Abstract
The aim of this study was to explore the biological functions of a tetraspanin family protein CD82 expressed aberrantly in chemotherapy-resistant CD34+/CD38− acute myelogenous leukemia (AML) cells. Microarray analysis of patient-isolated CD34+/CD38− AML cells revealed that the levels of anti-apoptotic protein BCL2L12 were downregulated after CD82 depletion by specific short hairpin RNA (shRNA). Western blot analysis indicated that BCL2L12 was aberrantly expressed in patient-isolated AML cells and AML cell lines. Furthermore, CD82 blockade by a specific antibody downregulated BCL2L12 in parallel with dephosphorylation of signal transducer and activator of transcription 5 (STAT5) and AKT, whereas pharmacological inhibition of STAT5 and AKT activation decreased BCL2L12 expression in leukemia cells. In addition, shRNA-mediated downregulation of BCL2L12 increased the levels of cleaved caspase-3 and suppressed proliferation of leukemia cells, impairing their engraftment in immunodeficient mice. Taken together, our results indicate that CD82 regulated BCL2L12 expression via STAT5A and AKT signaling and stimulated proliferation and engrafting of leukemia cells, suggesting that CD82 and BCL2L12 may be promising therapeutic targets in AML.
Similar content being viewed by others
Introduction
Acute myelogenous leukemia (AML) is the most common form of acute leukemia affecting adults1 and is initiated and maintained by a subset of self-renewing leukemia stem cells.2 CD34+/CD38− AML cells have been shown to fulfill the criteria for leukemia stem cells in vivo,3, 4 and recent studies on severely immunocompromised mice have found that even CD34− or CD38+ AML cells were able to reconstitute AML.5, 6 CD34+/CD38− AML cells are refractory to conventional chemotherapeutic agents such as cytarabine and anthracyclines that interfere with DNA synthesis and induce apoptosis primarily in replicating cells.7
The BCL2 family of proteins comprises pro-apoptotic members BAX, BAD, BID and BCL-Xs and anti-apoptotic members BCL-2, BCL-xL and BCL-W.8 Bcl2-like protein 12 (BCL2L12) is a newly identified member of the BCL2 family encoded by the BCL2L12 gene located on chromosome 19p13.3 between the IRF3 and PRMT1/HRMT1L2 genes and close to the RRAS oncogene;9, 10 it has 13 distinct transcripts resulting from alterative gene splicing.11 BCL2L12 contains a highly conserved BH2 domain, a BH3-like motif and a proline-rich region, and its classical BCL2L12 isoform 1 is expressed in various tissues including the breast, thymus, prostate, fetal liver, colon, placenta, pancreas, small intestine, spinal cord, kidney and bone marrow.12, 13
Cytoplasmic BCL2L12 has been shown to block post-mitochondrial apoptosis in glioblastoma14 via inhibition of caspase-3/7 activation, conferring apoptotic resistance and a tendency to necrosis in glial cells.15 BCL2L12 does not affect cytochrome c release or apoptosome-driven caspase-9 activation, but instead inhibits post-mitochondrial apoptosis signaling in glioblastoma at the level of effector caspase activation.16 Nuclear BCL2L12 interacts with the p53 tumor suppressor protein and inhibits p53-dependent apoptosis. Thus, BCL2L12 is a multifunctional protein that blocks apoptosis and induces necrosis via interaction with cytoplasmic and nuclear apoptotic pathways in glioblastoma cells.14, 15, 16, 17
Notably, the neural cell adhesion molecule CD56 activates nuclear translocation of nuclear factor-κB that increases levels of BCL2L12 in AML cells. Interestingly, high levels of CD56 associates with poor prognosis of AML.18 Moreover, BCL2L12 overexpression is associated with poor disease outcome in AML patients.19 High levels of BCL2L12 mRNA correlate with advanced clinical stage and shorter overall survival in patients with solid tumors and hematological malignancies, including chronic myeloid leukemia and chronic lymphocytic leukemia.19, 20, 21, 22, 23 These observations suggested that BCL2L12 may be chemotherapy response marker and chemotherapy target in various types of malignancies. However, the role of BCL2L12 in the maintenance of leukemia stem cells and AML resistance to conventional treatment has not been studied.
We recently compared protein expression profiles of CD34+/CD38− and CD34+/CD38+ cells freshly isolated from AML patients using isobaric tags for relative and absolute quantitation (iTRAQ) and detected aberrant CD82 expression in CD34+/CD38− AML cells.24 CD82 was found to negatively regulate matrix metalloproteinase 9 and promote the adherence of these cells to fibronectin in bone marrow microenvironment. Downregulation of CD82 has been shown to inhibit colony-forming ability of CD34+/CD38−AML cells, suggesting that CD82 supports CD34+/CD38− cell survival.24 However, the mechanism underlying CD82 pro-survival activity in AML cells remains obscure.
Here, we performed a functional and mechanistic analysis of the role of CD82 in AML cells and severely immunocompromised mice to elucidate its biological functions.
Materials and methods
Sample collection and isolation of CD34+/CD38− and CD34+/CD38+ AML cells
Each study participant provided informed written consent, and the study was approved by the Kochi University Institutional Review Board (Nankoku, Japan). Leukemia cells were isolated from the patients with AML (n=28, Table 1) classified according to the World Health Organization classification system as minimally differentiated AML (case 3), AML without maturation (cases 1, 7, 13 and 17), AML with maturation (cases 2, 11, 14, 18, 24, 27 and 28), acute myelomonocytic leukemia (cases 4, 12 and 25), acute monocytic leukemia (cases 15, 16 and 21), AML with myelodysplasia changes (cases 5, 6, 8, 9, 19 and 23) and therapy-related AML (cases 10, 20, 22 and 26). CD34+/CD38− and CD34+/CD38+ cells were purified by magnetic cell sorting using the CD34 MultiSort kit and CD38 MicroBead kit (Miltenyi Biotec GmbH, Bergisch Gladbach, Germany), as previously described.24
Isolation of CD34+/CD38−/CD82− and CD34+/CD38−/CD82+ AML cells and RT-PCR
Leukemia cells were stained with fluorescent-labeled antibodies fluorescein isothiocyanate-anti-CD34 (IM1870; Beckman Coulter, Brea, CA, USA), allophycocyanin-anti-CD38 (cat. no. 303509; BioLegend, San Diego, CA, USA) and phycoerythrin-anti CD82 (cat. no. 342104; BioLegend) and separated by cell sorting using a FACS Aria II instrument (BD Biosciences, Heidelberg, Germany). RNA was extracted using the CellAmp Direct RNA Prep Kit for reverse transcriptase-PCR (RT-PCR; Takara, Shiga, Japan) and reverse transcription was performed using the One Step PrimeScript RT-PCR Kit (Takara). Amplification was conducted using a StepOne plus system (Life Technology, CA, USA) at the following conditions: 42 °C for 5 min, 95 °C for 10 s, and 40 cycles at 95 °C for 5 s, 55 °C for 30 s and 72 °C for 1 min. The 18S gene was used as internal control. PCR primers are listed in Table 2.
Cell culture
Acute monocytic leukemia cell line MOLM13 carrying an internal tandem duplication (ITD) of the juxtamembrane domain of FLT3 (FLT3/ITD) was kindly provided by Dr Yoshinobu Matsuo (Fujisaki Cell Center, Okayama, Japan).25 Kasumi-1 cells carrying Asn822Lys c-Kit mutation were a kind gift from Dr Asou (Hiroshima University, Hiroshima, Japan). Leukemia cell lines HL60, NB4, U937 and THP-1 and FLT3/ITD-expressing MV4-11 were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA). Cell viability was determined after Trypan blue staining by light microscopy.
Pharmacological inhibition
The phosphatidylinositol 3-kinase (PI3K) inhibitor LY294002 was purchased from LC Laboratories (Woburn, MA, USA), and the Janus-associated kinase 2 inhibitor AZ960 was synthesized by AstraZeneca R&D (Osaka, Japan).26 MOLM13 and Kasumi-1 cells were treated with LY294002 (1, 5 and 10 μM) or AZ960 (1, 2 and 5 μM) for 48 h before the analysis of protein expression by western blotting.
Microarray hybridization
Total RNA was extracted using the single-step Trizol RNA extraction kit (Invitrogen, Carlsbad, CA, USA) according to the protocol, concentrated by isopropanol precipitation and column-purified using the QIAGEN RNeasy Mini Kit (QIAGEN, Venlo, Holland). RNA concentration was determined using a Nanodrop ND-1000 spectrophotometer (Thermo Scientific, Waltham, MA, USA) and RNA quality was evaluated using the Agilent 2100 Bioanalyzer expert software (Agilent Technologies, Santa Clara, CA, USA). RNA samples with the RNA integrity number ⩾6.0 were considered of a sufficient quality for gene expression profiling.
Cy3 and Cy5 dyes were used to label control and experimental complementary DNA (250 ng) according to the protocol of Miltenyi Biotec. Cy3- and Cy5-labeled complementary DNA was combined and hybridized for 17 h at 65 °C to the Agilent Whole Human Genome Oligo Microarrays 8 × 60K using a hybridization chamber and oven (Agilent Technologies). The microarrays were washed once with the Agilent Gene Expression Wash Buffer 1 for 1 min at room temperature followed by a second wash with preheated (37 °C) Agilent Gene Expression Wash Buffer 2 for 1 min. The last washing step was performed with acetonitrile. After vigorous washing, the hybridized microarrays were scanned using an Agilent DNA microarray scanner (Agilent Technologies). The resulting images were analyzed using the Rosetta Resolver gene expression data analysis system (Rosetta Biosoftware, Kirkland, WA, USA) (National Center for Biotechnology Information (NCBI); GSE64527).
Western blotting
Total cell proteins or nuclear protein were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (30 μg of total cell protein per lane) and western blot analysis was performed as described previously24 using the primary antibodies against phospho(p)-STAT5, p-AKT, AKT, caspase-3, cleaved caspase-3, poly ADP-ribose polymerase (Cell Signaling Technology, Beverly, MA, USA), STAT5, CD82, BAX, p53 (Santa Cruz Biotechnology, Santa Cruz, CA, USA), BCL2L12 and GAPDH (Abcam, Cambridge, UK). Band intensities were quantified using the ImageJ software (Wayne Rasband, NIH, Bethesda, MD, USA).
Flow cytometry analysis
The level of CD82 in leukemia cells was assessed by flow cytometry using phycoerythrin-anti CD82 (BioLegend).
RNA isolation and real-time RT-PCR
Total RNA was extracted from leukemia cells and reverse transcribed according to the manufacturer’s instructions (Takara). Real-time RT-PCR was performed using Power SYBR Green PCR Master Mix (Applied Biosystems, Warrington, UK) in a StepOne plus system at the following conditions: 95 °C for 10 min and 40 cycles at 95 °C for 15 s and 60 °C for 1 min. The expression of the 18S gene was used for normalization. The primers are listed in Table 2.
Lentiviral and plasmid vectors and cell transfection
Short hairpin RNA (shRNA) specific to human BCL2L12 was designed according to the human BCL2L12 gene sequence (NCBI accession number NM_138639). Human CD82-specific shRNA was synthesized based on the human CD82 transcript variant 2 (NCBI accession number NM_001024844). The control, BCL2L12, and CD82 shRNA lentiviral vectors, designed to coexpress GFP, were purchased from GeneCopoeia (Rockville, MD, USA). Lentiviral shRNA particles were produced in 293FT packaging cells (Invitrogen) using the Lenti-Pac FIV Expression Packaging Kit (GeneCopoeia). Three types of CD82 shRNA-expressing lentivirus particles were mixed and used for cell transduction in serum-free medium. After overnight incubation, fresh 10% fetal bovine serum-supplemented medium containing 1 μg/ml puromycin was added for 7 days. CD82 cDNA was purchased from the Mammalian Gene Collection (BC000726, GE Healthcare, Little Chalfont, Buckinghamshire, UK) and was used as the template for PCR. STAT5A cDNA (NM_003152) was synthesized by TAKARA BIO Inc. (Shiga, Japan). Gene products were cloned into the pLenti6.3/V5-TOPO vector (Invitrogen). The product was cloned into the pLenti6.3/V5-TOPO vector (Invitrogen). Lentiviral particles were produced using the viral power packaging system (Invitrogen) and transduced into MOLM13 cells as previously described.24 After 72 h, blasticidin (10 μg/ml; Invitrogen) was added to select for stably transduced cells. MOLM13 cells were transiently transfected with the Gag-AKT expression vector27 using the Fugene transfection reagent (Promega, Madison, WI, USA).
CD82 antibody binding
The binding of human anti-CD82 monoclonal antibody (53H5) (Santa Cruz Biotechnology) to cell surface of leukemia cells was examined by microscopy (OLYMPUS FV1000-D, Tokyo, Japan). The phycoerythrin-conjugate secondary antibody was obtained from BioLegend.
Apoptosis assay
Cellular apoptosis was assessed by the release of cytochrome c from mitochondria into cytosol using the Cytochrome c Releasing Apoptosis Assay Kit (BioVision, Milpitas, CA, USA) according to the manufacturer’s instructions.
Colony-forming assay
The colony-forming assay was performed with methylcellulose medium H4034 (StemCell Technologies, Vancouver, BC, Canada), as previously described.24
Bone marrow transplantation and engraftment
NOD.Cg-Rag1tm1Mom Il2rgtm1Wjl/SzJ mice (Stock Number, 007799) were purchased from the Jackson Laboratory (Bar Harbor, ME, USA)24 and bred in a pathogen-free environment in accordance with the guidelines of the Kochi University School of Medicine; 6-week-old female and male animals were utilized for the experiments. Human MOLM13 cells (5 × 106) transfected with control shRNA (n=14) or BCL2L12-specific shRNA1 (n=14) were injected via the tail vein and cell engraftment was analyzed using flow cytometry after staining of peripheral blood monoclonal cells with human CD82 phycoerythrin-conjugated monoclonal antibody (BioLegend).
Statistical analysis
All statistical analyses were performed using the statistical software GraphPad PRISM (GraphPad Software Inc., San Diego, CA, USA). Differences were analyzed by Student’s t-test and were considered significant at a P-value of <0.05 and highly significant at a P-value of <0.01. Mouse survival was calculated using the Kaplan–Meier method, and survival curves were compared by log-rank test.
Results
Gene expression profiles in CD34+/CD38− AML cells expressing CD82-specific shRNA
To explore the function of CD82 in CD34+/CD38− AML cells, CD34+/CD38− cells isolated from AML patients (n=3, cases 1–3) were transduced with CD82-specific shRNA and their gene expression profiles were compared with the cells transduced with control shRNA by microarray analysis. The threefold differences in more than two AML cases were considered as significant. Between CD82-depleted and control CD34+/CD38− AML cells, 995 genes were found differentially expressed in at least two AML samples (NCBI; GSE64527). Among them, anti-apoptotic BCL2 family genes BCL2L12 (NM_138639) and BCL2L2 (NM_004050) were the only genes downregulated in CD82-depleted CD34+/CD38− AML samples isolated from all three AML patients (Supplementary Table S1). We previously showed that CD34+/CD38− AML cells aberrantly expressed CD82.24 These observations suggested the possibility that BCL212 and BCL2L2 may be overexpressed by CD82 in leukemia cells. In addition, other researchers performed the comparison of mRNA expression between AML cells such as KG-1, U937 and THP-1 cells and normal monocytes isolated from healthy volunteers using microarray. They found that the expression of BCL2L12 but not BCL2L2 was significantly upregulated in AML cells (Table 3, GDS2251, http://www.ncbi.nlm.nih.gov/geoprofiles/29050864).28 Given that BCL2L12 overexpression was associated with poor outcome in AML,19 BCL2L12 may be an important gene involved in maintenance of survival of AML cells. We therefore investigated the relationship between CD82 and BCL2L12 in AML cells in this study.
CD82 regulates BCL2L12 expression in leukemia cells
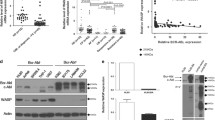
To validate the results of microarray (Supplementary Table S1), we performed real-time RT-PCR that showed that levels of BCL2L12 decreased by 0.3-fold in CD34+/CD38− AML cells after depletion of CD82 by half by shRNA (n=5, cases 4–7 and 19; Figure 1a and figure not shown),29 whereas forced expression of CD82 in CD34+/CD38+ AML cells by 10-fold (figure not shown)29 demonstrated a twofold increase in BCL2L12 (n=6, cases 1–3, 17, 19 and 20; Figure 1b). We also compared BCL2L12 mRNA levels in CD34+/CD38−/CD82+ and CD34+/CD38−/CD82− AML cells isolated from the patients (n=10, cases 1, 2, 6–9, 19, 22, 27 and 28). As expected, BCL2L12 expression was 10-fold higher in CD34+/CD38−/CD82+AML cells than their CD82-negative counterparts (Figure 1c). To explore the relationship between BCL2L12 and CD82, we used CD82-overexpressing MOLM13 and Kasumi-1 cells. Similarly, in CD82+ MOLM13 and Kasumi-1 cells, there was a twofold increase in BCL2L12 expression compared with CD82− cells (Figure 1d). These results suggested that CD82 might regulate BCL2L12 expression in leukemia cells.
The effect of CD82 on BCL2L12 expression in CD34+/CD38− AML cells. (a) CD34+/CD38− AML cells (n=5 patients) and (b) CD34+/CD38+ AML cells (n=6 patients) transduced with CD82 shRNA or CD82-expressing lentiviral particles were collected and analyzed for BCL2L12 mRNA expression by real-time RT-PCR. Each dot represents BCL2L12 level in each patient and the mean is indicated by the line. *P<0.05, **P<0.01. (c) CD34+/CD38−/CD82+ and CD34+/CD38−/CD82−AML cells (n=10 patients) were isolated by cell sorting and analyzed for BCL2L12 mRNA expression by real-time RT-PCR. (d) CD82+ and CD82− MOLM13 and Kasumi-1 cells were sorted and analyzed for BCL2L12 mRNA expression by real-time RT-PCR. *P<0.05.
CD82 and BCL2L12 expression in AML cells
We have previously shown that CD82 expression is upregulated in CD34+/CD38− AML cells (68±27%) compared with their CD34+/CD38+ counterparts (30±19%).24 Here, we found that BCL2L12 levels in CD34+/CD38− cells were higher than in CD34+/CD38+ cells (n=7, cases 1, 3, 8 and 23–26; Supplementary Figure S1A). A trend of positive correlation between CD82 and BCL2L12 mRNA levels was shown in CD34+/CD38− AML cells isolated from patients (r=0.73, Supplementary Figure S1B). Moreover, we compared CD82 and BCL2L12 expression in the cells isolated from AML patients (n=10, cases 10–19) and bone marrow mononuclear cells (BMMNCs) from healthy volunteers (n=4) by real-time RT-PCR. The levels of CD82 and BCL2L12 were 7757- and 13-fold higher, respectively, in AML cells than in BMMNCs (Figure 2a). The analysis of 10 AML patients showed a trend (r=0.36) of positive correlation between CD82 and BCL2L12 mRNA expression (Figure 2b). We next examined the protein levels of BCL2L12 in AML cells isolated from the patients (n=10, cases 10–19), BMMNCs isolated from a healthy volunteer (n=1) and AML cell lines by western blotting (Figure 2c). Consistent with mRNA data, BCL2L12 protein expression was higher in AML cells than in BMMNCs, although the levels varied among cell lines (Figure 2c). Similarly, flow cytometry analysis indicated that CD82 surface expression in AML cells was higher than in BMMNCs (Figure 2d). A trend of positive correlation between CD82 and BCL2L12 protein levels were shown in AML cells isolated from patients (r=0.3, Supplementary Figure S1C).
CD82 and BCL2L12 expression in AML cells. (a) CD82 and BCL2L12 mRNA levels in AML cells isolated from patients (n=10 patients) and BMMNCs isolated from healthy volunteers (n=4 patients). **P<0.01. (b) A scatter plot represents the correlation between CD82 and BCL2L12 mRNA expression in patients’ AML cells. (c, d) BMMNCs isolated from healthy volunteer (n=1), AML cells (n=10 patients), MOLM13, Kasumi-1, MV4-11, NB4, HL60, PL21, THP-1, U937 and MV4-11R248W cells were harvested and analyzed for BCL2L12 and BCL2 expression by western blotting and flow cytometry. Band intensities were quantified with ImageJ software (Wayne Rasband, NIH).
Molecular mechanisms underlying CD82 regulation of BCL2L12 expression in leukemia cells
We previously showed that CD82 regulated activation of signal transducer and activator of transcription 5 (STAT5) and AKT in AML cells.24 Thus, we examined whether CD82 modulated the expression of BCL2L12 via STAT5 or AKT signal pathway. The shRNA-mediated downregulation of CD82 in MOLM13 and Kasumi-1 cells attenuated BCL2L12 expression at both mRNA and protein levels and dephosphorylated both STAT5 and AKT as compared with control shRNA-transduced cells (Figures 3a and b). Moreover, the blockade of CD82 in these cells by a CD82-specific antibody decreased BCL2L12 expression at both the mRNA and protein levels and increased the expression of BAX and cleaved caspase-3 and phosphorylation of AKT and STAT5 (Figures 3c and d). Notably, forced expression of AKT or STAT5A increased BCL2L12 levels in MOLM13 cells and recovered BCL2L12 expression in the cells treated with CD82-specific shRNA (Figure 3e), confirming that CD82 regulated BCL2L12 levels via the AKT and STAT5 pathways. We next examined whether BCL2L12 expression would respond to pharmacological inhibition of AKT and STAT5 in leukemia cells and found that PI3K inhibitor LY294002 decreased both AKT phosphorylation and BCL2L12 expression (Figure 3f), whereas Janus-associated kinase 2 inhibitor AZ960 decreased STAT5 phosphorylation as well as BCL2L12 levels (Figure 3g). These results provide further support to our hypothesis that AKT and STAT5 signaling positively regulates BCL2L12 levels in leukemia cells.
The effect of CD82 on the expression of apoptotic proteins and phosphorylation of STAT5 and AKT in leukemia cell lines. (a, b) MOLM13 and Kasumi-1 cells transduced with CD82 shRNA were analyzed for BCL2L12 mRNA expression by real-time RT-PCR; *P<0.05, **P<0.01, (a) and for the expression of indicated proteins by western blotting (b). Band intensities were quantified using ImageJ software (Wayne Rasband, NIH). The number indicates the relative values of band intensity in CD82 shRNA-transduced cells compared with control shRNA-transduced cells. (c, d) MOLM13 and Kasumi-1 cells were treated with control IgG or CD82 antibody (0.5 and 1.0 μg/ml) for 96 h and analyzed for BCL2L12, BAX and cleaved caspase-3 mRNA levels by real time RT-PCR; *P<0.05, **P<0.01 (c) and for the expression of indicated proteins by western blotting (d). (e) MOLM13 cells expressing CD82-specific shRNA were transfected with AKT, STAT5A or vehicle for 72 h, harvested and analyzed for the expression of indicated proteins by western blotting. Each lane was loaded with 30 μg of total protein or nuclear protein. (f, g) MOLM13 and Kasumi-1 cells were treated with the inhibitor of PI3K/AKT signaling LY294002 or the inhibitor of Janus-associated kinase 2 (JAK2) kinase AZ960 for 48 h. The cells were harvested and analyzed for the expression of indicated proteins by western blotting.
The effect of BCL2L12 on apoptosis in leukemia cells
In Kasumi-1 and MOLM13 cells, BCL2L12-specific shRNAs decreased the expression of BCL2L12 and increased that of BAX and cleaved caspase-3 in parallel with the inhibition of cell survival, as evidenced by Trypan blue staining (Figures 4a–d). On the other hand, downregulation of BCL2L12 did not affect p53 expression and cytochrome c translocation from mitochondria into cytosol (Figures 4a–d).
BCL2L12 function in leukemia cells. (a) Kasumi-1 cells were transfected with control shRNA or BCL2L12-specific shRNA1 and shRNA2 and analyzed for the expression of indicated proteins by western blotting. (b) Kasumi-1 cells transfected with BCL2L12-specific shRNA1 and shRNA2 for 96 h were stained with Trypan blue, and the number of viable cells was counted under a light microscope. *P<0.05. (c) MOLM13 cells were transfected with control shRNA or BCL2L12-specific shRNA1 and analyzed for the expression of indicated proteins by western blotting. (d) MOLM13 cells transfected with control shRNA or BCL2L12-specific shRNA1 for 96 h were stained with Trypan blue, and the number of viable cells was counted under a light microscope. *P<0.05. (e) Colony-forming assay. CD34+/CD38− AML cells (n=3 patients) transduced with BCL2L12 shRNAs were cultured in methylcellulose medium for 16 days, and the colonies were counted. (f) CD34+/CD38+ AML cells (n=3 patients) transduced with BCL2L12 shRNA were collected and analyzed for BCL2L12 mRNA expression by real-time RT-PCR. Each dot represents BCL2L12 level in each patient and the mean is indicated by the line.
Then, we examined whether BCL2L12 regulated the survival of CD34+/CD38− AML cells. The results show that the shRNA-mediated downregulation of BCL2L12 in CD34+/CD38− AML cells decreased colony-forming ability of these cells by ∼50% compared with the control cells (n=3, cases 3, 19 and 21; Figures 4e and f).
BCL2L12 effects in vivo
We next explored BCL2L12 effects on the engraftment of AML cells in severely immunocompromised mice. MOLM13 cells (5 × 106) transduced with control shRNA or BCL2L12-specific shRNA1 were intravenously transplanted into NOD.Cg-Rag1tm1Mom Il2rgtm1Wjl/SzJ mice and their engraftment and animal survival were examined (Figure 5). At 3 weeks after transplantation, the expression of human CD82 in mouse peripheral blood monoclonal cells indicating the engraftment of MOLM13 cells was analyzed. The engraftment of MOLM13 cells with decreased BCL2L12 expression was significantly impaired compared with the control cells (BCL2L12 shRNA1, 38±12%; control shRNA, 59±16%; Figure 5a). In addition, Kaplan–Meier survival curves demonstrated that immunodeficient mice transplanted with BCL2L12-depleted MOLM13 cells survived longer than those transplanted with the control cells (P<0.01, Figure 5b).
The effect of BCL2L12 on the engraftment of leukemia cells in immunodeficient mice and mouse survival. MOLM13 cells transduced with control shRNA or BCL2L12-specific shRNA1 were transplanted into NOD.Cg-Rag1tm1Mom Il2rgtm1Wjl/SzJ mice via the tail vein (each group). (a) In 1–3 weeks after transplantation, peripheral blood was collected, incubated with CD82 antibodies and analyzed by flow cytometry. *P<0.05, **P<0.01. (b) Kaplan–Meier analysis of mouse survival (P<0.01).
Discussion
Microarray analysis and RT-PCR found that BCL2L12 was downregulated in CD82-depleted CD34+/CD38− AML samples isolated from AML patients (Supplementary Table S1 and Figure 1a). A trend of positive correlation between CD82 and BCL2L12 expression at mRNA and protein levels was noted in AML cells isolated from patients (Figure 2b and Supplementary Figure S1C), suggesting the possibility that CD82 might regulate expression of BCL2L12 in AML cells. In fact, downregulation of CD82 by specific shRNA or antibody attenuated BCL2L12 expression and reduced activation of STAT5 and AKT in AML cells, whereas overexpression of AKT or STAT5 restored BCL2L12 levels in CD82-deficient cells (Figure 3), suggesting that in leukemia cells, CD82 positively regulated BCL2L12 expression via AKT and STAT5 signaling (Figure 6).
Amounts of BCL2L12 mRNA and proteins were greater in AML cells than BMMNCs isolated from healthy volunteers (Figure 2). However, we were not able to compare levels of BCL2L12 between CD34+/CD38− AML cells and CD34+/CD38− BMMNCs isolated from healthy donors because of the paucity of the latter cells. Therefore, we could not reach the conclusion that levels of BCL2L12 were upregulated in AML cells compared with normal hematopoietic stem/progenitor cells.
We previously showed that CD82 activated AKT and STAT5 in leukemia cells.30 Long-term maintenance and expansion were impaired in STAT5-deficient leukemia stem cells and hematopoietic stem cells.31, 32 On the other hand, overexpression of STAT5 enhances the self-renewal potential of CD34+ cells.33 Similarly, activation of AKT by loss of PTEN (phosphatase and tensin homolog deleted on chromosome 10) induces self-renewal and expansion of hematopoietic stem cell via upregulation of anti-apoptotic Mcl-1.34 These observations suggested that activation of STAT5 and AKT is required for the self-renewal and expansion of primitive hematopoietic stem/progenitor as well as leukemia stem cells.
Downregulation of CD82 in CD34+/CD38− AML cells by an shRNA significantly impaired engraftment of these cells in severely immunocompromised mice.24 We have recently shown that CD82 negatively regulated matrix metalloproteinase 9 in AML cells, rendering these cells adherent to bone marrow microenvironment (Figure 6).24 In addition, CD82 activated STAT5 in association with an increase in production of IL-10 that decreased in the level of matrix metalloproteinase 9 in AML cells (Figure 6).24, 30 CD82 also stimulated expression of polycomb family protein EZH2 (enhancer of zeste homolog 2) via p38 mitogen-activated protein kinase pathway in AML cells, resulting in downregulation of PTEN in association with methylation of its promoter region (Figure 6).29 As upregulation of PTEN inactivates PI3K/AKT, CD82 may activate PI3K/AKT via downregulation of PTEN. Moreover, the use of CD82 monoclonal antibody in combination with cytarabine (AraC) significantly prolonged survival of immunodeficient mice bearing human AML cells.35 These results suggested that CD82 pathway may be biologically important target to treat patients with AML.
We found that the levels of CD82 mRNA and protein did not correlate in AML cells (n=10, r=0.2; Supplementary Figure S1d). One possible explanation might be related to the half-life of CD82 protein. Further studies are clearly required to verify the mechanisms by which expression of CD82 is regulated at the mRNA and protein levels.
Blockade of CD82 by the CD82 antibody did not increase p53 levels in leukemia cells (including FLT3/ITD-expressing AML cell line MV4-11 TP53 R248W (data not shown) that harbor a missense mutation CGG→TGG; R248W in the TP53 gene),36 suggesting that CD82 may induce apoptosis via a p53-independent pathway. Similarly, the downregulation of BCL2L12 did not affect p53 levels in MOLM13 cells, although it inhibited cell proliferation and increased the levels of pro-apoptotic factors such as cleaved caspase-3 and BAX. Although BCL2L12 inhibited apoptosis via the p53-dependent pathway in glioblastoma multiforme,16 our results demonstrate that in leukemia cells, BCL2L12 might exert pro-survival effects independently of p53 signaling. CD82/AKT and CD82/STAT5 signaling may inhibit apoptosis in association with the increase in BCL2L12 and decrease in BAX and cleaved caspase-3 levels, thus presenting a mechanism for chemotherapy resistance in AML.
Immunodeficient mice transplanted with BCL2L12-depleted MOLM13 cells demonstrated increased survival compared with those transplanted with control cells; however, they eventually died of leukemia, suggesting that BCL2L12 may partially mediate the survival of leukemia cells. We assume that not only BCL2L12 but also other BCL2 family members may play a role in supporting the survival of AML cells via the CD82 pathway.
BCL2L12 overexpression was associated with poor outcome in AML,19 although this study cohort was extremely small (n=28) and heterogeneous; five acute promyelocytic leukemia patients whose prognosis is favorable due to the treatment with all-trans retinoic acid37 were included in the study and all of them were negative for BCL2L12 expression.19 Intriguingly, high expression of BCL2L12 correlated with favorable outcome in gastric and colon cancer.13, 20 Further studies are clearly required to examine the association of BCL2L12 expression and prognosis of AML.
CD82 overexpression in AML cells enhances adhesion and regulate maintenance of AML cell within the bone marrow niche,24 but suppresses tumor invasion and metastasis and is associated with favorable prognosis in solid cancers such as lung, gastric, colon and cervix cancers.38, 39, 40, 41, 42 These findings suggest that high expression of CD82 and BCL2L12 in colon and gastric cancer may correlate with favorable prognosis, but in hematologic malignancies may be associated with poor outcome, indicating cancer type-specific biological functions of these molecules. Therefore, the manipulation of CD82 and BCL2L12 expression may constitute a therapeutic approach to AML treatment.
In conclusion, our results demonstrate that in leukemia cells, CD82 regulates BCL2L12 expression via STAT5 and AKT signaling, suggesting that BCL2L12 may be a promising therapeutic target to improve survival of AML patients.
References
Riva L, Luzi L, Pelicci PG . Genomics of acute myeloid leukemia: the next generation. Front Oncol 2012; 2: 40.
Bonnet D, Dick JE . Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med 1997; 3: 730–737.
Clarke MF, Dick JE, Dirks PB, Eaves CJ, Jamieson CH, Jones DL et al. Cancer stem cells-perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer Res 2006; 66: 9339–9344.
Ishikawa F, Yoshida S, Saito Y, Hijikata A, Kitamura H, Tanaka S et al. Chemotherapy-resistant human AML stem cells home to and engraft within the bone-marrow endosteal region. Nat Biotechnol 2007; 25: 1315–1321.
Taussig DC, Miraki-Moud F, Anjos-Afonso F, Pearce DJ, Allen K, Ridler C et al. Anti-CD38 antibody-mediated clearance of human repopulating cells masks the heterogeneity of leukemia-initiating cells. Blood 2008; 112: 568–575.
Sarry JE, Murphy K, Perry R, Sanchez PV, Secreto A, Keefer C et al. Human acute myelogenous leukemia stem cells are rare and heterogeneous when assayed in NOD/SCID/ IL2Rgc-deficient mice. J Clin Invest 2011; 121: 384–395.
Ikezoe T, Yang J, Nishioka C, Kojima S, Takeuchi A, Phillip Koeffler H et al. Inhibition of signal transducer and activator of transcription 5 by the inhibitor of janus kinases stimulates dormant human leukemia CD34+ /CD38- cells and sensitizes them to antileukemia agents. Int J Cancer 2011; 128: 2317–2325.
Adams JM, Cory S . The Bcl-2 protein family: arbiters of cell survival. Science 1998; 281: 1322–1326.
Floros KV, Thomadaki H, Florou D, Talieri M, Scorilas A . Alterations in mRNA expression of apoptosis-related genes BCL2, BAX, FAS, caspase-3, and the novel member BCL2L12 after treatment of human leukemic cell line HL60 with the antineoplastic agent etoposide. Ann NY Acad Sci 2006; 1090: 89–97.
Kitada S, Andersen J, Akar S, Zapata JM, Takayama S, Krajewski S et al. Expression of apoptosis-regulating proteins in chronic lymphocytic leukemia: correlations with In vitro and In vivo chemoresponses. Blood 1998; 91: 3379–3389.
Kontos CK, Scorilas A . Molecular cloning of novel alternatively spliced variants of BCL2L12, a new member of the BCL2 gene family, and their expression analysis in cancer cells. Gene 2012; 505: 153–166.
Scorilas A, Kyriakopoulou L, Yousef GM, Ashworth LK, Kwamie A, Diamandis EP . Molecular cloning, physical mapping, and expression analysis of a novel gene, BCL2L12, encoding a proline-rich protein with a highly conserved BH2 domain of the Bcl-2 family. Genomics 2001; 72: 217–221.
Kontos CK, Papadopoulos IN, Scorilas A . Quantitative expression analysis and prognostic significance of the novel apoptosis-related gene BCL2L12 in colon cancer. Biol Chem 2008; 389: 1467–1475.
Stegh AH, Brennan C, Mahoney JA, Forloney KL, Jenq HT, Luciano JP et al. Glioma oncoprotein Bcl2L12 inhibits the p53 tumor suppressor. Genes Dev 2010; 24: 2194–2204.
Stegh AH, Chin L, Louis DN, DePinho RA . What drives intense apoptosis resistance and propensity for necrosis in glioblastoma? A role for Bcl2L12 as a multifunctional cell death regulator. Cell Cycle 2008; 7: 2833–2839.
Stegh AH, Kim H, Bachoo RM, Forloney KL, Zhang J, Schulze H et al. Bcl2L12 inhibits post-mitochondrial apoptosis signaling in glioblastoma. Genes Dev 2007; 21: 98–111.
Stegh AH, Kesari S, Mahoney JE, Jenq HT, Forloney KL, Protopopov A et al. Bcl2L12-mediated inhibition of effector caspase-3 and caspase-7 via distinct mechanisms in glioblastoma. Proc Natl Acad Sci USA 2008; 105: 10703–10708.
Gattenloehner S, Chuvpilo S, Langebrake C, Reinhardt D, Müller-Hermelink HK, Serfling E et al. Novel RUNX1 isoforms determine the fate of acute myeloid leukemia cells by controlling CD56 expression. Blood 2007; 110: 2027–2033.
Thomadaki H, Floros KV, Pavlovic S, Tosic N, Gourgiotis D, Colovic M et al. Overexpression of the novel member of the BCL2 gene family, BCL2L12, is associated with the disease outcome in patients with acute myeloid leukemia. Clin Biochem 2012; 45: 1362–1367.
Florou D, Papadopoulos IN, Scorilas A . Molecular analysis and prognostic impact of the novel apoptotic gene BCL2L12 in gastric cancer. Biochem Biophys Res Commun 2010; 391: 214–218.
Mathioudaki K, Scorilas A, Papadokostopoulou A, Xynopoulos D, Arnogianaki N, Agnanti N et al. Expression analysis of BCL2L12, a new member of apoptosis-related genes, in colon cancer. Biol Chem 2004; 385: 779–783.
Karan-Djurasevic T, Palibrk V, Zukic B, Spasovski V, Glumac I, Colovic M et al. Expression of Bcl2L12 in chronic lymphocytic leukemia patients: association with clinical and molecular prognostic markers. Med Oncol 2013; 30: 405.
Papageorgiou SG, Kontos CK, Pappa V, Thomadaki H, Kontsioti F, Dervenoulas J et al. The novel member of the BCL2 gene family, BCL2L12, is substantially elevated in chronic lymphocytic leukemia patients, supporting its value as a significant biomarker. Oncologist 2011; 16: 1280–1291.
Nishioka C, Ikezoe T, Furihata M, Yang J, Serada S, Naka T et al. CD34+/CD38− acute myelogenous leukemia cells aberrantly express CD82 which regulates adhesion and survival of leukemia stem cells. Int J Cancer 2013; 132: 2006–2019.
Matsuo Y, MacLeod RA, Uphoff CC, Drexler HG, Nishizaki C, Katayama Y et al. Two acute monocytic leukemia (AML-M5a) cell lines (MOLM-13 and MOLM-14) with interclonal phenotypic heterogeneity showing MLL-AF9 fusion resulting from an occult chromosome insertion, ins(11;9)(q23;p22p23). Leukemia 1997; 11: 1469–1477.
Yang J, Ikezoe T, Nishioka C, Furihata M, Yokoyama A . AZ960, a novel Jak2 inhibitor, induces growth arrest and apoptosis in adult T-cell leukemia cells. Mol Cancer Ther 2010; 9: 3386–3395.
Burgering BM, Coffer PJ . Protein kinase B (c-Akt) in phosphatidylinositol-3-OH kinase signal transduction. Nature 1995; 376: 599–602.
Gebhard C, Schwarzfischer L, Pham TH, Schilling E, Klug M, Andreesen R et al. Genome-wide profiling of CpG methylation identifies novel targets of aberrant hypermethylation in myeloid leukemia. Cancer Res 2006; 66: 6118–6128.
Nishioka C, Ikezoe T, Yang J, Nobumoto A, Kataoka S, Tsuda M et al. CD82 regulates STAT5/IL-10 and supports survival of acute myelogenous leukemia cells. Int J Cancer 2014; 134: 55–64.
Nishioka C, Ikezoe T, Yang J, Yokoyama A . Tetraspanin family member, CD82, regulates expression of EZH2 via inactivation of p38 MAPK signaling in leukemia cells. PLoS One 2015; 10: e0125017.
Schepers H, van Gosliga D, Wierenga AT, Eggen BJ, Schuringa JJ, Vellenga E . STAT5 is required for long-term maintenance of normal and leukemic human stem/progenitor cells. Blood 2007; 110: 2880–2888.
Moore MA, Dorn DC, Schuringa JJ, Chung KY, Morrone G . Constitutive activation of Flt3 and STAT5A enhances self-renewal and alters differentiation of hematopoietic stem cells. Exp Hematol 2007; 35: 105–116.
Schuringa JJ, Chung KY, Morrone G, Moore MA . Constitutive activation of STAT5A promotes human hematopoietic stem cell self-renewal and erythroid differentiation. J Exp Med 2004; 200: 623–635.
Perry JM, He XC, Sugimura R, Grindley JC, Haug JS, Ding S et al. Cooperation between both Wnt/{beta}-catenin and PTEN/PI3K/Akt signaling promotes primitive hematopoietic stem cell self-renewal and expansion. Genes Dev 2011; 25: 1928–1942.
Nishioka C, Ikezoe T, Yokoyama A . Blockade of CD82 by a monoclonal antibody potentiates anti-leukemia effects of AraC in vivo. Cancer Med 2015; e-pub ahead of print 3 July 2015; doi:10.1002/cam4.482.
Nishioka C, Ikezoe T, Yang J, Udaka K, Yokoyama A . Simultaneous inhibition of DNA methyltransferase and histone deacetylase induces p53-independent apoptosis via down-regulation of Mcl-1 in acute myelogenous leukemia cells. Leuk Res 2011; 35: 932–939.
Yamamoto M, Okada K, Akiyama H, Kurosu T, Miura O . Evaluation of the efficacy of maintenance therapy for low-to-intermediate-risk acute promyelocytic leukemia in molecular remission: a retrospective single-institution study. Mol Clin Oncol 2015; 3: 449–453.
Dong JT, Lamb PW, Rinker-Schaeffer CW, Vukanovic J, Ichikawa T, Isaacs JT et al. KAI1, a metastasis suppressor gene for prostate cancer on human chromosome 11p11.2. Science 1995; 268: 884–886.
Adachi M, Taki T, Ieki Y, Huang CL, Higashiyama M, Miyake M . Correlation of KAI1/CD82 gene expression with good prognosis in patients with non-small cell lung cancer. Cancer Res 1996; 56: 1751–1755.
Maurer CA, Graber HU, Friess H, Beyermann B, Willi D, Netzer P et al. Reduced expression of the metastasis suppressor gene KAI1 in advanced colon cancer and its metastases. Surgery 1999; 126: 869–880.
Liu FS, Chen JT, Dong JT, Hsieh YT, Lin AJ, Ho ES et al. KAI1 metastasis suppressor gene is frequently down-regulated in cervical carcinoma. Am J Pathol 2001; 159: 1629–1634.
Knoener M, Krech T, Puls F, Lehmann U, Kreipe H, Christgen M . Limited value of KAI1/CD82 protein expression as a prognostic marker in human gastric cancer. Dis Markers 2012; 32: 337–342.
Acknowledgements
This work was supported in part by Takeda Science Foundation (to CN) and JSPS KAKENHI Grant Number 14446690 (to CN).
Author contributions
Takayuki Ikezoe contributed to the concept and design, interpreted and analyzed the data and wrote the article. Chie Nishioka performed all experiments and wrote the article. Atsuya Nobumoto and Masayuki Tsuda provided technical support. Akihito Yokoyama provided critical revision and intellectual content.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Rights and permissions
About this article
Cite this article
Nishioka, C., Ikezoe, T., Takeuchi, A. et al. The novel function of CD82 and its impact on BCL2L12 via AKT/STAT5 signal pathway in acute myelogenous leukemia cells. Leukemia 29, 2296–2306 (2015). https://doi.org/10.1038/leu.2015.219
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2015.219
- Springer Nature Limited
This article is cited by
-
Evaluation of the p53 pathway in polycystic ovarian syndrome pathogenesis and apoptosis enhancement in human granulosa cells through transcriptome data analysis
Scientific Reports (2023)
-
Identification of key gene signatures for the overall survival of ovarian cancer
Journal of Ovarian Research (2022)
-
A KDM4A-PAF1-mediated epigenomic network is essential for acute myeloid leukemia cell self-renewal and survival
Cell Death & Disease (2021)
-
Tetraspanin CD82 drives acute myeloid leukemia chemoresistance by modulating protein kinase C alpha and β1 integrin activation
Oncogene (2020)
-
CD82 supports survival of childhood acute myeloid leukemia cells via activation of Wnt/β-catenin signaling pathway
Pediatric Research (2019)