, is a novel regulator of BCR/ABL-mediated leukemogenesis. EMBO J. 1998;17:4442–55.
Trotta R, Vignudelli T, Candini O, Intine RV, Pecorari L, Guerzoni C, et al. BCR/ABL activates mdm2 mRNA translation via the La antigen. Cancer Cell. 2003;3:145–60.
Perrotti D, Neviani P. From mRNA metabolism to cancer therapy: chronic myelogenous leukemia shows the way. Clin Cancer Res. 2007;13:1638–42.
Ito T, Kwon HY, Zimdahl B, Congdon KL, Blum J, Lento WE, et al. Regulation of myeloid leukaemia by the cell-fate determinant Musashi. Nature. 2010;466:765–8.
Akindahunsi AA, Bandiera A, Manzini G. Vertebrate 2xRBD hnRNP proteins: a comparative analysis of genome, mRNA and protein sequences. Comput Biol Chem. 2005;29:13–23.
Golan-Gerstl R, Cohen M, Shilo A, Suh SS, Bakàcs A, Coppola L, et al. Splicing factor hnRNP A2/B1 regulates tumor suppressor gene splicing and is an oncogenic driver in glioblastoma. Cancer Res. 2011;71:4464–72.
Deneault E, Cellot S, Faubert A, Laverdure JP, Fréchette M, Chagraoui J, et al. A functional screen to identify novel effectors of hematopoietic stem cell activity. Cell. 2009;137:369–79.
Hunger SP, Galili N, Carroll AJ, Crist WM, Link MP, Cleary ML. The t(1;19)(q23; p13) results in consistent fusion of E2A and PBX1 coding sequences in acute lymphoblastic leukemias. Blood. 1991;77:687–93.
Kamps MP, Look AT, Baltimore D. The human t(1;19) translocation in pre-B ALL produces multiple nuclear E2A-Pbx1 fusion proteins with differing transforming potentials. Genes Dev. 1991;5:358–68.
Dedera DA, Waller EK, LeBrun DP, Sen-Majumdar A, Stevens ME, Barsh GS, et al. Chimeric homeobox gene E2A-PBX1 induces proliferation, apoptosis, and malignant lymphomas in transgenic mice. Cell. 1993;74:833–43.
Ficara F, Murphy MJ, Lin M, Cleary ML. Pbx1 regulates self-renewal of long-term hematopoietic stem cells by maintaining their quiescence. Cell Stem Cell. 2008;2:484–96.
Zhang X, Ma W, Cui J, Yao H, Zhou H, Ge Y, et al. Regulation of p21 by TWIST2 contributes to its tumor-suppressor function in human acute myeloid leukemia. Oncogene. 2015;34:3000–10.
Acknowledgements
This work was supported by National Natural Science Foundation of China (Nos. 31771579, 31371392 to YZ, 81400113 to HZ, 81500119 to XZ, and 81800151 to WM), the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), and the Innovation Capability Development Project of Jiangsu Province (No. BM2015004).
Author information
Authors and Affiliations
Contributions
DJ, PZ, and WM performed most experimental work and PZ initiated the experimental study. YF, WX, YW, and XZ provided critical technical supports. HZ and YZ conceived the project and designed the study. H.Z. also supervised the quality of the clinical samples. YZ, HZ, DJ, PZ, and WM wrote the manuscript. All authors have read and approved this manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Ji, D., Zhang, P., Ma, W. et al. Oncogenic heterogeneous nuclear ribonucleoprotein D-like modulates the growth and imatinib response of human chronic myeloid leukemia CD34+ cells via pre-B-cell leukemia homeobox 1. Oncogene 39, 443–453 (2020). https://doi.org/10.1038/s41388-019-0998-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-019-0998-9
- Springer Nature Limited
This article is cited by
-
miR-181a plays the tumor-suppressor role in chronic myeloid leukemia CD34 + cells partially via SERPINE1
Cellular and Molecular Life Sciences (2024)
-
A conserved ZFX/WNT3 axis modulates the growth and imatinib response of chronic myeloid leukemia stem/progenitor cells
Cellular & Molecular Biology Letters (2023)
-
The splicing factor RBM17 drives leukemic stem cell maintenance by evading nonsense-mediated decay of pro-leukemic factors
Nature Communications (2022)