Abstract
lysA gene encoding meso-diaminopimelic acid (DAP) decarboxylase enzyme that catalyzes l-lysine biosynthesis in the aspartate pathway in Streptomyces clavuligerus was overexpressed, and its effects on cephamycin C (CephC), clavulanic acid (CA), and tunicamycin productions were investigated. Multicopy expression of lysA gene under the control of glpF promoter (glpFp) in S. clavuligerus pCOlysA led to higher expression levels ranging from 2- to 6-fold increase at both lysA gene and CephC biosynthetic gene cluster at T36 and T48 of TSBG fermentation. These results accorded well with CephC production. Thus, 1.86- and 3.14-fold higher volumetric as well as 1.26- and 1.71-fold increased specific CephC yields were recorded in S. clavuligerus pCOlysA in comparison with the wild-type and its control strain, respectively, at 48th h. Increasing the expression of lysA provided 4.3 times more tunicamycin yields in the recombinant strain. These findings suggested that lysA overexpression in S. clavuligerus made the strain more productive for CephC and tunicamycin. The results also supported the presence of complex interactions among antibiotic biosynthesis pathways in S. clavuligerus.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Streptomyces clavuligerus produces several medically important secondary metabolites such as the highly potent β-lactam antibiotic cephamycin C (CephC); clavulanic acid (CA), a β-lactamase inhibitor; and tunicamycin, a glycosylation inhibitor. Of these secondary metabolites, CephC is a naturally occurring cephamycin antibiotic and potent against penicillin-resistant bacteria via a methoxyl group at the C-7 position of the cephalosporin core. CephC is highly stable at neutral pH and used as a raw material to synthesize semisynthetic antibiotics such as cefoxitin, cefmetazole, temocillin, and cefotetan [1, 2]. CA is a β-lactamase inhibitor used in combination with β-lactam antibiotics such as amoxicillin (Augmentin™) and ticarcillin (Timentin™) against β-lactamase-producing bacteria [3]. Non-β-lactam compounds such as holomycin with pyrrolidine structure and tunicamycin with a glucosamine-containing structure are also produced by S. clavuligerus [2, 4]. Tunicamycin is a fatty acyl nucleoside-type antibiotic and composed of a 14-membered gene cluster lacking cluster-situated regulator (CSR) [5].
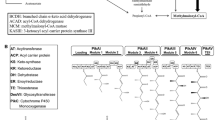
CephC biosynthesis pathway starts with the non-ribosomal condensation of l-α-aminoadipic acid (α-AAA), l-cysteine, and l-valine precursors to form delta-(l-alpha-aminoadipyl)-l-cysteinyl-d-valine (LLD-ACV). α-AAA is a catabolic product of l-lysine produced from the lysine branch of the aspartate metabolic pathway (Fig. 1) [7, 8]. l-Lysine is produced through decarboxylation of the cell wall precursor meso-diaminopimelate, catalyzed by the diaminopimelate (DAP) decarboxylase (E.C.4.1.1.20). This enzyme is encoded by lysA gene in the aspartate pathway. Then, l-lysine is converted to α-AAA in two steps of the CephC pathway by the enzymes encoded by lat and pcd genes [9, 10]. Therefore, availability of α-AAA is a limiting step for precursor flux from the aspartate pathway and for β-lactam biosynthesis [8, 11].
Aspartate pathway and its link to CephC pathway in S. clavuligerus [6]
Fermentation optimization or metabolic engineering approaches, or both, were used to overproduce CephC in the cell. The addition of l-lysine, its product, i.e., α-AAA, or its precursor, i.e., diaminopimelate, stimulated the production of CephC in S. clavuligerus [2, 11, 12]. Similarly, supplementation of the fermentation medium with l-lysine or diamines putrescine, cadaverine, and diaminopropane also enhanced cephamycin production in Nocardia lactamdurans, another cephamycin producer actinomycete [13, 14].
In addition to precursor supply to increase CephC yield in S. clavuligerus, there are some genetic engineering studies targeting biosynthetic or regulatory genes to improve the CephC production. For instance, Özcengiz et al. [15] introduced multicopy ask (aspartokinase) gene in the aspartate pathway into S. clavuligerus, leading to 3.3-fold higher specific CephC production in the recombinant strain. In addition, ccaR overproduction in S. clavuligerus resulted in 6.1-fold higher production of specific CephC [16]. Recently, it has been reported that overexpression of pimM gene from Streptomyces natalensis in S. clavuligerus resulted in a remarkable increase in CephC as well as CA and tunicamycin yields [17].
In S. clavuligerus, CephC and CA are synthesized simultaneously through different metabolic pathways [18]. Several reports indicated the positive impact of inducers and precursors as well as carbon or nitrogen sources in the fermentation media for the production of either compound, CephC or CA [19]. For instance, the culture supplemented with soluble starch as carbon source or cottonseed extract as complex nitrogen source enhanced CephC production while the glycerol feeding did not cause an increase in maximum CephC production [20]. In contrast, the glycerol feeding strongly enhanced CA as previously reported [21]. Interestingly, the variable concentration of 1,3-diaminopropane in the culture changes that connected production in the favor of either compound [13, 22].
The lysA gene of S. clavuligerus NRRL 3585 was previously PCR-amplified and characterized, and its sequence was deposited to the GenBank with the accession number of DQ078790.1 [6]. This study aimed at homologous multicopy expression of lysA gene in S. clavuligerus to determine its effect in the expression of CephC gene cluster and in the CephC yield. In order to see the effect of this manipulation more extensively, CA and tunicamycin productions were also evaluated in the lysA-overexpressed S. clavuligerus cells.
Materials and methods
Bacterial strains, plasmids, media, and culture conditions
Bacterial strains and plasmids used are given in Table 1. S. clavuligerus strains were grown in tryptic soy broth (TSB) medium at 28 °C and 220 rpm until the 1/10 diluted culture reached an optical density (OD600) of approximately 0.6. These seed cultures were used to inoculate (5%, v/v) 500-ml triple-baffled flasks containing 100 ml of TSB medium supplemented with 0.5% glycerol (to induce the pSPG glpF promoter, glpFp). The cultures were grown in triplicate, and duplicate samples were taken to quantify growth and antibiotic concentration. Cultures of S. clavuligerus pCOlysA and S. clavuligerus pSPG were supplemented with apramycin (50 μg/ml) to maintain the multicopy plasmids. To propagate Escherichia coli (E. coli), Luria broth (LB) or agar plates (LA) were used, and the strains were incubated at 37 °C. Antibiotics (ampicillin [100 μg/ml], kanamycin [25 μg/ml], chloramphenicol [25 μg/ml]) were added when needed. E. coli ESS 3235, Klebsiella pneumoniae ATCC 29665, and Bacillus subtilis ATCC 6633 strains were grown in LB at 30 °C and 200 rpm to an OD600 value of 0.9–1.0 before use in the CephC, CA, and tunicamycin bioassays, respectively. Mannitol soy flour (MS) agar [27] was used to grow exconjugants after conjugation.
Construction of S. clavuligerus pCOlysA and S. clavuligerus pSPG
The promoterless lysA gene was isolated by PCR as 1400-bp fragment from S. clavuligerus genomic DNA using the specific primers listed in Table 2. The lysA amplicon was subcloned to pGEM-T® Easy to give pCO5. Next, the lysA gene was released from pCO5 with NdeI-SpeI and ligated to downstream of glpF promoter in pSPG. The constructions were verified by restriction digestion, PCR, and sequencing of the PCR-amplified fragment. pSPG and pCOlysA were introduced by transformation into the methylation-deficient E. coli ET12567/pUZ8002 to bypass Streptomyces restriction barrier. Intergeneric conjugation between Streptomyces and E. coli was carried out as described by Flett et al. [28]. Exconjugants grown on MS agar were transferred to TSA containing appropriate antibiotics, and they were allowed to grow for up to 4 days at 28 °C. The presence of pSPG and pCOlysA in S. clavuligerus exconjugants was confirmed by PCR using Apr primers (Table 2) designed to amplify the apramycin resistance gene. The strain S. clavuligerus carrying the empty plasmid pSPG was the control strain in this work.
Nucleotide sequencing
The DNA sequencing was provided by BGI (Europe) through the Genoks Company using BigDye Cycle Sequencing Kit V3.1 (Applied Biosystems) of the Applied Biosystem and the chain termination method.
Fermentation and growth determination
Fermentation experiments were performed by growing three biological replicates of S. clavuligerus pSPG, S. clavuligerus pCOlysA, and S. clavuligerus NRRL3585 in the TSBG medium for 72 h. Two milliliters of samples taken from cultures with 12-h intervals was used for DNA quantification [29] and CephC and CA bioassays [16]. In the case of tunicamycin, 5 ml of the cultures were taken at every 12 h.
Antibiotic bioassays
CephC and CA concentrations were determined using culture supernatants while tunicamycin was extracted directly from the cultures prior to bioassay experiments [30, 31]. CephC, CA, and tunicamycin were quantified using the agar plate diffusion method [26]. CephC concentrations were calculated as cephalosporin C equivalents using a standard of cephalosporin C (Sigma). Potassium clavulanate (Sigma) and tunicamycin (Sigma) were used as standards to calculate the related antibiotic concentration in the samples.
RNA isolation and gene expression analyses
RNA was isolated from samples taken from cultures at 24th and 36th h of growth. RNA was purified using GeneJET RNA Purification Kit (Thermo Scientific) according to the manufacturer’s instructions. In addition, Phase Lock Gel Heavy (5 PRIME) columns were used to reduce sample loss during phenol extraction. DNase treatment of the isolated RNA samples was performed using a DNA-free™ kit (Thermo Scientific). RNA amount was determined by the NanoDrop® ND-2000 spectrophotometer (Thermo Scientific, Wilmington). High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) was used for cDNA synthesis. Reactions with SsoAdvanced™ SYBR® Green Supermix (Bio-Rad) for qRT-PCR analysis were performed on CFX96 Real-Time PCR (Bio-Rad). The primers designed previously [16] were used for qRT-PCR analysis. A standard qRT-PCR reaction included the following components: reaction buffer (5×) 4 μl; forward primer (10 μM), 0.4 μl; reverse primer (10 μM), 0.4 μl; cDNA, 1 μl; dH2O, 14.2 μl, in a final volume of 20 μl. The PCR reactions started with an initial denaturation step (30 s at 95 °C) and proceeded with 40 cycles of amplification (5 s at 95 °C and 30 s at 60 °C). In the qRT-PCR experiments, three biological and two technical replicates were used for each sample. hrdB (RNA polymerase sigma factor A) was used as the internal reference gene [16, 32]. The ΔΔCt method was chosen for calculation of the relative expression value of the genes [33].
Statistical analysis of qRT-PCR data
Two-way repeated ANOVA with Bonferroni’s post hoc testing was performed on the normalized gene expression to check whether expression data were statistically different between S. clavuligerus NRRL3585 (wild strain) and S. clavuligerus pCOlysA or between S. clavuligerus pCOlysA and S. clavuligerus pSPG. The GraphPad Prism software (GraphPad Software, San Diego, CA, USA) was used for statistical evaluation of qRT-PCR data. Error bars represent 95% confidence intervals. The statistical significance value in the graphs is indicated as ***p < 0.001, **p < 0.01, and *p < 0.05.
Results
Construction of S. clavuligerus pCOlysA
In this study, we constructed the S. clavuligerus pCOlysA strain for lysA overexpression. The presence of pCOlysA plasmid (Fig. 2a) in the recombinant strain was verified by PCR (Fig. 2b). The phenotypic observation of the recombinant strain together with the control strains is also provided in Fig. 3. As shown, the recombinant pCOlysA strain had similar mycelium formation and sporulation pattern as the wild type.
Vector map of recombinant pCOlysA constructed using Vector NTI Advance® 11.5 (a). PCR verification of S. clavuligerus pCOlysA (b). M, O’GeneRuler 1 kb plus DNA ladder (#SM1343); 1, negative control PCR in which S. clavuligerus genomic DNA was used as template for amplification of aac(3)IV-oriT amplicon; 2, aac(3)IV-oriT amplicon obtained by using S. clavuligerus pCOlysA DNA and AprF-R primer pair; 3, the positive control PCR using S. clavuligerus DNA as template for amplification of the lysA gene; 4, the lysA amplicon obtained using S. clavuligerus pCOlysA DNA as template and lysAF-R primer pair; 5, the negative control PCR using pSPG plasmid as template for amplification of the lysA gene
Analysis of the effect of lysA in the CephC biosynthesis genes
Multiple expressions of the lysA gene encoding meso-diaminopimelate decarboxylase under the control of glpFp in the pSPG replication vector in S. clavuligerus pCOlysA led to 1.5-fold (p < 0.001) increase in its own relative expression at T36. On the other hand, the relative expression values of lat gene, involved in the conversion of l-lysine to α-AAA in the CephC biosynthesis step, were 2.41 and 2 (T24 and T36), respectively, and were statistically significant (p < 0.001 and p < 0.01). At both sampling hours, the expression levels of pcbAB, cefD, and cefF biosynthetic genes showed a statistically significant increase ranging from 4 to 6 in S. clavuligerus pCOlysA compared with those of the control strain. The expression levels of other biosynthetic genes, i.e., pcbC, cefE, cmcH, and cmcI, showed 2- to 4-fold increase in T36 compared with those of the control strain (p < 0.01 or p < 0.001). Expression of the ccaR regulatory gene located in the pathway increased 2.85-fold, especially in T24, as well (p < 0.001).
Interestingly, the relative expression value of orf10, a gene of unknown function, compared with that of the control strain was determined as 2.85 (at T36). The orf10 gene is expressed in the ccaR–orf10–blp–lat–pcbAB–pcbC polycistronic transcript [34], and its higher expression might also be related to CcaR regulation. The pbpA, a gene encoding putative low molecular weight penicillin-binding protein (PBP), plays a role with pcbR providing autoresistance to β-lactam antibiotics. Therefore, in S. clavuligerus pCOlysA, relative expression values of pbpA and pcbR were found to be 2.15 (p < 0.05) and 1.60 (p < 0.001) at 36 h compared with those of the control strain. Another gene of resistance, the bla, was expressed 3.64 times more in S. clavuligerus pCOlysA than that in the control strain (p < 0.001). The blp gene, probably coding for a β-lactamase inhibitor protein, showed 1.32-fold higher expression value at 36th h. However, there was no statistically significant difference in the relative expression value of pcd, cmcJ, and cmcT genes in both hours (Fig. 4).
qRT-PCR results of the lysA and CephC gene cluster in S. clavuligerus pCOlysA and S. clavuligerus pSPG control strain compared with those in S. clavuligerus NRRL 3585. The gene expression values in the wild strain were taken as 1 (***p < 0.001, **p < 0.01, *p < 0.05). The genes that have p > 0.05 do not have any asterisk
Comparison of antibiotic (CephC, CA, and tunicamycin) production
Growth of S. clavuligerus pCOlysA was slightly higher at T72 than those of the other strains, and then gradually decreased depending on time (Fig. 5). When CephC production was compared, the highest volumetric CephC production belonged to S. clavuligerus pCOlysA as 100.75 μg/ml at 48th h. At the same time, the production values of S. clavuligerus NRRL 3585 and S. clavuligerus pSPG were 54.04 and 32.05 μg/ml, respectively. Thus, the multiple expressions of the lysA gene in S. clavuligerus pCOlysA were 1.86-fold more compared with those of the wild-type strain and 3.14 times more compared with those of the control strain. However, when the specific CephC productions were compared, the highest value was reached by S. clavuligerus pCOlysA at 48th h as in volumetric production. S. clavuligerus pCOlysA produced 1.29- and 1.71-fold higher specific CephC in comparison with those of wild-type and control strains (Fig. 6).
Regarding CA yields, S. clavuligerus pCOlysA provided the highest volumetric CA production as 114.87 μg/ml at T48, corresponding to 1.72-fold more yield compared with that of S. clavuligerus NRRL 3585. However, multiple expressions of the lysA gene under the control of glpFp in the recombinant strain did not have a positive effect on specific CA production as being almost the same in both strains.
In the case of tunicamycin, its production delayed to late hours of incubation, especially after T84. The highest production of tunicamycin was provided by S. clavuligerus pCOlysA at 120th h as 451.7 μg/ml, and this value corresponded to at least 4.5 times more volumetric tunicamycin than those of the controls. Similarly, the specific antibiotic yield of S. clavuligerus pCOlysA was very high at T108 and T120, as well. The multiple expressions of lysA in S. clavuligerus resulted in a very productive tunicamycin overproducer strain.
Discussion
There have been many examples of strain improvement strategies applied to S. clavuligerus aiming high levels of antibiotic production. Such strategies range from media optimization to genetic engineering approaches. However, there are limited reports in regard to enhancing CephC yields through the manipulation of aspartate pathway in S. clavuligerus. Yılmaz et al. [35] disrupted hom gene encoding homoserine dehydrogenase in the aspartate pathway in S. clavuligerus to see its effect in CephC yields. They showed that the hom mutant exerted 4.3-fold specific CephC production in chemically defined media. Later, the same group overexpressed multicopy of the ask gene, encodes aspartokinase involved in the first step of aspartate pathway in CephC biosynthesis, in S. clavuligerus NRRL 3585 and its hom mutant [15]. They reported that ask overexpression in the wild type resulted in an average of 3- and 1.8-fold increase in specific and volumetric CephC production in TSB and modified chemically defined medium, respectively, while hom-minus background significantly decreased CephC yields in both media probably due to enzyme overproduction or metabolic burden.
The deduced amino acid sequence of S. clavuligerus lysA gene exhibited significant homology to known DAP decarboxylases, especially those of actinobacteria such as Streptomyces coelicolor, Corynebacterium efficiens, and Mycobacterium tuberculosis [6]. LysR encoded by the lysR gene is located immediately upstream from lysA and was found to be a positive regulator in E. coli [36]. The lysR gene has also been determined by homology in S. coelicolor (U37580, GenBank). LysR-type transcriptional regulators (LTTRs) act on gene expressions in a similar way to the model regulator lysR that controls lysA expression in Enterobacteriaceae. Co-inducers are important for the function of LTTRs and probably contribute to a feedback loop. In that regulation, a product or intermediate of a given metabolic/synthesis pathway which is generally activated by an LTTR acts as the co-inducer required for transcriptional activation or repression [37, 38]. In our study, we applied a rational approach by homologous expression of the lysA gene under the control of glpFp in TSBG in S. clavuligerus. The lysA overexpression in the recombinant S. clavuligerus pCOlysA strain led to only 1.5-fold upregulation in its own expression level while the CephC gene cluster expression levels increased by 2- to 6-fold, indicating such a feedback loop could be possible in the control of lysA expression in the recombinant strain. Regarding increased CephC cluster expression levels, the expression results were in agreement with the production of CephC, which was almost 2-fold increase in the recombinant strain compared with that of the wild type.
CcaR is a common CSR of CephC and CA located at the CephC gene cluster. Several studies indicated a high correlation between CephC and CA. In addition to CSR regulation, pleiotropic and nutritional regulation mechanisms are also effective in the production of both metabolites. S. clavuligerus CcaR activator functions as a positive regulator in the CephC gene cluster by binding to cefD-cmcI bidirectional promoter, its own promoter, lat and cefF promoter regions [34]. The ccaR overexpression under the control of pglpF in TSBG in S. clavuligerus resulted in 6.1-fold increased specific CephC production compared with the wild type [16]. In addition, multicopy expression of ccaR under glpFp resulted in significantly improved industrial high-titer CA producer S. clavuligerus strains [39]. Thus, the increased expression of biosynthetic genes in S. clavuligerus pCOlysA might be indirectly associated with positive effect of the lysA overexpression on CcaR.
Cavallieri et al. [19] showed by flux balance analysis that lysine and maltose addition to chemically defined media increased both CephC and CA productions in S. clavuligerus. It was also shown that the addition of glycerol or other vegetable oils promotes CA production [40, 41]. In our study, the lysA overexpression and glycerol addition did not affect CA yields in the recombinant strain.
Tunicamycin is composed of a uracil, an N-acetylglucosamine (GlcNac), an unusual 11-carbon 2 aminoaldose sugar called tunicamine, and an amid-linked fatty acid. The unique tunicamine 11-carbon dialdose sugar backbone arises from a 5-carbon furanose precursor derived from uridine and a 6-carbon N-acetylamino-pyranose precursor derived from UDP-GlcNac. The α,β-1″,11′-glycosidic linkage between tunicamine and GlcNac is specific for this metabolite [31, 42]. Therefore, tunicamycin biosynthesis might improve with the supplementation of 5- and 6-carbon carbohydrate precursors or uridine, which might considered bottlenecks for tunicamycin biosynthesis as stated by Martínez-Burgo et al. [17]. They reported that tunicamycin overproduction in the pimM-overexpressed S. clavuligerus was not due to overexpression of tunicamycin biosynthetic gene cluster but might be a result of an increase in the supply of tunicamycin precursors.
Apart from this study [17], the tunicamycin-related studies mostly focused on structural and functional elucidation. Its regulation and interaction with other antibiotic metabolic pathways are still unknown. The absence of a CSR in the tunicamycin gene cluster makes its regulation mechanism more attractive [5, 43,44,45,46]. However, we have recently proposed a possible role of BldG pleiotropic regulator for tunicamycin in S. clavuligerus through qRT-PCR and bioassay studies by using bldG mutant [25] and overexpressed strains [47]. Consequently, this manipulation in S. clavuligerus through multicopy expression of the lysA gene under the control of inducible glycerol promoter led to a significant increase in the tunicamycin titer. This is the first report indicating that a manipulation of the aspartate pathway resulted in an elevated level of tunicamycin in the recombinant strain although the exact underlying mechanism has not known yet.
References
Brites LM, Oliveira LM, Barboza M (2013) Kinetic study on cephamycin C degradation. Appl Biochem Biotechnol 171:2121–2128. https://doi.org/10.1007/s12010-013-0502-x
Leite CA, Cavallieri AP, Araujo ML (2013) Enhancing effect of lysine combined with other compounds on cephamycin C production in Streptomyces clavuligerus. BMC Microbiol 13(1):296
Toro L, Pinilla L, Avignone-Rossa C, Ríos-Estepa R (2018) An enhanced genome-scale metabolic reconstruction of Streptomyces clavuligerus identifies novel strain improvement strategies. Bioprocess Biosyst Eng 41(5):657–669. https://doi.org/10.1007/s00449-018-1900-9
De La Fuente A, Lorenzana LM, Martin JM, Liras P (2002) Mutants of Streptomyces clavuligerus with disruptions in different genes for clavulanic acid biosynthesis produce large amounts of holomycin: possible cross-regulation of two unrelated secondary metabolic pathways. J Bacteriol 184(23):6559–6565. https://doi.org/10.1128/JB.184.23.6559-6565.2002
Wyszynski FJ, Hesketh AR, Bibb MJ, Davis BG (2010) Dissecting tunicamycin biosynthesis by genome mining: cloning and heterologous expression of a minimal gene cluster. Chem Sci 1(5):581–589
Yağcıoğlu Ç (2004) Cloning and characterization of Streptomyces clavuligerus meso-diaminopimelate decarboxylase (lysA) gene. Dissertation, Middle East Technical University
Kern BA, Hendlin D, Inamine E (1980) L-lysine-ε-aminotransferase involved in cephamycin C synthesis in Streptomyces lactamdurans. Antimicrob Agents Chemother 17:679–685
Tunca S, Yılmaz EI, Piret J, Liras P, Özcengiz G (2004) Cloning, characterization and heterologous expression of the aspartokinase and aspartate semialdehyde dehydrogenase genes of cephamycin C-producer Streptomyces clavuligerus. Res Microbiol 155(7):525–534
Romero J, Martín JF, Liras P, Demain AL, Rius N (1997) Partial purification, characterization and nitrogen regulation of the lysine epsilon-aminotransferase of Streptomyces clavuligerus. J Ind Microbiol Biotechnol 18(4):241–246
Pérez-Llarena FJ, Rodríguez-García A, Enguita FJ, Martín JF, Liras P (1998) The pcd gene encoding piperideine-6-carboxylate dehydrogenase involved in biosynthesis of α-aminoadipic acid is located in the cephamycin cluster of Streptomyces clavuligerus. J Bacteriol 180(17):4753–4756
Rius N, Maeda K, Demain AL (1996) Induction of L-lysine-ε-aminotransferase by L-lysine in Streptomyces clavuligerus, producer of cephalosporins. FEMS Microbiol Lett 144:207–211
Özcengiz G, Demain AL (2013) Recent advances in the biosynthesis of penicillins, cephalosporins and clavams and its regulation. Biotechnol Adv 31(2):287–311. https://doi.org/10.1016/j.biotechadv.2012.12.001
Leitão AL, Enguita FJ, De La Fuente JL, Liras P, Martin JF (1999) Inducing effect of diamines on transcription of the cephamycin C genes from the lat and pcbAB promoters in Nocardia lactamdurans. J Bacteriol 181(8):2379–2384
Leitão AL, Enguita FJ, Martín JF, Santos Oliveira JF (2001) Effect of exogenous lysine on the expression of early cephamycin C biosynthetic genes and antibiotic production in Nocardia lactamdurans MA4213. Appl Microbiol Biotechnol 56:670–675. https://doi.org/10.1007/s002530100715
Özcengiz G, Okay S, Ünsaldı E, Taşkın B, Liras P, Piret J (2010) Homologous expression of aspartokinase (ask) gene in Streptomyces clavuligerus and its hom-deleted mutant: effects on cephamycin C production. Bioengineered Bugs 1(3):191–197
Kurt A, Álvarez-Álvarez R, Liras P, Özcengiz G (2013) Role of the cmcH–ccaR intergenic region and ccaR overexpression in cephamycin C biosynthesis in Streptomyces clavuligerus. Appl Microbiol Biotechnol 97(13):5869–5880
Martínez-Burgo Y, Santos-Aberturas J, Rodríguez-García A, Barreales EG, Tormo JR, Truman AW, Reyes F, Aparicio JF, Liras P (2019) Activation of secondary metabolite gene clusters in Streptomyces clavuligerus by the PimM regulator of Streptomyces natalensis. Front Microbiol 10:580. https://doi.org/10.3389/fmicb.2019.00580
Alvarez-Álvarez R, Rodríguez-García A, Santamarta I, Pérez-Redondo R, Prieto-Domínguez A, Martínez-Burgo Y, Liras P (2014) Transcriptomic analysis of Streptomyces clavuligerus ΔccaR::tsr: effects of the cephamycin C-clavulanic acid cluster regulator CcaR on global regulation. Microb Biotechnol 7:221–231
Cavallieri AP, Baptista AS, Leite CA, da Costa Araujo MLG (2016) A case study in flux balance analysis: lysine, a cephamycin C precursor, can also increase clavulanic acid production. Biochem Eng J 112:42–53
Bellão C, Antonio T, Araujo MLGC, Badino AC (2013) Production of clavulanic acid and cephamycin C by Streptomyces clavuligerus under different fed-batch conditions. Braz J Chem Eng 30:257–266. https://doi.org/10.1590/S0104-66322013000200004
Saudagar PS, Singhal RS (2007) Optimization of nutritional requirements and feeding strategies for clavulanic acid production by Streptomyces clavuligerus. Bioresour Technol 98:2010–2017
Leite CA, Cavallieri AP, Baptista AS, Araujo ML (2016) Dissociation of cephamycin C and clavulanic acid biosynthesis by 1,3-diaminopropane in Streptomyces clavuligerus. FEMS Microbiol Lett 363(1):fnv215. https://doi.org/10.1093/femsle/fnv215
Kieser T, Bibb M, Buttner M, Chater K, Hopwood D (2000) Practical Streptomyces genetics. The John Innes Foundation, John Innes Centre, Norwich
Hu WS, Braña AF, Demain AL (1984) Carbon source regulation of cephem antibiotic production by resting cells of Streptomyces clavuligerus and its reversal by protein synthesis inhibitors. Enzyme Microb Tech 6(4):155–160. https://doi.org/10.1016/0141-0229(84)90023-1
Bignell DR, Tahlan K, Colvin KR, Jensen SE, Leskiw BK (2005) Expression of CcaR, encoding the positive activator of cephamycin C and clavulanic acid production in Streptomyces clavuligerus, is dependent on BldG. Antimicrob Agents Chemother 49(4):1529–1541
Romero J, Liras P, Martín JF (1984) Dissociation of cephamycin and clavulanic acid biosynthesis in Streptomyces clavuligerus. Appl Microbiol Biotechnol 20:318–325
Hobbs G, Frazer CM, Gardner DCJ, Cullum JA, Oliver SG (1989) Dispersed growth of Streptomyces in liquid culture. Appl Microbiol Biotechnol 31:272–277. https://doi.org/10.1007/s00253-018-8748-4
Flett F, Mersinias V, Smith CP (1997) High efficiency intergeneric conjugal transfer of plasmid DNA from Escherichia coli to methyl DNA-restricting streptomycetes. FEMS Microbiol Lett 155:223–229
Burton K (1968) Determination of DNA concentration with diphenylamine. Methods Enzymol 12:163–166
Tsvetanova BC, Price NP (2001) Liquid chromatography-electrospray mass spectrometry of tunicamycin-type antibiotics. Anal Biochem 289(2):147–156. https://doi.org/10.1006/abio.2000.4952
Tsvetanova BC, Kiemle DJ, Price NP (2002) Biosynthesis of tunicamycin and metabolic origin of the 11-carbon dialdose sugar, tunicamine. J Biol Chem 277(38):35289–35296
Aigle B, Wietzorrek A, Takano E, Bibb MJ (2000) A single amino acid substitution in region of the principal sigma factor of Streptomyces coelicolor A3(2) results in pleiotropic loss of antibiotic production. Mol Microbiol 37:995–1004
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25:402–408
Santamarta I, López-García MT, Kurt A, Nárdiz N, Alvarez-Álvarez R, Pérez-Redondo R, Martín JF, Liras P (2011) Characterization of DNA-binding sequences for CcaR in the cephamycin-clavulanic acid supercluster of Streptomyces clavuligerus. Mol Microbiol 81(4):968–981. https://doi.org/10.1111/j.1365-2958.2011.07743.x
Yılmaz EI, Çaydasi AK, Özcengiz G (2008) Targeted disruption of homoserine dehydrogenase gene and its effect on cephamycin C production in Streptomyces clavuligerus. J Ind Microbiol Biotechnol 35:1–7
Stragier P, Richaud F, Borne F, Patte JC (1983) Regulation of diaminopimelate decarboxylase synthesis in Escherichia coli. I. Identification of a lysR gene encoding an activator of the lysA gene. J Mol Biol 168:307–320
Tropel D, van der Meer JR (2004) Bacterial transcriptional regulators for degradation pathways of aromatic compounds. Microbiol Mol Biol Rev 68:474–500. https://doi.org/10.1128/MMBR.68.3.474-500.2004
Maddocks SE, Oyston PC (2008) Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology 154:3609–3623. https://doi.org/10.1099/mic.0.2008/022772-0
Kurt-Kızıldoğan A, Vanlı-Jaccard G, Mutlu A, Sertdemir G, Özcengiz G (2017) Genetic engineering of an industrial strain of Streptomyces clavuligerus for further enhancement of clavulanic acid production. Turk J Biol 41:342–353. https://doi.org/10.3906/biy-1608-17
Ser HL, Law JWF, Chaiyakunapruk N, Jacob SA, Jacob SA, Palanisamy UD, Chan KG, Goh BH, Lee LH (2016) Fermentation conditions that affect clavulanic acid production in Streptomyces clavuligerus: a systematic review. Front Microbiol 7:522. https://doi.org/10.3389/fmicb.2016.00522
Kurt Kızıldoğan A (2017) The effect of different vegetable oils on clavulanic acid production in Streptomyces clavuligerus. Anadolu J Agr Sci 32(2):223–228. https://doi.org/10.7161/omuanajas.287668
Price NP, Tsvetanova B (2007) Biosynthesis of the tunicamycins: a review. J Antibiot (Tokyo) 60(8):485–491
Winn M, Goss RJ, Kimura KI, Bugg TD (2010) Antimicrobial nucleoside antibiotics targeting cell wall assembly: recent advances in structure– function studies and nucleoside biosynthesis. Nat Prod Rep 27(2):279–304
Karki S, Kwon SY, Kwon HJ (2011) Cloning of tunicamycin biosynthetic gene cluster from Streptomyces chartreusis NRRL3882. J Korean Soc Appl Biol Chem 54(1):136–140
Wyszynski FJ, Lee SS, Yabe T, Wang H, Gomez-Escribano JP, Bibb MJ, Lee SJ, Davies GJ, Davis BG (2012) Biosynthesis of the tunicamycin antibiotics proceeds via unique exo-glycal intermediates. Nat Chem 4(7):539–546
Widdick D, Royer SF, Wang H, Vior NM, Gomez-Escribano JP, Davis BG, Bibb MJ (2018) Analysis of the tunicamycin biosynthetic gene cluster of Streptomyces chartreusis reveals new insights into tunicamycin production and immunity. Antimicrob Agents Chemother 62:e00130–e00118. https://doi.org/10.1128/AAC.00130-18
Baş L, Otur Ç, Kurt-Kızıldoğan A Enhanced tunicamycin biosynthesis in BldG overexpressed Streptomyces clavuligerus. Appl Biochem Microbiol (in press)
Acknowledgments
We acknowledge Sezer Okay for critical reading of the manuscript.
Funding
This study was supported by Ondokuz Mayıs University Research Fund (Samsun, Turkey) with the project ID number of PYO.ZRT.1902-A.15.001.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Responsible Editor: Gisele Monteiro.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Otur, Ç., Kurt-Kızıldoğan, A. Homologous expression of lysA encoding diaminopimelic acid (DAP) decarboxylase reveals increased antibiotic production in Streptomyces clavuligerus. Braz J Microbiol 51, 547–556 (2020). https://doi.org/10.1007/s42770-019-00202-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42770-019-00202-2