Abstract
Breast cancer is a deadly, noncommunicable disease that affects women worldwide. Early detection is crucial in providing effective treatment and improving survival rates. Towards that, a novel DualNet-deep-learning-classifier model for efficient and accurate breast cancer detection is proposed. The proposed model includes five major phases: “pre-processing, segmentation, feature extraction, feature selection, and breast cancer detection”. The pre-processing phase involves noise removal via Wiener filtering and image contrast enhancement via a contrast stretching approach. Then, from the pre-processed mammogram images, the ROI region is identified using the new gradient-based watershed segmentation approach. Subsequently, from the identified ROI regions the texture [grey level run-length matrix (GLRLM), multi-threshold rotation invariant LBP (MT-RILBP) (proposed)], color (color correlogram), and shape features (Zernike moment) are extracted; and among the extracted features, the optimal features are chosen using a hybrid optimization model-FlyBird optimization algorithm (FBO), which incorporates both the “fruit fly optimization algorithm and bird mating optimizer”. The breast cancer classification phase uses a DualNet-deep-learning-classifiers approach that includes “long short-term memory networks (LSTM), convolutional spiking neural networks (CSNN), and a new optimized autoencoder (OptAuto)”. The LSTM and CSNN are trained using the identified optimal features. The outcome from LSTM and CSNN is fed as input to OptAuto, wherein the outcome regarding the presence/absence of breast cancer is identified. Moreover, the weight function of the autoencoder is tuned using the new FBO. The proposed model is evaluated in terms of “sensitivity, accuracy, specificity, precision, TPR, FPR, TNR, F1-score, and recall”. Overall, the proposed model holds promise for accurate and efficient breast cancer detection, with potential for future clinical applications.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Cancer of the breast is the most common cause of death among women worldwide. Early discovery and precise diagnosis can lead to a full recovery and prevent fatalities [1]. The likelihood of survival for cancer is considerably increased by early diagnosis. Sadly, pathological analysis is a challenging, time-consuming process requiring in-depth understanding [2]. Radiologists recommend using digital mammograms, ultrasounds, and MRIs among other breast cancer detection methods. Due to its low cost, ease, and superior results for early detection, mammography is a widely utilized technique. Mammogram images can be used in computer-aided diagnosis systems to provide useful data about “breast density, shape, and anomalies such as calcifications and masses”, aiding in early identification. “Mammography” is the most effective method of early breast cancer detection [3]. “Mammograms” is a type of X-ray that is used to detect breast cancer in women aged 50–70. They are incredibly good at finding little cancers and giving precise results [4].
To avoid tiredness and errors, research has been done to automatically identify cancer cells in mammography images using image processing and computer vision. Technologies built on artificial intelligence have been launched to improve detection speed and reduce human error. These techniques make it easier and more accurate for radiology specialists to identify the condition [5]. To identify malignant cancers, a classifier is trained using the structural, morphological, and texture aspects of the extracted region of interest or the complete image. To differentiate between benign and malignant cancers, the nuclei must first be segmented. Modern techniques for segmentation include watershed segmentation, the threshold method, active contours, and regional expansion. Segmentation methods can be supervised or unsupervised [6]. A breast cancer grading technique, Bayesian classifier, and domain knowledge structural constraints can be used to develop gland and nuclei segmentation [7]. Patch combining and semantic classification are used in a technique for segmenting breast cancers. The image can then be improved by histogram equalization, bilateral filtering, and pyramid mean shift filtering [8]. By lowering false-negative and false-positive rates, modified VGG can increase the effectiveness of mammography analysis, making it a useful tool for radiologists [9]. Using mammography and ultrasound pictures, a “deep convolutional neural network (CNN)” breast cancer classification model has now been developed. The models consist of four convolutional layers and one fully linked layer that enables the automatic extraction of standout characteristics with fewer adjustable parameters [10]. By combining context data and high-resolution characteristics, STAN, a new deep learning architecture, outperforms conventional methods for segmenting tiny breast cancers [11]. The SHA-MTL is a multi-task learning model for image segmentation and binary classification in breast ultrasound [12]. “Skin, fibro glandular tissue, mass, and fatty tissue” may all be distinguished in 3D breast ultrasound images using a CNN [13]. Using only five learnable layers and effective feature extraction, a deep CNN model might be used to automatically classify breast cancer from mammography and ultrasound pictures [14]. For classifying cancer-normal instances on mammograms, a CNN architecture was created employing a revised classifier model and accelerated feature learning [15].
The major contribution of this research work is:
-
To introduce a new multi-threshold rotation invariant LBP (MT-RILBP)-based texture feature extraction model.
-
To select the optimal features using the new hybrid optimization model-FlyBird optimization algorithm (FBO), which incorporates both the “Fruit Fly optimization algorithm (FOA) and bird mating optimizer (BMO)”.
-
To design a new DualNet-deep-learning-classifiers approach with “long short-term memory networks (LSTM), convolutional spiking neural networks (CSNN), and a new optimized autoencoder (OptAuto)”.
-
To fine-tune the weight of the optimized autoencoder (OptAuto) using the new FBO.
The rest of this paper is arranged as: Section “Literature Review” portrays the literature review of the works under the subject. Section “Proposed Methodology for Breast Cancer Prediction Using DualNet Deep Learning Classifiers” portrays the proposed methodology for breast cancer detection. Section “Results and Discussion” discusses the recorded results. This paper is concluded in Section “Conclusion”.
Literature Review
In 2021, Salama and Aly et al. [16] presented a framework for segmenting and classifying breast cancer images using three datasets and evaluated using various models, such as “InceptionV3, DenseNet121, ResNet50, VGG16, and MobileNetV2”. The breast area in mammography images was segmented using the “modified U-Net model”. To get over the death of tagged data.
In 2019, Vijayarajeswari et al. [17] classified mammograms by feature extraction using the Hough transform. To accomplish this, support vector machines were utilized to classify the mammography images after the Hough transform to detect features of a specific shape. The findings demonstrated that the suggested method successfully identified abnormal mammography pictures, with a higher degree of accuracy being attained with the application of the SVM classifier. Ninety-five mammograms from a dataset were used to evaluate the approach.
In 2021, AlGhamdi and Mottaleb et al. [18] presented a model to identify whether patches corresponded to the same mass. The network was made up of a feature extraction component that used tied dense blocks and a three-layer nearby patch matching component, including a cross-input neighborhood differences layer that defined a summary of the neighborhood differences.
In 2020, Sha et al. [19] suggested to use of a variety of techniques, such as image noise reduction, the “grasshopper optimization algorithm, and CNN-based optimal image segmentation” were used to identify the cancerous area in mammogram images, and its effectiveness was compared to ten other cutting-edge techniques. The outcomes demonstrated that the suggested strategy increased precision and reduced computational expense.
In 2021, Zebari et al. [20] proposed a novel approach to classify breast cancer using mammography pictures. It employs wavelet transform and an improved FD technique to extract features and uses it to calculate ROI, using a combination of thresholding and machine learning. The classification process makes use of five classifiers, and the output is then fused.
In 2022, Maqsood et al. [21] developed a deep learning system using an "end-to-end" training method, to recognize mammography screening images for breast cancer. To extract texture features, the method used a modified contrast enhancement technique with a transferable texture CNN with an energy layer. Using the deep properties of different “CNN models”, the performance of TTCNN was examined. In three datasets, the approach was tested, and it outperformed existing techniques with an average accuracy of 97.49%. According to the study, deep learning algorithms can enhance clinical tools for Breast cancer detection at an early stage.
In 2022, Li et al. [22] proposed using a multi-input deep learning network to automatically classify breast cancer. To preserve contextual information during pooling, the network simultaneously analyzed four images of each breast, each with a unique set of characteristics. The technique lowers the likelihood of incorrect diagnoses and avoidable biopsies, assisting physicians in correctly identifying breast cancer from numerous CESM images.
In 2021, Kavitha et al. [23] introduced OMLTS-DLCN, a new digital mammogram-based breast cancer diagnosis model. The model uses a back-propagation neural network for classification, for “segmentation, optimal Kapur’s based multilevel thresholding with shell game optimization algorithm” was used.
In 2019, Khan et al. [24] suggested a Mammography classification using a CAD system according to Multi-View Feature Fusion. Anomaly classification is based on mass/calcification and malignancy, and the system had three steps. For each view, CNN-based feature extraction models were applied, and the retrieved features were then merged to get the final forecast. The system had four views of mammograms for training, and it outperformed single-view-based methods for mammography classification using accuracy.
In 2020, Song et al. [25] proposed a deep learning-based network which has been used during the feature extraction to extract the features of CESM images. A test was run to assess the performance of a dataset of internal CESM images using a framework with three stages: input, image feature extraction, and classification. 760 pictures from 95 patients made up the collection.
Problem Statement
Manual mammography examination can be time-consuming and error-prone when looking for breast cancer. An infrared photo collection is analyzed to identify breast cancer images using a variety of pre-processing approaches. One of these methods involves eliminating the pectoral muscle area before feature extraction, because it has less intensity variation than the tumor. Thermal mapping is then used to identify cancerous regions, and cancer is found in these pictures using feature selection and classification methods. Although classification divides the data into non-cancerous and malignant portions, feature selection identifies the most pertinent features from the database. The need for more effective and accessible breast cancer screening methods is highlighted by the fact that the present techniques are time-consuming, costly, and necessitate additional labor for radiologists to operate the equipment.
Proposed Methodology for Breast Cancer Prediction Using DualNet Deep Learning Classifiers
Early pre-processing of the mammograms makes a difference in how distinct desired items are from unwanted background noise. The variable being measured here is intensity, because mammographic images have low contrast and it is difficult to distinguish between masses in them; pre-processing is done. Comparing pectoral muscle intensity to cancer intensity, there is typically little variance. The proposed methodology for the DualNet-deep-learning-classifier model for efficient and accurate breast cancer detection can be described in the following steps:
-
Pre-processing: The mammogram images are pre-processed using Wiener filtering and contrast stretching to reduce noise and enhance image contrast.
-
Segmentation: The pre-processed mammogram images are segmented using the Gradient-based watershed segmentation approach to identify the ROI regions.
-
Feature extraction: Subsequently, from the identified ROI regions the texture [Grey level run-length matrix (GLRLM), multi-threshold rotation invariant LBP (MT-RILBP) (proposed)], color (color correlogram), and shape features (Zernike moment) are extracted.
-
Feature selection: The extracted features are fed to FBO that incorporates both the FOA and the BMO to select the optimal features.
-
Breast cancer detection: The breast cancer classification phase uses a DualNet-deep-learning-classifiers approach that includes LSTM, CSNN, and a new optimized autoencoder (OptAuto). The LSTM and CSNN are trained using the identified optimal features. The outcome from LSTM and CSNN is fed as input to OptAuto to identify the presence/absence of breast cancer.
-
Evaluation: The proposed model is evaluated using metrics, such as sensitivity, accuracy, specificity, precision, TPR, FPR, TNR, F1-score, and recall to assess the accuracy and efficiency of breast cancer detection.
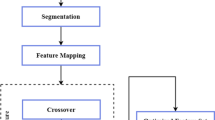
Overall, the proposed methodology involves pre-processing the mammogram images to reduce noise and enhance contrast, segmenting the images using improved watershed segmentation, extracting and selecting optimal features, and using a DualNet-deep-learning-classifier approach for breast cancer detection. The model holds promise for accurate and efficient breast cancer detection with the potential for future clinical applications (Fig. 1).
Pre-processing
The pre-processing stage's primary goal is to remove noise from the mammogram and improve the image's contrast enhancement. Therefore, in research, raw mammogram images are pre-processed using Wiener filtering (for noise removal) and contrast stretching (for image contrast enhancement).
Wiener Filtering
The Wiener filter technique is a statistical approach to remove noise from each pixel in an image. It carries out the best possible exchange between noise smoothing and inverse filtering. It is the ideal filter to use for reducing the overall MSE during the noise-smoothing process. By placing an MSE restriction between the estimates and the original image, it attempts to construct an image. Wiener filters analyze frequency domain data, but they may fail to recover noise-damaged frequency components. The minimum Wiener filter function in the frequency domain is described by Eq. (1), and the minimized error is shown by Eq. (2). The inversion of blurring and additive noise is instantly removed by the Wiener filter
where the degradation function is \(f(t,n)\); the complex conjugate of \(f\) is \({\left|f\left(t,n\right)\right|}^{2}\),\({f}^{*}\left(t,n\right)=f\left(t,n\right){ f}^{*}\left(t,n\right),\) noise's power spectrum is \(f\left(t,n\right); {W}_{O}(t,n)\), and the unaltered image's power spectrum is \({W}_{G}(t,n)\).
Contrast Stretching
Contrast stretching, also known as normalizing, is used in the second phase. Adjusting the range of intensity values is a technique for stretching images that improves image quality.
-
1.
Initially, before computing the gradient, the dermo copy images are modified using the Sobel edge filter while maintaining a 3 × 3 kernel size.
-
2.
Dividing the grey image into several equal-sized blocks (4, 8, 12,…) and rearranging them in ascending order based on gradient intensities. Weights are now changed for each block following the gradient's strength. This method is described by Eq. (3)
$$\psi \left(h,j\right)=\left\{\begin{array}{c}{\varrho }_{\omega }^{s1} if {\vartheta }_{C}\left(h,j\right)\le g{t}_{1}; \\ {\varrho }_{\omega }^{s2} g{t}_{1}<{\vartheta }_{C}\left(h,j\right)\le g{t}_{2}\\ {\varrho }_{\omega }^{s3} g{t}_{1}<{\vartheta }_{C}\left(h,j\right)\le g{t}_{3};\\ {\varrho }_{\omega }^{s4} otherwise\end{array}\right.$$(3)where \(g{t}_{I}\) is the threshold for gradient intervals and \({\varrho }_{\omega }^{sI}\) (I = 1,…, 4) are statistical weight coefficients.
-
3.
Cumulative weighted grey values are now calculated for each block according to Eq. (4).
$${C}_{r}\left(y\right)=\sum_{I=1}^{4}{\varrho }_{\omega }^{gI}{V}_{I}\left(y\right),$$(4)
where \({V}_{I}(y)\) represents the sum of the grey-level pixel values for each block I.
For an optimum solution, three factors including the maximum region extraction, block size, and weighting criteria should all be considered. Yet, the informative regions are between 25 and 75%; therefore, choosing 12 blocks with an aspect ratio of 8:3 considers the minimum value of 25%. The weights are assigned according to the number of edge points, Epi, for each block, as shown in Eq. (5) for this purpose
\({D}_{{\text{max}}}^{L}\) max is the block with the greatest number of edges. Morphological operations, which are a sequence of connecting stages used before feature vector creation to provide the greatest amount of differentiability between foreground and background, are used. Closing, filling, and reconstruction are uses of morphological processes that effectively operate on all segment’s pictures. These adjustments to make to all photographs to make the objects stand out more against the background.
ROI Identification
Breast cancer detection requires the use of segmentation techniques. Through image analysis, segmentation which comprises detection, feature extraction, classification, and treatment plays a crucial role. Physicians use segmentation to calculate the amount of breast tissue for planning treatments. Subsequently, from the pre-processed data, the ROI is identified via Improved Watershed segmentation.
Gradient-Based Watershed Segmentation (Proposed)
One of the main limitations of the watershed segmentation technique is that it can lead to over-segmentation, where regions that should be merged are split into multiple segments. This can be addressed by adding gradient-based segmentation techniques within the watershed model. These techniques involve using gradient information to identify boundaries and separate regions based on their gradient values. Gradient information-based watershed segmentation is a popular image-processing technique for identifying regions of interest (ROIs) in an image. This method is based on the analysis of the gradient information of the image, which can be used to identify the boundaries between different regions. In this technique, the gradient image is first computed from the input image, and then, the watershed segmentation algorithm is applied to the gradient image to obtain the ROIs. Here is a workflow for gradient information-based watershed segmentation in image processing with some expressions:
-
Step 1:
Load the image: Load the pre-processed image \({{\text{img}}}_{n}^{{\text{pre}}}\) into memory.
-
Step 2:
Convert to grayscale: Convert the input image to grayscale gray_img = img.convert('\({{\text{img}}}_{n}^{{\text{pre}}}\)'). The grayscale image is pointed as \({{\text{img}}}_{n}^{{\text{gray}}}\).
-
Step 3:
Compute gradient: Compute the gradient of the grayscale image \({{\text{img}}}_{n}^{{\text{gray}}}\). This is done using a Canny edge detector (\({G}_{{\text{mag}}}\), \({G}_{{\text{dir}}}\))
$${G}_{{\text{mag}}}\left(x,y\right)={\text{sqrt}}({{G}_{{\text{mag}}}x\left(x,y\right)}^{2}+{{G}_{{\text{mag}}}y\left(x,y\right)}^{2}$$(6)$${G}_{{\text{dir}}}\left(x,y\right)={\text{atan}}2({G}_{-}{y\left(x,y\right)}^{2}+{{G}_{-}x\left(x,y\right)}^{2}.$$(7) -
Step 4:
Thresholding: Threshold \((T)\) the gradient image to obtain a binary image that contains the edges and boundaries of the objects in the image
$$T\left(x,y\right)=1;if {G}_{{\text{mag}}}\left(x,y\right)>T$$(8)$$T\left(x,y\right)={\text{otherwise}}.$$ -
Step 5:
Morphological operations: Apply reconstruction filter operations \({(T}_{{\text{morph}} })\) such as erosion and dilation to the binary image to remove noise and fill gaps in the edges.
-
Step 6:
Morphological reconstruction filter: When the contour edge information is lost, the region contour shifts in position, despite the standard pre-smoothing filter's ease of use in improving outcomes in decreasing noise and irregular details. Because of the characteristics of the chest cancer in the mammogram image, the target's edge contour information can be successfully retained during reconstruction filtering. It also does not cause a positional shift in the visible area's contour in the rebuilt image. The following describes morphological reconstruction:
$${R}_{n+1}=\left({R}_{n} \oplus tl\right)\bigcap w,$$(9)where \({R}_{tl}\) is a morphological reconstruction of the mask images acquired from the marker image \(w\), where \(tl\) is the structural component responsible for the enlargement of the marker image; \(\alpha\) it also represents the original image used as a mask; and \(Rn\) and is the final iteration's output image. The marker image \(a\)'s initial iteration is \({R}_{E}\). When \({R}_{n+1}={R}_{n}\), Eq. (6) is iterated till the end.
When using morphological reconstruction or closed reconstruction, which can only remove one noise or detail from an image, a shift in the position of the target contour has been easily possible. Hence, by employing the hybrid opening and closure reconstruction process, texture details, as well as shading noise, can be removed simultaneously, and image morphology reconstruction helps to improve the boundary information while decreasing the number of pseudo-minimum values
A morphological corrosion procedure is \(\ominus\) shown in Eq. (10).
The closed operation reconstruction is described in Eq. (11) as
Equation (12) gives the following definition of the closed operation reconstruction:
-
Step 7:
Marker generation: Generate markers for the watershed segmentation algorithm. This is done using regional minima/maxima. Breast cancermammogram images that have undergone morphological reconstruction filtering still contain a few small value points in the image that are not suppressed, because they are unrelated to the target object, and the segmentation results contain a large number of meaningless regions that can be used for marker extraction. Fix it. When using the watershed segmentation method on a target image, imply the target region's minimum value in the gradient image and mask any additional minimum values to minimize the number of meaningless regions in the segmentation result. Only the target area's minimum value used may be preserved to reduce the likelihood of over-segmentation issues. As a result, the soiled-proposes morphological-bases extend minimum transform technique h is used to address the issue of regional minimum value labeling. This technique's main challenge is choosing the threshold h, which, when established, effectively eliminates the local minimum with a depth of less than h. The h-minima approach has the advantage of allowing for direct threshold determination. The constant threshold h, insufficient flexibility, and single adaption are the disadvantages. The threshold h can be chosen using the adaptive acquisition method to prevent the impact of artificial setting elements. The threshold h divides the target image into two groups, If h is the value of the threshold, then pixels in the target class \({T}_{e}\) of the grayscale of \(\left\{\mathrm{0,1},\dots h\right\}\) T 1 is a backdrop class that includes pixels in the \(h+1,h+2,\dots ,L-1\) grayscale. Discover the target class \({T}_{e}\) and the background class \({T}_{1}\)'s intra or inter-class variance.
Target class \({T}_{e}\) occurrence likelihood:
$${Z}_{e}=\sum_{I=e}^{h}{V}_{I};$$(13)background class \({T}_{1}\) occurrence likelihood
$${Z}_{1}= \sum_{I=h+1}^{L-1}{V}_{I}.$$(14)The target class \({T}_{e}\)'s average value is
$${\mu }_{e}=\frac{\sum_{I=e}^{h}I{V}_{I}}{{z}_{e}}.$$(15)Background class \({T}_{1}\) average
$${\mu }_{1}= \frac{\sum_{I=h+1}^{L-1}I{V}_{I}}{{z}_{1}}$$(16)$$h={{\text{argmin}}}_{0 \le h<L}\left\{{\sigma }_{Z}^{2}\right\}$$(17)$$h={{\text{argmin}}}_{0 \le h<L}\left\{{\sigma }_{n}^{2}\right\}.$$(18) -
Step 8:
Watershed segmentation: Perform the watershed segmentation algorithm on the input image using the generated markers. The above-mentioned “Otsu method” is used to calculate the threshold H, and the reconstructed gradient image. To avoid the appearance of meaningless minima, Gse is recovered using the extended minimum transform method. As a result, the marker appears at a low value. Following the acquisition of the local minimum value marker image linked to the target region, the gradient image is modified using Soiled's minim imposition technique, so that other pixel values become consistent as needed. This ensures that the local minimum occurs only at the marked position and eliminates any other local minimum regions. This is shown in Eq. (19)
$${g}^{{\text{mark}}}={\text{imimposemin}}({g}_{tl},{T}_{1}\vdots {T}_{e}),$$(19)where \({g}^{{\text{mark}}}\) denotes the updated gradient image and \(\mathrm{imimpose min}\) denotes the forced minimum operation suggested by Soiled. Following the minimal value forced minimum operation, the watershed segmentation method is then applied to the gradient image \({g}^{{\text{mark}}}\), which is shown in Eq. (20)
$${g}_{{\text{ws}}}={\text{watershade}} \left({g}^{{\text{mark}}}\right),$$(20)where \(Watershed\) represents using marker value; for the marking completion, an image of the relevant target area could be obtained \({g}_{{\text{ws}}}\).
-
Step 9:
Post-processing: Apply post-processing techniques such as merging or filtering to the segmented image to remove over-segmentation and refine the segmentation results.
-
Step 10:
Output: Save the segmented image (ROI-identified image)\({{\text{img}}}_{n}^{{\text{ROI}}}\) as output. This is the identified ROI region.
Feature Extraction
The process of extracting relevant and useful information or features from pre-processed images is referred to as feature extraction.
Texture Feature
Texture features refer to the visual attributes of a surface or material, including its pattern, roughness, smoothness, and other surface characteristics. These characteristics are used in different types of applications, such as “computer vision, image processing, and machine learning”, to classify and analyze images based on their texture.
Grey-Level Run Length Matrix (GLRLM)
The “grey level run length technique” works by calculating the number of grey-level runs of varying lengths. A grey-level run is a collection of adjacent image points that have similar grey-level values.\(N\) seems to be a run-length matrix as follows: \(N (k,j)\), \(k\) signifies the number of runs with grey-level intensity pixels, and j the length of a run on a particular orientation. Matrix \(N\) is defined as \(z by y\), where \(c\) is the image's maximum grey level and b is the image's longest possible run length. The orientation is described by a displacement vector \(k(c,b)\), where c and b are the displacements for the x and y axes, respectively. In this methodology run, texture is defined in four directions (0°, 45°, 90°, and 135°), and four run-length matrices are produced as a result.
“GLRLM” is used to derive seven features: “short run emphasis (SRE), long run emphasis (LRE), grey level non-uniformity (GLN), run-length non-uniformity (RLN), run percentage (RP), low grey-level run emphasis (LGRE), and high grey-level run emphasis (HGRE)”.
Multi-threshold Rotation Invariant LBP (MT-RILBP) (Proposed)
Local binary patterns (LBP) is a popular method for describing the texture of an image. It works by comparing each pixel with its surrounding pixels and assigning a binary code to each pixel based on the comparison results. The binary codes are then used to create a histogram of the texture features in the image. However, the standard LBP method is sensitive to rotation, which means that the resulting histogram may be different for the same texture if the image is rotated. To address this issue, an improved rotation invariant LBP (IRI-LBP) method aims to generate a histogram that is invariant to rotation. Multi-threshold rotation invariant LBP (MT-RILBP) is an extension of the standard LBP and its variants that aims to address the limitations of previous rotation invariant LBP methods. The novel concept in MT-RILBP is the use of a novel rotation invariant coding scheme that takes into account the gradient information of the image in addition to the texture information. Another important concept in MT-RILBP is the use of multiple thresholds for generating LBP patterns. The traditional LBP uses a single threshold to determine whether a neighbor pixel is brighter or darker than the center pixel. However, this threshold may not be optimal for all images and textures. To address this limitation, MT-RILBP uses multiple thresholds to generate multiple LBP patterns for each pixel. IRI-LBP uses multiple thresholds to generate multiple LBP patterns for each pixel. Specifically, for each pixel in an image, MT-RILBP computes several LBP codes with different thresholds. Let us assume that we use \({T}_{i}\) thresholds, denoted as \({t}_{1}, {t}_{2},\dots\), and \(P\) neighbors to compute the LBP codes. For each threshold, a binary code \({C}_{i}\) is assigned to each pixel \(P\) based on whether its \(P\) neighbors are brighter or darker than the center pixel, using the following formula:
When \(U\left({{\text{LBP}}}_{K,I}\right)\le 2\), LBP is defined as \({{\text{LBP}}}_{K,I}^{u2}\) with \(K(K-1) + 2\) discriminative patterns. Although the histogram spectrum feature can be shortened using a uniform pattern, this processing method is feasible. Experiments and observations show that uniform LBPs are fundamental texture properties that comprise the vast majority of patterns, accounting for up to 90% of the time. Furthermore, no matter how the LBP is rotated, its structure remains the same, implying that the original and rotated LBPs have the same order and bitwise 0/1 changes. The rotation invariant texture description is obtained by
\({\text{ROR}}\left(c,k\right)\) indicates the rotation of the LBP code \(c\), \(k\) times around the center pixel, where \({\text{ri}}\) denotes rotation invariance. In other words, the LBP with the smallest decimal value serves as a stand-in for other LBPs in the same family. The uniform rotation invariant LBP.\({\mathrm{ LBP}}_{K,I}^{{\text{riu}}2}\) can be calculated as follows:
Rotation invariant uniform pattern with \(K+ 2\) discriminative patterns is denoted by \({\text{riu}}2\). As a result, the texture spectrum histogram's dimension has also been greatly reduced. The texture spectrum histogram \({S}_{{\text{original}}}\) can be obtained by gathering statistics on the frequency of occurrences \({{\text{LBP}}}_{K,I}^{{\text{riu}}2}\) at every possible pixel position in the image.
Color Feature
Color features are image features that focus on analyzing the colors present in an image. By analyzing the colors, computers can extract useful information about an image's content for tasks such as object recognition, image retrieval, and image segmentation. However, color features can be sensitive to changes in lighting conditions and color variations. As a result, they are often used in conjunction with other types of image features. Despite these challenges, color features remain an important tool for analyzing images, particularly in applications such as content-based image retrieval. As technology advances, we can expect even more sophisticated techniques for analyzing color features.
Color Correlogram
A color correlogram is a type of color feature that analyzes the spatial relationships between colors in an image. Specifically, it computes the frequency of occurrence of color pairs separated by a certain distance in an image. By analyzing the spatial relationships between colors, a computer can extract useful information about an image's content, which can be used for tasks such as image retrieval and segmentation. Color correlograms are particularly useful for analyzing textures in images and are often used in conjunction with other types of image features to enhance the precision of image analysis algorithms (Fig. 2).
Let \(w \in \left[m\right]\) n be a fixed a priori distance. The correlogram of \(I\) is then defined for \(r,q \in \left[n\right],r\in \left[w\right]\)
Given any pixel of color \({x}_{r}\) in the image,\({\gamma }_{{x}_{r,}{x}_{q}}^{\left(r\right)}\) calculates the likelihood that a pixel located r pixels away from the given pixel are of color \({x}_{q}\). The correlogram's size is \(O\left({n}^{2}w\right).\) Only spatial correlation between identical colors is captured by the auto correlogram of \(I\)
These data are a subset of the correlogram and takes up only \(O(nw)\) space.
We must address the following issue when deciding on \(w\) to define the correlogram. A large \(w\) would necessitate costly computation and large storage requirements. A small \(w\) could jeopardize the quality.
Shape Feature
Shape features are image features that focus on analyzing the shape and geometric properties of objects in an image. By analyzing the shape features, a computer can extract useful information about the objects' characteristics and use it for tasks, such as object recognition, image retrieval, and segmentation (Fig. 3).
Zernike Moment
The Zernike moments of order m with repetition n for a continuous image function f(c, b) which vanishes outside the unit circle have been calculated
where \(m\) isa nonnegative integer and \(n\) an integer \(m-|n|\) that is both nonnegative and even. The complex-valued functions \({E}_{mn}\left(c,b\right)\) are defined as follows:
where \(\rho\) and \(\theta\) is just the unit's polar coordinates disc \({I}_{mn}\) and \(\rho\) are (Zernike polynomials) given by
\({I}_{m,-n}\left(\rho \right)= {I}_{mn}\left(\rho \right)\) are the polynomials orthogonal as well as satisfy
with
The origin is set to the image's center to compute the Zernike moments, and the pixel coordinates are mapped to the unit circle range. It is possible to compute a discretized original image function \(f(c,b)\) whose moments are the same as those of \(f(c,b)\) up to the specified order. We can reconstruct \(\widehat{f}(c,b)\) as a result of the orthogonality of the Zernike basis
Hybrid Optimization Model for Optimal Feature Selection
The optimization algorithm to use from a group of many algorithms that accomplish the same optimization is selected by hybrid optimizations dynamically at compile time. For each section of code being optimized, they employ a heuristic to forecast the best algorithm. The hybridization of different optimization algorithms has become a common practice in recent years. In this case, we will discuss the hybridization of the FOA and BMO for optimal feature selection for breast cancer classification. FOA is a population-based optimization algorithm that is inspired by the foraging behavior of fruit flies. It has been successfully applied in various optimization problems, including feature selection. BMO, on the other hand, is a recently developed algorithm that is based on the mating behavior of birds. It has shown promising results in solving optimization problems, including feature selection. To hybridize these two algorithms, we can take advantage of their strengths and combine them to overcome their weaknesses. FOA is good at exploring the search space and avoiding getting stuck in local optima, while BMO is good at exploiting the search space by focusing on promising regions. As per the proposed approach, the FOA model is induced with the BMO model (Fig. 4).
Mathematically, the proposal can be given as:
-
Step 1:
Initialization: Initialize the population of fruit flies and birds randomly. The current solution is denoted as \(t\).
-
Step 2:
Evaluate: Evaluate the fitness of each solution as \(Fit={\text{min}}\left(Error\right)\).
-
Step 3:
Selection: Select two birds (male and female) randomly from the population. Males and females so make up the two genders in a bird society. The birds with the most genetic promise in civilization are those that are female. There are two groups of ladies. Males are split into three types, with the females being parthenogenetic and polyandrous and the men being monogamous, polygynous, and promiscuous.
-
Step 4:
Mating: Perform mating between the selected birds to create a new offspring bird. A male typically mates with just one female in a two-parent mating pattern known as monogamy. To decide which of the girls to choose as his mate, each guy a probabilistic method is used to rate the quality of females. A higher chance of selection exists for female birds with superior DNA. Equation (39) depicts how two chosen parents can create a new brood
$$\overrightarrow{{x}_{i}}=\overrightarrow{x}+w\times \overrightarrow{r}\times \left({\overrightarrow{x}}^{i}-\overrightarrow{x}\right);$$(38)$$\begin{gathered} c = a\, {\text{random integer number between}} 1 {\text{and }} n \hfill \\ if\, r_1 > mcf;\,x_{{\text{brood}}} \left( c \right) = l\left( c \right) - r_2 \times \left( {l\left( c \right) - u\left( c \right)} \right), \hfill \\ \end{gathered}$$(39)where \(n\) is the problem dimension, \(w\) is a time-varying weight that is used to alter the selected female, \(\overrightarrow{r}\) is a one-dimensional vector \(1\times d\) with each element being a random number between 0 and 1 distributed, and this random vector impacts the respective element of (\(\overrightarrow{{x}^{i}}-\overrightarrow{x)} , u\) and l are the lower and upper limits of the elements. The first part of Eq. (39) suggests that each male bird seeks a deserving female to mate with to create new genes to raise a high-quality brood. Then, with a probability of \(1- mcf\), the male bird seeks to increase the quality of his offspring by altering one of its genes.
Mutation (proposed): Perform mu where \(n\) is the problem dimension, \(w\) is a time-varying weight that is used to alter the selected female, \(\overrightarrow{r}\) is a one-dimensional vector \(1\times d\) with each element being a random number between 0 and 1 distributed, and this random vector impacts the respective element of (\(\overrightarrow{{x}^{i}}-\overrightarrow{x)} , u\) and l are the lower and upper limits of the elements. The first part of Eq. (39) suggests that each male bird seeks a deserving female to mate with to create new genes to raise a high-quality brood. Then, with a probability of \(1- mcf\), the male bird seeks to increase the quality of his offspring by altering one of its genes.
tation on the offspring bird to increase its diversity. The offspring generated are updated based on the levy flight mechanism of levy flight enhanced FOA (LFOA). The “Levy flight mechanism” is frequently utilized to enhance metaheuristics. Levy statistics is the name given to the phenomenon. In essence, stochastic non-Gaussian walks make up the LF (Fig. 5). Relative to the Levy stable distribution, its step value is scattered. The following serves as a representation of the Levy distribution
\(\beta\) represents step length \((s)\) is a crucial Levy index for adjusting stability.
-
Step 5:
Evaluate: Evaluate the fitness of the offspring bird.
-
Step 6:
Replacement: Replace the worst bird in the population with the offspring bird.
Breast Cancer Classification via DualNet-Deep-Learning-Classifier Model
The breast cancer classification phase uses a DualNet-deep-learning-classifiers approach that includes “LSTM, CSNN, and a new optimized autoencoder (OptAuto)”. The LSTM and CSNN are trained using the identified optimal features. The outcome from LSTM and CSNN is fed as input to OptAuto, wherein the outcome regarding the presence/absence of breast cancer is identified. Moreover, the weight function of the autoencoder is tuned using the new FBO (Fig. 6).
Convolution-Spiking Neural Networks
Since neurons are arranged in layers in ANNs, the SNN's architecture is comparable. With a fully connected technique, neurons in adjacent layers are linked. A neuron's output is produced immediately following the ANN propagation spatial information layer by layer; it receives presynaptic input from neurons in the layer above it. A spike train time series describing the output patterns of SNN neurons contrasts with how neurons interpret information in the spatial domain. The leaky integrate and fire model is one of three widely used neuron models that currently exist to explain the spiking behavior of SNNs (LIF). Given its straightforward hardware implementation and low computational complexity, LIF is known to be the neuron model that is utilized the most frequently (Fig. 7). The following equation describes how the neuron's membrane potential changes \((u)\) in the LIF model:
where \(I(t)\) stands for presynaptic input, which is defined by the spiking activity of incoming synaptic routes at time t and synaptic weights, and \({u}_{i}\left(t\right)\) is the neuron's membrane potential \(i \mathrm{at time} t\). The presynaptic input is equal to the sum of the synaptic weights \({w}_{1}\) and \({w}_{2}\) when the first two incoming synaptic routes are stimulated simultaneously. Until it crosses a predetermined threshold, the membrane potential \(u\) continuously updates. The membrane potential u is then reset after neuron I fire the spike.
Long Short-Term Memory
It will be possible to store and convert the memory of an LSTM cell in the cell state from input to output. An LSTM cell is made up of the “input gate, forget gate, update gate, and the output gate”. The forget gate, as the name implies, selects information to the input gate and selects information to be incorporated the input gate feeds information into the neuron, the update gate updates the cell, and the output gate generates new long-term memory from previous memory units. The LSTM's four key elements will function and interact uniquely as it receives long-term memory, short-term memory, and input sequence at a particular time step, and generates new short-term memory, long-term memory, and output sequence during the same time step (Fig. 8). The input gate, which may be mathematically expressed as the following, determines which data must be supplied to the cell:
The vectors are multiplied element by element by the operator "\(*\)".
The forget gate, which complies with the following mathematical definition, controls which prior memory information is ignored:
The update gate, which is theoretically represented by the following formula, modifies the cell state:
The output gate updates the previous time step's hidden layer, which can also update the output as provided by the previous time step
Optimized Autoencoder (OptAuto)
“Deep autoencoder” is a strong unsupervised feature representation strategy with several layers that are hidden. The fact that hidden layer parameters motivate the neural idea of data learning is automatically learned by the provided data rather than being generated manually. With the help of DAE, we were inspired to discover the Video sequence's time axis characteristics. The high-dimensional deep features are compressed to low dimensions with a small error during the transformation. Deep characteristics from a series of frames are extracted and learned using an effective four-layered architecture, along with hidden patterns and frame-to-frame changes. To make the autoencoder's time complexity less complicated, high-dimensional data are reduced by a half factor. High computational complexity is obtained by compressing large dimensional data using numerous deep layers and short steps. Via "hierarchical grouping" or "part-whole decomposition" in the incoming data, the DAE learns. The stacked autoencoder's early stages capture changes and features of the first order in the raw input data. In contrast, the intermediate layers are taught features of the second order which correspond to first-order feature patterns. In light of this, we contend that the suggested DAE successfully picks up on the variations and recurring patterns of human behavior in video sequences.
Both of these processes make up the autoencoder. Following some nonlinearity functions, such as the sigmoid and relu described in Eq. (48), comes encoding, which involves multiplying data by weights, adding biases, and then encoding. After that, the data are decoded to the same number of inputs as in Eq. (49), which is the second step. Reducing the mean squared error to nearly zero, the weights are modified using a back-propagation algorithm
According to Eqs. (50) and (51), the first hidden layer of the stacked autoencoder receives input \(x\), while the other receives input from the network's previously hidden layer. Here, \("n"\) stands for the number of encoding layers, and \({w}^{l},{x}^{l},{b}^{l}\) stand for the relevant layer's data, weights, and biases, respectively
The weight \({w}^{l}{x}^{l}\) of the autoencoder is optimized via a new hybrid optimization model, to enhance the detection accuracy of the model.
Results and Discussion
Experimental Setup
The proposed breast cancer diagnosis model has been implemented in MATLAB. The evaluation has been made using the data collected from: database 1: https://www.kaggle.com/datasets/awsaf49/cbis-ddsm-breast-cancer-image-dataset; and Database 2:https://www.kaggle.com/datasets/kmader/mias-mammography. The proposed model is evaluated in terms of “sensitivity, accuracy, specificity, precision, TPR, FPR, TNR, F1-Score, and Recall”. The proposed model has been tested by varying the learning rate from 70, 80, and 90, respectively.
Overall Performance Analysis of the Proposed Model for Database 1 and Database 2
The Table 1 provides a performance of various models on a given dataset-1. The proposed model achieved the highest accuracy of 0.938221, outperforming all other models, including FOA, BMO, CHO, and MSA. The suggested model's precision is also the highest among all models, with a value of 0.978307. The model's sensitivity and specificity are 0.942722 and 0.963072, respectively, indicating a well-balanced model. Additionally, the proposed model's F-measure of 0.953127 and MCC of 0.908756 are the highest among all models. The proposed model also has the lowest FPR of 0.062134, indicating the lowest rate of false positives. Overall, the proposed model is the most accurate and precise model among all the models evaluated in this study.
Overall Performance Analysis of the Proposed Model for Database 2
Table 2 shows the metrics of various models’ performance on a given dataset, using a learning rate of 1. With an accuracy of 0.947704, the proposed model outperformed all other models, CHO was next, with an accuracy of 0.897489. The proposed model also had the highest precision of 0.988190, indicating a low false-positive rate. Furthermore, the model has high values of 0.952246 and 0.972801 for sensitivity and specificity, respectively, indicating a well-balanced model. The proposed model's F-measure and MCC are 0.962754 and 0.917932, respectively, which are the highest among all models. The model also has the lowest FPR value of 0.062761, indicating the lowest rate of false positives. Overall, the proposed model is the most accurate and precise model among all the models evaluated in this study, with a well-balanced performance on sensitivity and specificity.
Table 1 shows the overall performance analysis of the proposed model for database 1. The proposed model achieved an accuracy of 0.938221, which is higher than the accuracies achieved by the FOA, BMO, CHO, and MSA models. The precision, sensitivity, and specificity of the proposed model are also higher than the other models. The F-Measure and MCC of the proposed model are 0.953127 and 0.908756, respectively, which are also higher than the other models. These results suggest that the proposed model outperforms the other models in terms of accuracy and various other metrics. In terms of false-positive rate (FPR) and false-negative rate (FNR), the proposed model has the lowest FPR (0.062134) among all the models, indicating that the proposed model can identify benign cases accurately. The FNR of the proposed model is also lower than the other models except for the CHO model, indicating that the proposed model can detect malignant cases with higher accuracy. The results suggest that the proposed model has a higher potential for identifying both benign and malignant cases. Table 2 shows the overall performance analysis of the proposed model for database 2. The proposed model achieved an accuracy of 0.947704, which is higher than the accuracies achieved by the FOA, BMO, CHO, and MSA models. The precision, sensitivity, and specificity of the proposed model are also higher than the other models. The F-Measure and MCC of the proposed model are 0.962754 and 0.917932, respectively, which are also higher than the other models. These results suggest that the proposed model outperforms the other models in terms of accuracy and various other metrics. In terms of false-positive rate (FPR) and false-negative rate (FNR), the proposed model has the lowest FPR (0.062761) among all the models, indicating that the proposed model can identify benign cases accurately. The FNR of the proposed model is also lower than the other models, indicating that the proposed model can detect malignant cases with higher accuracy. The results suggest that the proposed model has a higher potential for identifying both benign and malignant cases. The results from both tables indicate that the proposed model outperforms the other models in terms of accuracy, sensitivity, specificity, precision, F-Measure, and MCC. The proposed model has the lowest FPR among all the models, indicating that the proposed model can identify benign cases with higher accuracy. The FNR of the proposed model is also lower than the other models, indicating that the proposed model can detect malignant cases with higher accuracy. Overall, the proposed breast cancer diagnosis model is effective in identifying both benign and malignant cases of breast cancer with higher accuracy than the other models. The results suggest that the proposed model has the potential to aid in the early detection and diagnosis of breast cancer, which can significantly improve patient outcomes and survival rate (Fig. 9).
Performance Analysis for Varying Training Rates
Based on the given metrics in Table 3, it appears that for all performance metrics, the suggested technique generates the most effective outcomes. Specifically, the proposed method has the highest accuracy (0.942957), sensitivity (0.947478), precision (0.983248), and specificity (0.947478) among all methods compared (0.967942), F-measure (0.957936), MCC (0.913339), NPV (0.967942), and the lowest false-positive rate (0.062448) and false-negative rate (0.082905). On the other hand, the FOA method has the lowest specificity (0.932254) and the highest false-negative rate (0.130284), while the CHO method has the lowest accuracy (0.827437) and the highest false-positive rate (0.139336). The BMO and MSA methods have relatively good performance but are outperformed by the proposed method in most measures. Overall, based on these metrics, it appears that the suggested technique is the best-performing method among those considered (Fig. 10).
Conclusion
In this research work, a novel deep learning approach has been introduced. The proposed model includes five major phases: “Pre-processing, segmentation, feature extraction, feature selection, and breast cancer detection”. The pre-processing phase involves noise removal via Wiener filtering and image contrast enhancement via contrast stretching approach. Then, from the pre-processed mammogram images, the ROI region is identified using the new gradient-based watershed segmentation approach. Subsequently, from the identified ROI regions, the texture [Grey level run-length matrix (GLRLM), Multi-Threshold Rotation invariant LBP (MT-RILBP) (Proposed)], color (Color correlogram), and shape features (Zernike Moment) are extracted; and among the extracted features, the optimal features are chosen using a hybrid optimization model-FBO, which incorporates both the “FOA and BMO”. The breast cancer classification phase uses a DualNet-deep-learning-classifiers approach that includes “LSTM, CSNN, and a new optimized autoencoder (OptAuto)”. The LSTM and CSNN are trained using the identified optimal features. The outcome from LSTM and CSNN is fed as input to OptAuto, wherein the outcome regarding the presence/absence of breast cancer is identified. Moreover, the weight function of the autoencoder is tuned using the new FBO. The proposed model is evaluated in terms of “sensitivity, accuracy, specificity, precision, TPR, FPR, TNR, F1-Score, and Recall”.
Data availability
The dataset generated and analyzed during the current study are available from the corresponding author on reasonable request.
References
Jahangeer GSB, Rajkumar TD. Early detection of breast cancer using a hybrid of series network and VGG-16. Multim Tools Appl. 2021;80:7853–86.
Khened M, Kori A, Rajkumar H, Krishnamurthi G, Srinivasan B. A generalized deep learning framework for whole-slide image segmentation and analysis. Sci Rep. 2021;11(1):1–14.
Al-Tam RM, Al-Hejri AM, Narangale SM, Samee NA, Mahmoud NF, Al-Masni MA, Al-Antari MA. A hybrid workflow of residual convolutional transformer encoder for breast cancer classification using digital X-ray mammograms. Biomedicines. 2022;10(11):2971.
Ibrahim A, Mohammed S, Ali HA, Hussein SE. Breast cancer segmentation from thermal images based on chaotic salp swarm algorithm. IEEE Access. 2020;8:122121–34.
Liu Q, Liu Z, Yong S, Jia K, Razmjooy N. Computer-aided breast cancer diagnosis based on image segmentation and interval analysis. Automatika. 2020;61(3):496–506.
Yu C, Chen H, Li Y, Peng Y, Li J, Yang F. Breast cancer classification in pathological images based on hybrid features. Multim Tools Appl. 2019;78:21325–45.
Ho DJ, Yarlagadda DV, D’Alfonso TM, Hanna MG, Grabenstetter A, Ntiamoah P, Brogi E, Tan LK, Fuchs TJ. Deep multi-magnification networks for multi-class breast cancer image segmentation. Comput Med Imaging Graph. 2021;88: 101866.
Huang Q, Huang Y, Luo Y, Yuan F, Li X. Segmentation of breast ultrasound image with semantic classification of superpixels. Med Image Anal. 2020;61: 101657.
Khamparia A, Bharati S, Podder P, Gupta D, Khanna A, Phung TK, Thanh DN. Diagnosis of breast cancer based on modern mammography using hybrid transfer learning. Multidimension Syst Signal Process. 2021;32:747–65.
Muduli D, Dash R, Majhi B. Automated diagnosis of breast cancer using multi-modal datasets: a deep convolution neural network based approach. Biomed Signal Process Control. 2022;71: 102825.
Shareef B, Xian M, Vakanski A. Stan: Small tumor-aware network for breast ultrasound image segmentation. In: IEEE 17th International Symposium on Biomedical Imaging (ISBI); 2020. p. 1–5.
Zhang G, Zhao K, Hong Y, Qiu X, Zhang K, Wei B. SHA-MTL: soft and hard attention multi-task learning for automated breast cancer ultrasound image segmentation and classification. Int J Comput Assist Radiol Surg. 2021;16:1719–25.
Xu Y, Wang Y, Yuan J, Cheng Q, Wang X, Carson PL. Medical breast ultrasound image segmentation by machine learning. Ultrasonics. 2019;91:1–9.
Xu Y, Hou S, Wang X, Li D, Lu L. A Medical Image Segmentation Method Based on Improved UNet 3+ Network. Diagnostics. 2023;13(3):576.
Leena Nesamani S, Nirmala Sugirtha Rajini S, Josphine MS, Jacinth Salome J. Deep learning-based mammogram classification for breast cancer diagnosis using multi-level support vector machine. In: Komanapalli VLN, Sivakumaran N, Hampannavar S, editors. Advances in Automation, Signal Processing, Instrumentation, and Control: Select Proceedings of i-CASIC 2020. Singapore: Springer; 2021. p. 371–83.
Salama WM, Aly MH. Deep learning in mammography images segmentation and classification: automated CNN approach. Alex Eng J. 2021;60(5):4701–9.
Vijayarajeswari R, Parthasarathy P, Vivekanandan S, Basha AA. Classification of mammogram for early detection of breast cancer using SVM classifier and Hough transform. Measurement. 2019;146:800–5.
AlGhamdi M, Abdel-Mottaleb M. DV-DCNN: Dual-view deep convolutional neural network for matching detected masses in mammograms. Comput Methods Programs Biomed. 2021;207: 106152.
Sha Z, Hu L, Rouyendegh BD. Deep learning and optimization algorithms for automatic breast cancer detection. Int J Imaging Syst Technol. 2020;30(2):495–506.
Zebari DA, Ibrahim DA, Zeebaree DQ, Mohammed MA, Haron H, Zebari NA, Damaševičius R, Maskeliūnas R. Breast cancer detection using mammogram images with improved multi-fractal dimension approach and feature fusion. Appl Sci. 2021;11(24):12122.
Maqsood S, Damaševičius R, Maskeliūnas R. TTCNN: A breast cancer detection and classification towards computer-aided diagnosis using digital mammography in early stages. Appl Sci. 2022;12(7):3273.
Li X, Cui J, Song J, Jia M, Zou Z, Ding G, Zheng Y. Contextual features and information bottleneck-based multi-input network for breast cancer classification from contrast-enhanced spectral mammography. Diagnostics. 2022;12(12):3133.
Kavitha T, Mathai PP, Karthikeyan C, Ashok M, Kohar R, Avanija J, Neelakandan S. Deep learning based capsule neural network model for breast cancer diagnosis using mammogram images. Interdiscip Sci. 2022;14:113–29.
Khan HN, Shahid AR, Raza B, Dar AH, Alquhayz H. Multi-view feature fusion based four views model for mammogram classification using convolutional neural network. IEEE Access. 2019;7:165724–33.
Song J, Zheng Y, Zakir Ullah M, Wang J, Jiang Y, Xu C, Zou Z, Ding G. Multiview multimodal network for breast cancer diagnosis in contrast-enhanced spectral mammography images. Int J Comput Assist Radiol Surg. 2021;16(6):979–88.
Acknowledgements
The Vijayanagara Sri Krishnadevaraya University, Ballari provided the facilities needed to conduct the research, which the authors gratefully acknowledged.
Funding
No funding received for this research.
Author information
Authors and Affiliations
Contributions
Both authors worked together to implement and evaluate the outcome of the research work.
Corresponding author
Ethics declarations
Conflict of interest
No conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the topical collection “Advances in Computational Approaches for Image Processing, Wireless Networks, Cloud Applications and Network Security” guest edited by P. Raviraj, Maode Ma and Roopashree H R.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kadadevarmath, J., Reddy, A.P. Improved Watershed Segmentation and DualNet Deep Learning Classifiers for Breast Cancer Classification. SN COMPUT. SCI. 5, 458 (2024). https://doi.org/10.1007/s42979-024-02642-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42979-024-02642-6