Abstract
The genus Streptomyces have been highly regarded for their important source of natural products. Combined with the technology of genome sequencing and mining, we could identify the active ingredients from fermentation broth quickly. Here, we report on Streptomyces sp. strain fd1-xmd, which was isolated from a soil sample collected in Shanghai. Interestingly, the fermentation broth derived from this strain demonstrated broad-spectrum antimicrobial activity against gram-positive bacteria, gram-negative bacteria, and eukaryotes. To identify the antimicrobial substances and their biosynthetic gene clusters, we sequenced the fd1-xmd strain and obtained a genome 7,929,999 bp in length. The average GC content of the chromosome was 72.5 mol%. Knockout experiments demonstrated that out of eight biosynthetic gene clusters we could identify, two are responsible for the biosynthesis of the antibiotics streptothricin (ST) and tunicamycin (TM). The ST biosynthetic gene cluster from fd1-xmd was verified via successful heterologous expression in Streptomyces coelicolor M1146. ST production had a yield of up to 0.5 g/L after the optimization of culture conditions. This study describes a novel producer of ST and TM and outlines the complete process undertaken for Streptomyces sp. strain fd1-xmd genome mining.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Approximately half of bioactive microbial metabolites are isolated from microbes, and nearly 70% of these products are derived from Actinobacteria (Berdy 2005). The genus Streptomyces, belonging to the family Streptomycetaceae, is the largest genus in the domain Bacteria and currently encompasses 823 species and 38 subspecies with validly published names at the time of writing (http://www.bacterio.net/streptomyces.html). This genus is aerobic and gram-positive, and most members possess aerial and substrate mycelia (Waksman and Henrici 1943). Streptomyces members also are characterized by complex, regulated secondary metabolism networks and produce many valuable products, such as antibiotics, enzyme inhibitors, vitamins, antitumor agents, and antifungal compounds. Because Streptomyces has important commercial and medicinal value in medicine, agriculture, food, chemical industry, and other fields, research of Streptomyces remains important (Goodfellow and Fiedler 2010; Labeda et al. 2012).
The majority of secondary metabolites are rarely found and tested due to their instability and presence in only trace amounts. Many members of Streptomyces have been exploited for antibiotic production by genome sequencing means. Sequencing of the Streptomyces coelicolor A3(2) genome presents a typical case: more than 20 clusters coding for known or predicted secondary metabolites were detected. These metabolites hold a great deal of value for the fields of medicine and agriculture (Bentley et al. 2002).
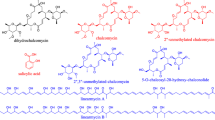
Streptothricins (STs) are a group of natural products featuring a unique streptolidine base and a 10-carbamoylated gulosamine, such as nourseothricins, albothricin, and LL-AC541 (Borders et al. 1970; Ohba et al. 1986; Romer et al. 1986; Vantamelen et al. 1961). Three new 12-carbamoylated STs have been isolated from the Streptomyces sp. I08A 1776 culture broth in 2012, supplement the members of ST family (Gan et al. 2012). The biosynthetic pathway of the carbamoylated D-GulN moiety was also clear in 2015 (Guo et al. 2015). ST was first isolated from Streptomyces lavendulae grown in simple medium in flasks (Waksman 1943; Waksman and Woodruff 1942). Then, many members of the ST family have been identified, including STs A to F, ST X, with different numbers of residues of L-β-lysine homopolymer chains, as well as members with similar chemical structures (Ji et al. 2007; Khokhlov and Reshetov 1964; Kim et al. 1994). The active ingredient in the S. lavendulae broth was identified as ST F, the first member of the ST family (Kusumoto et al. 1982a, b). Consider the distribution of antibiotic biosynthetic pathways; ST is found in about 10% of random soil Actinomycetes and gets a lot of attention because of a broad spectrum of antibacterial activities against gram-positive, gram-negative bacteria, and pathogenic fungi (Baltz 2007; Waksman 1943; Waksman and Woodruff 1942). It inhibits protein biosynthesis in prokaryotic cells by inhibiting polypeptide synthesis via ribosome interactions (Haupt et al. 1980; Inamori et al. 1990; Takemoto et al. 1980). ST is not currently used therapeutically in humans due to its inherent toxicity, including nephrotoxicity (Hartl et al. 1986; Hoffmann et al. 1986a, b; Witte 2000). Although this molecule elicits delayed toxicity, it exerts a real and significant impact on agriculture. Zhongshengmycin, of which ST is one of the main components, brought great economic value to China in the 1990s (Zhu et al. 2002). ST resistance was also used to be selection marker in plant cell research (Jelenska et al. 2000).
Tunicamycin (TM) was first discovered in Streptomyces lysosuperificus, and this glucosamine-containing antibiotic acts against gram-positive bacteria, yeast, fungi, and viruses (Kenig and Reading 1979; Takatsuki et al. 1971). Takatsuki published the structure of TM in 1977. TM mixtures contain four homologous antibiotics, TMs A–D, which differ in the length of the carbon chain of the trans α/β-unsaturated iso-fatty acid component (Takatsuki et al. 1977). TM specifically inhibits the dolichol pyrophosphate-mediated glycosylation of asparaginyl residues in glycoproteins. This molecule is also a peptidoglycan and a nucleoside glycolipid inhibitor, which blocks the transfer of GlcNAc-P from UDP-GlcNAc to dolichol phosphate (BillotKlein et al. 1997; Olden et al. 1979). TM is widely applied and is used to treat cancer by inducing endoplasmic reticulum stress (Xu et al. 2009).
In this study, we obtained a complete genome sequence of Streptomyces sp. strain fd1-xmd, which was isolated from soil in Shanghai. We identified the ability of this strain to produce ST and TM by performing genome mining and liquid chromatograph-mass spectrometer (LC-MS), generating knockout mutants and inducing heterologous expression. Simultaneously, biosynthetic ST and TM gene clusters were found by performing bioinformatic analysis of our experimental data. Through this study, we provide a preliminary procedure for the mining of known natural bioactive products with genome sequence and a resource material for analyzing the biosynthesis of the antibiotics ST and TM.
Materials and methods
Isolation, purification, and morphologic observation of strain fd1-xmd
Strain fd1-xmd was isolated from a soil sample collected at the campus of Fudan University, Shanghai, China, in 2013. The strain was recovered on Gause’s synthetic agar medium (starch 20 g, KNO3 1 g, K2HPO4 0.5 g, MgSO4 0.25 g, NaCl 0.5 g, FeSO4·7H2O, agar 15 g, deionized water 1 L, pH 7.2–7.4) (Gause et al. 1983) and was purified on glucose-yeast-maltose medium (GYM; glucose 4 g, yeast extract 4 g, malt extract 10 g, CaCO3 2 g, agar 15 g, deionized water 1 L, pH 7.2–7.4) (Shima et al. 1996) at 28 °C. A small piece of cover glass was inserted into the Gause’s synthetic agar medium after fd1-xmd spore was coated on the plate evenly. Spore chain morphology and spore surface ornamentation were observed from the cover glass after incubation at 28 °C for 14 days using a cold-field emission scanning electron microscope (New Generation SU8010, SU8000 Series, Hitachi, Japan).
Genome sequencing, mining, and bioinformatic analysis
The fd1-xmd genome was extracted using a bacterial genomic DNA isolation kit (Generay, Shanghai). This genome was sequenced by PacBio RSII system, gaining 400,388 reads with average length 2476 bp. All reads were assembled by HGAP program providing a 124-fold coverage of the whole genome. Then, the initial genome with 21 contigs was amended by Illumina PE reads with 148-fold coverage based on bowtie2 and SAMtools software (Langmead et al. 2009; Li et al. 2009). Last Sanger-based sequencing was employed to facilitate gap closing, and final sequence assembly was conducted using the Phred/Phrap/Consed package. Relationships between contigs were visualized using the ContigScape plugin (Tang et al. 2013). This genome annotation was based on the NCBI Prokaryotic Genome Annotation Pipeline (PGAP) (Tatusova et al. 2016). Potential secondary metabolites of strain fd1-xmd were analyzed using the secondary metabolite predict tool antiSMASH (Blin et al. 2013). Phylogenetic trees were constructed with the neighbor-joining method using MEGA5 software package (Saitou and Nei 1987; Tamura et al. 2011) and was evaluated using the bootstrap resampling method with 1000 repeats (Felsenstein 1985). Circular genome maps were generated using GenomeViz and DNAplotter (Carver et al. 2009; Ghai et al. 2004). Comparative analyses of gene clusters were based on the Artemis and ACT tools (Carver et al. 2008). Genome-wide colinearity analysis was performed using gepard (Krumsiek et al. 2007).
Construction of fd1-xmd gene cluster deletion mutants
All bacterial strains and plasmids used in this study are listed (Supporting Information, Table S1). Plasmids pFTB100-10 and pFTB100-27 were used to knock out the TM and ST biosynthetic gene clusters, respectively. The backbone vector was cloned from the plasmid pFDZ100 using the primer pair pFTB100-F/R. The resistance genes were cloned from pZB101 using the primer pair aphII-F/R. The homologous arms of two clusters were cloned from the genome of fd1-xmd using the primers 10-up-F/R, 10-down-F/R, 27-up-F/R, and 27-down-F/R. An additional eight plasmids, pFTB100-10-Apra, pFTB100-29-Apra, pFTB100-39-Apra, pFTB100-42-Apra, pFTB100-45-Apra, pFTB100-59-Apra, pFTB100-8-1-Apra, and pFTB100-8-2-Apra, were constructed for second-round knockout experiments. The double mutant plasmids differed simply in the alteration from kanamycin to apramycin using the primer pair aac-F/R and pSET152 as the template. The 16 primer pairs used to amplify the homologous arms of gene clusters are listed (Supporting Information, Table S2). The vector, two homologous arms, and the resistance gene were ligated using an In-Fusion Cloning Kit (ClonExpress MultiS One-Step Cloning Kit, Vazyme, Nanjing). The plasmids were transferred to strain fd1-xmd through conjugation with the help of Escherichia coli ET 12567 (Flett et al. 1997). Viable knockout clones grew on a mannitol soya flour agar (MS; soya flour 20 g, mannitol 20 g, agar 15 g, deionized water 1 L, pH 7.2–7.4) plate in the presence of kanamycin or apramycin (Kieser et al. 2000). Knockout was confirmed by performing PCR analysis using 2× Taq Master Mix (Novoprotein Scientific, Shanghai).
Construction of the plasmid containing complete streptothricin biosynthetic gene cluster
Two plasmids, p27-clone-up and p27-clone-down, containing homologous arms of the ST biosynthetic gene cluster and attB/attP sites of phage PhiBT1 were built to obtain the complete gene cluster from the fd1-xmd genome. The construction approach followed methods that described by Dai et al. (2015). The upstream DNA fragment (2015 bp) and downstream DNA fragment (2016 bp) of the ST gene cluster were amplified using the purified fd1-xmd genome as a template and 27-clone-up-F/R and 27-clone-down-F/R as primers. A MunI restriction site was added to the upstream PCR product via the primer 27-clone-up-F. MunI and XbaI restriction sites were added to the downstream PCR product via the primers 27-clone-down-F/R. Then, the upstream DNA fragment containing the MunI restriction site was ligated to plasmid pMD19-T (TaKaRa, Dalian) to form a circular plasmid named pT-stre-up. Plasmid pT-stre-up was cut into two fragments using the restriction enzymes MunI and XbaI. The 4699-bp fragment was chosen for the following steps. Plasmid pBAC-DZY101 was digested with the NheI and EcoRI restriction endonucleases. The 9500-bp enzyme-digested product derived from pBAC-DZY101 was purified and ligated to the 4699-bp fragment using T4 DNA ligase (Promega, Beijing). The downstream DNA fragment was also digested with XbaI and MunI and then ligated to a 3129-bp DNA fragment digested with XbaI and EcoRI from plasmid pCY104-SET152 using T4 DNA ligase. The newly obtained circular plasmids were transformed into E. coli DH10B cells. The final products isolated from transformed cells were named p27-clone-up and p27-clone-down.
Then, these two plasmids were introduced into fd1-xmd. Following homologous recombination, p27-clone-up and p27-clone-down were inserted into the upstream and downstream of ST gene cluster, respectively, obtained the strain fd1-xmd-up-down. The genome of fd1-xmd-up-down was extracted and reacted with integrase PhiBT1 for half an hour at 30 °C to cyclize the DNA between attB-attP sites (Zhang et al. 2008). The reaction mixture was transformed into DH10B to obtain the plasmid pDRX-27 containing the complete ST biosynthetic gene cluster.
Shake flask fermentation
Spores of S. coelicolor M1146, the mutant carrying the ST gene cluster, strain fd1-xmd, and the knockout mutants were inoculated in 3 mL of Luria broth (LB; tryptone 10 g, yeast extract 5 g, NaCl 10 g, deionized water 1 L) at 30 °C with shaking (220 rpm) (Gomez-Escribano and Bibb 2011). After 48 h, the cultures were transferred into 100 mL of rich tryptone soya broth medium (TSB; 45 g TSB, deionized water 1 L) (OXOID, UK) containing bead rings in 250-mL shake flasks at 30 °C with shaking (200 rpm) for 2 days.
Antibacterial assay
ST and TM standard samples were purchased from the Enzo Life Sciences and Sangon Biotech, respectively. Different concentrations of the standard samples were diluted with deionized water or methanol. Strain Micrococcus luteus, Bacillus cereus, Saccharomyces cerevisiae, Escherichia coli, Staphylococcus aureus, Mycobacterium smegmatis, and Rhizopus nigricans were used as indicator stains for antibacterial bioactivity tests involving ST and TM. Micrococcus luteus, Bacillus cereus, and Escherichia coli were cultured in LB liquid medium at 37 °C. Saccharomyces cerevisiae was cultured in YPD liquid medium (BD, USA) at 30 °C. Staphylococcus aureus and Rhizopus nigricans were cultured in nutrient broth medium (BD, USA) at 37 and 30 °C, respectively. Mycobacterium smegmatis was cultured in 7H9/10 medium (BD, USA) at 37 °C. Approximately 500 μL of the indicator strain broth was evenly mixed with appropriate agar medium in square petri dishes. Spores of Rhizopus nigricans were coated on the surface of agar plate evenly. Several pieces of round filter paper were placed on the agar plate surfaces. Fermentation broth samples obtained from fd1-xmd, M1146, and other mutant strains were centrifuged, and then, 10–30 μL of the supernatant and the standard samples were slowly dropped onto the dry filter paper pieces. Inhibition zones were observed after incubating the plates at 37 or 30 °C for 12 h. We determined inhibition zone size using a Vernier caliper.
Initial isolation and identification of streptothricin and tunicamycin
HP20 resins were used for the initial isolation of ST and TM from fermentation broth of strain fd1-xmd. Then, 100 mL of the supernatant of fermentation broth was incubated with 1% HP20 resins shaking at 30 °C, 200 rpm. After 12 h, the resins were collected by standard sieves (100 mesh) and analyzed with 10 mL of methanol at 30 °C, 200 rpm for 12 h.
QTRAP 5500 Instrument (AB Sciex, USA) and LC-30A system (Shimadzu, Japan) were used for ST qualitative analysis. Test samples were separated with solution A (ultrapure water with 1% formic acid) and B (acetonitrile with 1% formic acid) at a flow rate of 0.3 mL/min. The column was Waters Acquity UPLC C18 BEH 2.1 × 150 mm, with a 1.7-μm particle diameter. The injection volume was 10.00 μL each time. The characteristic peaks (m/z) of ST F were 503.400 and 171.100. For ST ESI analysis, UltiMate™ 3000 (Thermo, USA) and Compact™ (BRUKER, Germany) were used under the column Agilent Poroshell 120 EC C18 4.6 × 100 mm, with a 2.7-μm particle diameter. The column was programmed to run maintaining 10% B for 10 min.
1290 Infinity (Agilent Technologies, USA) and 6530 Accurate-Mass Q-TOF LC/MS (Agilent Technologies, USA) were used for TM ESI analysis. Waters Acquity UPLC HSS T3 2.1 × 100 mm, with a 1.8-μm particle diameter, and a water/acetonitrile gradient were used as mobile phase. The gradient started with 2% acetonitrile for 1 min. The percent of acetonitrile was increased to 98% in the next 6 min. The flow rate of the mobile phase was 0.5 mL/min, and the injection volume was 10.00 μL each time. The characteristic peak (m/z) of TM A was 817.4077, TM B was 931.4234, TM C was 845.4390, and TM D was 859.4547.
Accession number
The genome sequence of Streptomyces sp. strain fd1-xmd has been deposited in the GenBank/EMBL/DDBJ database under accession number CP019798.
Results
Characteristics of Streptomyces sp. strain fd1-xmd
Streptomyces sp. strain fd1-xmd, isolated in Shanghai, is aerobic and gram-positive with a milky-white aerial mycelium and a light yellow substrate mycelium. The color of spores is purple gray. And the strain makes light yellow pigment that diffuses in the agar (Fig. 1a, b). It grows well on Gause’s synthetic medium at 28 °C. A colony is visible 1 week after inoculation under appropriate culture conditions. The aerial mycelium is clearly differentiated into straight segmented chains of spores with a smooth surface as observed under a scanning electron microscope (Fig. 1c, d). Wild-type fd1-xmd produces biologically active substances that inhibit the growth of Micrococcus luteus, Bacillus cereus, and Saccharomyces cerevisiae. Strain fd1-xmd was deposited at CGMCC (China General Microbiological Culture Collection Center) under the accession CGMCC 4.7344.
Complete genome sequence of strain Streptomyces sp. strain fd1-xmd and phylogenetic analysis
We report the annotated complete genome sequence of strain fd1-xmd, which is 7,929,999 bp in length (Fig. 2a). The genomic DNA GC content of this strain is 72.5 mol%. The entire genome contains 6996 coding sequences with an average length of 984 bp, 72 tRNA genes, and 7 complete rRNA operons. To identify strain fd1-xmd, neighbor-joining tree based on the nearly complete 16S rRNA gene, gyrB gene, and recA gene sequences was constructed (Fig. 3, Fig. S1). The 16S rRNA analysis showed that the isolated strain clustered with Streptomyces amritsarensis JCM 19660T, S. globosus LMG 19896T, S. toxytricini NBRC 12823T, S. yangpensis fd2-tbT, S. flavotricini NBRC 12770T, S. racemochromogenes NRRL B-5430T, and S. katrae NBRC 13447T. The gyrB analysis showed that fd1-xmd clustered with Streptomyces katrae S3, Streptomyces goshikiensis ATCC 23914, and Streptomyces sp. Mg1. The recA analysis showed that fd1-xmd clustered with Streptomyces goshikiensis NRRL B-5428, Streptomyces sp. Mg1, Streptomyces nojiriensis AS 4.1897, and Streptomyces katrae S3. In conclusion, we consider that the closest strain to fd1-xmd is Streptomyces katrae and Streptomyces sp. Mg1 in phylogenetic classification.
The complete genome of Streptomyces sp. strain fd1-xmd and comparison with Streptomyces sp. Mg1. a From outside to inside: predicted genes color-coded based on COG categories of the forward and reverse strands, gene clusters, RNA, GC content, and GC skew. b Genome-wide colinearity analysis between Streptomyces sp. strain fd1-xmd and Streptomyces sp. Mg1
Neighbor-joining tree based on analysis of the nearly complete 16S rRNA gene sequence showing phylogenetic relationship between strain fd1-xmd and other nearby members of the genus Streptomyces. Kitasatospora nipponensis HKI 0315T was used as an outgroup. GenBank accession numbers are shown in brackets. Bootstrap support for each node is indicated as a percentage calculated from 1000 randomly resampled datasets. Bar, 0.005 substitutions per nucleotide position. Streptomyces sp. Mg1, shown in bond, is the most homologous strain with strain fd1-xmd between all Streptomyces strains with complete genome
Twenty-five putative gene clusters that produce different types of natural products were predicted (Table 1), specifically four NRPSs, one T1-PKS, one T2-PKS, one T3-PKS, three siderophores, one lassopeptide, four terpenes, one melanin-terpene, one butyrolactone, one lantipeptide, one bacteriocin, one bacteriocin-NRPS, one nucleoside, one T1pks-lantipeptide, one butyrolactone-T1PKS, one terpene-NRPS, and one T1PKS-NRPS. Most of these biosynthetic gene clusters were located at both ends of the chromosome, which are non-core regions ranging from 0 to 1.5 Mb and 6.0 to 7.9 Mb. These results are consistent with our colinearity analysis of the complete genomes of Streptomyces sp. strain fd1-xmd and Streptomyces sp. Mg1 (CP011664.1); these two strains share greater consistency from 1.5 to 6.0 Mb (Fig. 2b). Also, we found that Streptomyces sp. Mg1 is the most closely related complete genome with strain fd1-xmd based on blast against nt database with long fragment sequence.
Identification of the streptothricin and tunicamycin biosynthetic gene clusters
To identify the antibacterial substances produced in the fermentation broth of strain fd1-xmd, a series of mutant strains were constructed. We found that the active ingredient has high water solubility since we isolated strain fd1-xmd. And NRPS products are more likely to show water solubility than PKS and other gene products. We choose eight possible gene clusters for first and second-round knocking-out experiments according to better water solubility and high integrity. These biosynthetic gene clusters encode TM (named as 10 in this paper), ST (named as 27), and six other NRPSs (named as 29, 39, 42, 45, 59, 8-1/8-2, respectively; two deletion mutant strains 8-1/8-2 were constructed because the eighth T1PKS-NRPS biosynthetic gene cluster is too large). We deleted the key synthetase genes B1K54_RS324595, B1K54_RS324600, and B1K54_RS324605 in the ST gene cluster, which caused the inhibition zone on the Micrococcus luteus plate, Saccharomyces cerevisiae plate, and Escherichia coli plates to disappear, indicating that the active substance inhibiting the growth of Micrococcus luteus and Saccharomyces cerevisiae was ST. However, the inhibition zone for the ST gene cluster minus mutant strain fd1-xmd-ko27 became smaller on the Bacillus cereus plate but did not disappear. We then knocked out the key genes B1K54_RS26950 and B1K54_RS26955 in the TM gene cluster as well as other predicted NRPSs in the ST gene cluster knockout mutants (Fig. 4). We tested the inhibition zones caused by fermentation broth samples derived from all mutant and wild-type strains. Inhibition zone sizes for both the TM and ST gene cluster knockout strains decreased significantly compared with the zone for the ST gene cluster knockout strain. However, the zones for the other six mutants did not obviously change.
The inhibition zone of fd1-xmd and other knockout mutant strains on Micrococcus luteus (a), Bacillus cereus (b), Saccharomyces cerevisiae (c), and Escherichia coli (d) plates. First line: wild-type fd1-xmd; second line: the left three fd1-xmd-ko10, the right three fd1-xmd-ko27; third line: the left three fd1-xmd-ko27-10, the right three fd1-xmd-ko27-29; fourth line: the left three fd1-xmd-ko27–39, the right three fd1-xmd-ko27-42; fifth line: the left three fd1-xmd-ko27-45, the right three fd1-xmd-ko27-59; sixth line: the left three fd1-xmd-ko27-8-1, the right three fd1-xmd-ko27-8-2. We added 10 μL of liquid fermentation broth to each filter paper on Micrococcus luteus and Bacillus cereus plates and 20 μL on Saccharomyces cerevisiae plate. Each mutant strain has three replications with the except of the wild-type strain, which has six replications
In order to identify the antimicrobial substances more deeply, we used HP20 resins to isolate ST and TM from fermentation broth of strain fd1-xmd and other three knockout strains. From the result of antibacterial assay shown in Fig. S2, we could draw the conclusion that HP20 resins absorb TM but no ST. ESI analysis of concentrated methanol extract of HP20 resins from fd1-xmd and supernatant of fd1-xmd fermentation broth absorbed by HP20 resins shows that ST-F, TM-A, TM-B, TM-C, and TM-D were detected successfully. Thus, these results indicate that the observed biological activity is due to the production of TM and ST.
Bioinformatic analysis of the streptothricin and tunicamycin biosynthetic gene clusters in fd1-xmd and other related strains
After genome annotation and gene cluster prediction, the ST and TM biosynthetic gene clusters were identified. We selected from a wide range of putative members of the corresponding biosynthetic clusters. The sequence of the ST gene cluster of strain fd1-xmd exhibited high homology to the sequences AB684619 (Streptomyces rochei NBRC 12908), KC935381 (Streptomyces sp. TP-A0356), and KF498701 (Streptomyces lavendulae subsp. lavendulae strain BCRC 12163) (Fig. 5a) (Chang et al. 2014; Li et al. 2013; Maruyama et al. 2012). We identified 21 ORFs ranging from B1K54_RS32535 to B1K54_RS32635 that completed the ST gene cluster, which is similar to AB684619 and KC935381 but differs from KF498701 in that it lacks the stnS gene.
Comparison of the ST and TM biosynthetic gene cluster. a Homology analysis of the ST biosynthetic gene cluster among strain fd1-xmd, Streptomyces rochei, Streptomyces sp. TP-A0356, and Streptomyces lavendulae subsp. lavendulae. b Homology analysis of the TM biosynthetic gene cluster among fd1-xmd, Streptomyces chartreusis strain NRRL 3882, and Streptomyces clavuligerus strain ATCC 27064
The similarity between the sequence of the TM gene cluster of strain fd1-xmd and HQ111437 (Streptomyces chartreusis strain NRRL 3882) was far greater than the similarity between the tunicamycin gene cluster and CM000913 (Streptomyces clavuligerus ATCC 27064) (Fig. 5b) (Chen et al. 2010; Medema et al. 2010). The TM gene cluster of strain fd1-xmd contains 12 ORFs that appear to lie in a single operon ranging from B1K54_RS26960 to B1K54_RS27015.
Isolation and heterologous expression of the streptothricin gene cluster
The plasmid pDRX-27 containing the complete ST gene cluster was constructed, with a length of 41,818 bp (Fig. 6a). Plasmids obtained from two colonies were digested with PstI and contained the correct digested products (Fig. 6b). Then, plasmids pZB101 (Zhang et al. 2013) and pDRX-27 was transferred into Streptomyces coelicolor M1146 through conjugation in turn. Recombined by the pZB101 encoded PhiC31 integrase, pDRX-27 was inserted into the genome of M1146. These viable colonies were picked and validated. The newly obtained strain with complete ST gene cluster was named M1146-stre. The fermentation broth from M1146 and M1146-stre was used to test ST bioactivity. We clearly observed inhibition zones on the Micrococcus luteus, Bacillus cereus, Staphylococcus aureus, Mycobacterium smegmatis, Escherichia coli, and Rhizopus nigricans plates (Fig. 6c, d, Fig. S3). We also obtained an ST yield of nearly 0.5 g/L, based on the size of the inhibition zone (Supporting Information, Table S3). A peak of ST F at 1.80 min was observed in the LC-MS trace for the M1146-stre fermentation sample, which was the same peak observed for the standard sample (Fig. 6e). Based on these results, we conclude that fd1-xmd contains a complete ST gene cluster, and the product is bioactive.
Heterologous expression of the ST biosynthetic gene cluster in Streptomyces coelicolor M1146. a Map of the plasmid pDRX-27 in which heterologous expression of the ST biosynthetic gene cluster was performed. b Restriction enzyme map of pDRX-27 under PstI. The inhibition zones obtained with different concentrations of ST standard samples and mutant strains on Micrococcus luteus (c) and Bacillus cereus (d) plates. First line: concentrations of the standard samples from the left to the right are 0.2, 0.5, 1.0, 2.0, and 5.0 g/L. We added 10 μL of each standard sample to each filter paper. Second line: wild-type Streptomyces coelicolor M1146. We added 10 μL of liquid fermentation broth to each filter paper. Lines 3 to 5 represent three replicates for which 10, 20, and 30 μL of liquid fermentation broth derived from M1146-stre were added to each filter paper. e LC-MS analysis of ST production in S. coelicolor M1146 and M1146-stre. The first line is the standard sample
Discussion
With the development of sequencing technologies and specifically high-throughput sequencing instruments, whole-genome sequencing of microbes has become very common and convenient. Complete genome sequences are quickly constructed by obtaining long reads using PacBio sequencing technology and filling in gaps with short reads obtained using Illumina sequencing technology. The price of sequencing is also acceptable to most labs. The use of genome sequencing technology to explore novel and unknown microbial resources carried high potential. An increasing number of natural products were determined from uncultured microorganisms through genome sequencing and mining. On the other hand, some kinds of bioactive products could not be identified because the very low yield, genome sequencing, and subsequent bioinformatic analysis may solve the problem. In addition to identifying new types of antibiotics and other bioactive substances, important strain information may also be obtained from complete genome sequences. Additionally, it is possible to modify the metabolic networks of microorganisms based on their genomic information. Strain fd1-xmd produces bioactive substances that inhibit the growth of gram-positive bacteria, gram-negative bacteria, and eukaryotes. These characteristics intrigued all of us. After performing sequencing and assembly, we reported the complete genome sequence of strain fd1-xmd and identified two complete functional gene clusters: TM and ST.
An interesting point is that strain fd1-xmd has seven complete rRNA operons, but more than half strains of Streptomyces have only six complete rRNA operons. This can also be a reference for studies of systematics and evolution of genus Streptomyces.
The inhibition zone of Bacillus cereus did not disappear when key genes in the TM and ST gene clusters were completely knocked out. This indicates that there may exist other unknown bioactive natural products in the fermentation broth of strain fd1-xmd. We will explore these bioactive substances of strain fd1-xmd by performing genome mining and experiments in the future. Another significant point to be considered is that an obvious peak at 1.50 min was observed in the LC-MS trace for the M1146-stre fermentation sample. More experimental work will be done in follow-up study.
Heterologous expression of the ST gene cluster from strain fd1-xmd in S. coelicolor M1146 was successfully achieved with a yield as high as 0.5 g/L. Bead rings play a great role on the production. The yield is only half of the normal level without the bead rings when fermentation. Aerial mycelium and substrate mycelium of Streptomyces may form mycelial pellets in the liquid fermentation medium. Bead rings could smash the mycelial pellets and benefit the growth of strain fd1-xmd. It is noteworthy that in other studies, the production of ST was achieved after 7 to 12 days of growth, whereas good production levels of heterologous expression were obtained after only 2 days in our study. This discovery can greatly reduce the production cycle of ST and suggest that strain M1146-stre be a very good producer of ST.
In conclusion, we isolated Streptomyces sp. strain fd1-xmd and demonstrated the ability of this strain to naturally produce TM and ST based on the results of an antibacterial assay and LC-MS. A combined PacBio-Illumina approach was implemented to obtain the complete genome sequence of Streptomyces sp. strain fd1-xmd. We identified the complete TM and ST biosynthetic gene clusters of fd1-xmd and compared them with other related strains. We also utilized an additional genetic source for TM and ST production. We believe that this work represents a complete scientific study, from strain isolation, to genome sequencing and mining, to gene cluster knockout, and finally to successful heterologous expression. Thus, this study provides a preliminary procedure for the mining of known natural bioactive products combine with genome sequence.
References
Baltz RH (2007) Antimicrobials from actinomycetes: back to the future. Microbe 2(3):125–131
Bentley SD, Chater KF, Cerdeno-Tarraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, Bateman A, Brown S, Chandra G, Chen CW, Collins M, Cronin A, Fraser A, Goble A, Hidalgo J, Hornsby T, Howarth S, Huang CH, Kieser T, Larke L, Murphy L, Oliver K, O’Neil S, Rabbinowitsch E, Rajandream MA, Rutherford K, Rutter S, Seeger K, Saunders D, Sharp S, Squares R, Squares S, Taylor K, Warren T, Wietzorrek A, Woodward J, Barrell BG, Parkhill J, Hopwood DA (2002) Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417(6885):141–147. https://doi.org/10.1038/417141a
Berdy J (2005) Bioactive microbial metabolites. J Antibiot 58(1):1–26. https://doi.org/10.1038/ja.2005.1
BillotKlein D, Shlaes D, Bryant D, Bell D, Legrand R, Gutmann L, van Heijenoort J (1997) Presence of UDP-N-acetylmuramyl-hexapeptides and -heptapeptides in enterococci and staphylococci after treatment with ramoplanin, tunicamycin, or vancomycin. J Bacteriol 179(15):4684–4688. https://doi.org/10.1128/jb.179.15.4684-4688.1997
Blin K, Medema MH, Kazempour D, Fischbach MA, Breitling R, Takano E, Weber T (2013) antiSMASH 2.0—a versatile platform for genome mining of secondary metabolite producers. Nucleic Acids Res 41(W1):W204–W212. https://doi.org/10.1093/nar/gkt449
Borders DB, Sax KJ, Lancaste JE, Hausmann WK, Mitscher LA, Wetzel ER, Patterson EL (1970) Structures of Ll-Ac541 and Ll-Ab664—new streptothricin-type antibiotics. Tetrahedron 26(13):3123–3133. https://doi.org/10.1016/S0040-4020(01)92895-9
Carver T, Berriman M, Tivey A, Patel C, Bohme U, Barrell BG, Parkhill J, Rajandream MA (2008) Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics 24(23):2672–2676. https://doi.org/10.1093/bioinformatics/btn529
Carver T, Thomson N, Bleasby A, Berriman M, Parkhill J (2009) DNAPlotter: circular and linear interactive genome visualization. Bioinformatics 25(1):119–120. https://doi.org/10.1093/bioinformatics/btn578
Chang CY, Lyu SY, Liu YC, Hsu NS, Wu CC, Tang CF, Lin KH, Ho JY, Wu CJ, Tsai MD, Li TL (2014) Biosynthesis of streptolidine involved two unexpected intermediates produced by a dihydroxylase and a cyclase through unusual mechanisms. Angew Chem Int Edit 53(7):1943–1948. https://doi.org/10.1002/anie.201307989
Chen WQ, Qu DJ, Zhai LP, Tao MF, Wang YM, Lin SJ, Price NPJ, Deng ZX (2010) Characterization of the tunicamycin gene cluster unveiling unique steps involved in its biosynthesis. Protein Cell 1(12):1093–1105. https://doi.org/10.1007/s13238-010-0127-6
Dai RX, Zhang B, Zhao GP, Ding XM (2015) Site-specific recombination for cloning of large DNA fragments in vitro. Eng Life Sci 15(6):655–659. https://doi.org/10.1002/elsc.201400267
Felsenstein J (1985) Confidence-limits on phylogenies—an approach using the bootstrap. Evolution 39(4):783–791. https://doi.org/10.1111/j.1558-5646.1985.tb00420.x
Flett F, Mersinias V, Smith CP (1997) High efficiency intergeneric conjugal transfer of plasmid DNA from Escherichia coli to methyl DNA-restricting streptomycetes. FEMS Microbiol Lett 155(2):223–229. https://doi.org/10.1111/j.1574-6968.1997.tb13882.x
Gan ML, Zheng XD, Liu YF, Guan Y, Xiao CL (2012) Three new 12-carbamoylated streptothricins from Streptomyces sp I08A 1776. Bioorg Med Chem Lett 22(19):6151–6154. https://doi.org/10.1016/j.bmcl.2012.08.003
Gause GF, Preobrazhenskaya TP, Sveshnikova MA, Terekhova LP, Maximova TS (1983) A guide for the determination of Actinomycetes. Nauka, Moscow (in Russian)
Ghai R, Hain T, Chakraborty T (2004) GenomeViz: visualizing microbial genomes. BMC Bioinf 5(1):198. https://doi.org/10.1186/1471-2105-5-198
Gomez-Escribano JP, Bibb MJ (2011) Engineering Streptomyces coelicolor for heterologous expression of secondary metabolite gene clusters. Microb Biotechnol 4(2):207–215. https://doi.org/10.1111/j.1751-7915.2010.00219.x
Goodfellow M, Fiedler HP (2010) A guide to successful bioprospecting: informed by actinobacterial systematics. Antonie Van Leeuwenhoek 98(2):119–142. https://doi.org/10.1007/s10482-010-9460-2
Guo Z, Li J, Qin H, Wang M, Lv X, Li X, Chen Y (2015) Biosynthesis of the carbamoylated D-gulosamine moiety of streptothricins: involvement of a guanidino-N-glycosyltransferase and an N-acetyl-D-gulosamine deacetylase. Angew Chem Int Edit 54(17):5175–5178. https://doi.org/10.1002/anie.201412190
Hartl A, Guttner J, Stockel U, Hoffmann H (1986) Acute and subchronic toxicity of nourseothricin in laboratory animals. Arch Exp Vet 40(5):727–735
Haupt I, Jonak J, Rychlik I, Thrum H (1980) Action of streptothricin-F on ribosomal functions. J Antibiot 33(6):636–641. https://doi.org/10.7164/antibiotics.33.636
Hoffmann H, Hartl A, Bocker H, Kuhnel HJ, Hesse G, Flemming J (1986a) Pharmacokinetics of nourseothricin in laboratory animals. Arch Exp Vet 40(5):699–709
Hoffmann H, Kirchner E, Knappe H, Hillesheim HG, Hartl A, Hubler D, Chemnitius KH, Morgenstern E, Grupe R (1986b) Pharmacological action profile of nourseothricin. Arch Exp Vet Med 40(5):710–720
Inamori Y, Amino H, Tsuboi M, Yamaguchi S, Tsujibo H (1990) Biological activities of racemomycin-B, beta-lysine rich streptothricin antibiotic, the main component of Streptomyces-Lavendulae Op-2. Chem Pharm Bull 38(8):2296–2298. https://doi.org/10.1248/cpb.38.2296
Jelenska J, Tietze E, Tempe J, Brevet J (2000) Streptothricin resistance as a novel selectable marker for transgenic plant cells. Plant Cell Rep 19(3):298–303. https://doi.org/10.1007/s002990050016
Ji ZQ, Wang MA, Zhang JW, Wei SO, Wu WJ (2007) Two new members of streptothricin class antibiotics from Streptomyces qinlingensis sp nov. J Antibiot 60(12):739–744. https://doi.org/10.1038/ja.2007.96
Kenig M, Reading C (1979) Holomycin and an antibiotic (Mm-19290) related to tunicamycin, metabolites of Streptomyces-Clavuligerus. J Antibiot 32(6):549–554. https://doi.org/10.7164/antibiotics.32.549
Khokhlov AS, Reshetov PD (1964) Chromatography of streptothricins on carboxymethylcellulose. J Chromatogr 14(3):495–496. https://doi.org/10.1016/S0021-9673(00)86662-5
Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA (2000) Practical Streptomyces genetics. John Innes Foundation, Norwich
Kim BT, Lee JY, Lee YY, Kim OY, Chu JH, Goo YM (1994) N-Methylstreptothricin-D—a new streptothricin-group antibiotic from a Streptomyces spp. J Antibiot 47(11):1333–1336. https://doi.org/10.7164/antibiotics.47.1333
Krumsiek J, Arnold R, Rattei T (2007) Gepard: a rapid and sensitive tool for creating dotplots on genome scale. Bioinformatics 23(8):1026–1028. https://doi.org/10.1093/bioinformatics/btm039
Kusumoto S, Imaoka S, Kambayashi Y, Shiba T (1982a) Total synthesis of antibiotic streptothricin-F. Tetrahedron Lett 23(29):2961–2964. https://doi.org/10.1016/S0040-4039(00)87506-1
Kusumoto S, Kambayashi Y, Imaoka S, Shima K, Shiba T (1982b) Total chemical-structure of streptothricin. J Antibiot 35(7):925–927. https://doi.org/10.7164/antibiotics.35.925
Labeda DP, Goodfellow M, Brown R, Ward AC, Lanoot B, Vanncanneyt M, Swings J, Kim SB, Liu Z, Chun J, Tamura T, Oguchi A, Kikuchi T, Kikuchi H, Nishii T, Tsuji K, Yamaguchi Y, Tase A, Takahashi M, Sakane T, Suzuki KI, Hatano K (2012) Phylogenetic study of the species within the family Streptomycetaceae. Antonie Van Leeuwenhoek 101(1):73–104. https://doi.org/10.1007/s10482-011-9656-0
Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10(3):R25. https://doi.org/10.1186/gb-2009-10-3-r25
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Proc GPD (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25(16):2078–2079. https://doi.org/10.1093/bioinformatics/btp352
Li JE, Guo ZY, Huang W, Meng XX, Ai GM, Tang GL, Chen YH (2013) Mining of a streptothricin gene cluster from Streptomyces sp TP-A0356 genome via heterologous expression. Sci China Life Sci 56(7):619–627. https://doi.org/10.1007/s11427-013-4504-2
Maruyama C, Toyoda J, Kato Y, Izumikawa M, Takagi M, Shin-ya K, Katano H, Utagawa T, Hamano Y (2012) A stand-alone adenylation domain forms amide bonds in streptothricin biosynthesis. Nat Chem Biol 8(9):791–797. https://doi.org/10.1038/Nchembio.1040
Medema MH, Trefzer A, Kovalchuk A, van den Berg M, Muller U, Heijne W, Wu LA, Alam MT, Ronning CM, Nierman WC, Bovenberg RAL, Breitling R, Takano E (2010) The sequence of a 1.8-Mb bacterial linear plasmid reveals a rich evolutionary reservoir of secondary metabolic pathways. Genome Biol Evol 2:212–224. https://doi.org/10.1093/gbe/evq013
Ohba K, Nakayama H, Furihata K, Furihata K, Shimazu A, Seto H, Otake N, Yang ZZ, Xu LS, Xu WS (1986) Albothricin, a new streptothricin antibiotic. J Antibiot 39(6):872–875. https://doi.org/10.7164/antibiotics.39.872
Olden K, Pratt RM, Jaworski C, Yamada KM (1979) Evidence for role of glycoprotein carbohydrates in membrane-transport-specific inhibition by tunicamycin. Proc Natl Acad Sci U S A 76(2):791–795. https://doi.org/10.1073/pnas.76.2.791
Romer W, Hesse G, Miosga N, Fricke H (1986) Chemical determination of the streptothricin antibiotic nourseothricin. Arch Exp. Vet Med 40(5):693–698
Saitou N, Nei M (1987) The neighbor-joining method—a new method for reconstructing phylogenetic trees. Mol Biol Evol 4(4):406–425
Shima J, Penyige A, Ochi K (1996) Changes in patterns of ADP-ribosylated proteins during differentiation of Streptomyces coelicolor A3(2) and its developmental mutants. J Bacteriol 178(13):3785–3790. https://doi.org/10.1128/jb.178.13.3785-3790.1996
Takatsuki A, Arima K, Tamura G (1971) Tunicamycin, a new antibiotic. 1. Isolation and characterization of tunicamycin. J Antibiot 24(4):215–223. https://doi.org/10.7164/antibiotics.24.215
Takatsuki A, Kawamura K, Okina M, Kodama Y, Ito T, Tamura G (1977) Structural elucidation of tunicamycin. 2. Structure of tunicamycin. Agric Biol Chem Tokyo 41(11):2307–2309
Takemoto T, Inamori Y, Kato Y, Kubo M, Morimoto K, Morisaka K, Sakai M, Sawada Y, Taniyama H (1980) Physiological-activity of streptothricin antibiotics. Chem Pharm Bull 28(10):2884–2891. https://doi.org/10.1248/cpb.28.2884
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28(10):2731–2739. https://doi.org/10.1093/molbev/msr121
Tang B, Wang Q, Yang MJ, Xie F, Zhu YQ, Zhuo Y, Wang SY, Gao H, Ding XM, Zhang LX, Zhao GP, Zheng HJ (2013) ContigScape: a Cytoscape plugin facilitating microbial genome gap closing. BMC Genomics 14(1):289. https://doi.org/10.1186/1471-2164-14-289
Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J (2016) NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res 44(14):6614–6624. https://doi.org/10.1093/nar/gkw569
Vantamelen E, Whitfield GB, Dyer JR, Carter HE, Whaley HA (1961) Constitution of the streptolin-streptothricin group of Streptomyces antibiotics. J Am Chem Soc 83(20):4295–4296. https://doi.org/10.1021/Ja01481a051
Waksman SA (1943) Production and activity of streptothricin. J Bacteriol 46(3):299–310
Waksman SA, Henrici AT (1943) The nomenclature and classification of the actinomycetes. J Bacteriol 46(4):337–341
Waksman SA, Woodruff HB (1942) Streptothricin, a new selective bacteriostatic and bactericidal agent active against gram-negative bacteria. P Soc Exp Biol Med 49(2):207–210. https://doi.org/10.3181/00379727-49-13515
Witte W (2000) Selective pressure by antibiotic use in livestock. Int J Antimicrob Agents 16:S19–S24
Xu HL, Inagaki Y, Seyama Y, Sugawara Y, Kokudo N, Nakata M, Wang FS, Tang W (2009) Expression of KL-6 mucin, a human MUC1 mucin, in intrahepatic cholangiocarcinoma and its potential involvement in tumor cell adhesion and invasion. Life Sci 85(9–10):395–400
Zhang L, Ou XJ, Zhao GP, Ding XM (2008) Highly efficient in vitro site-specific recombination system based on Streptomyces phage phi BT1 integrase. J Bacteriol 190(19):6392–6397. https://doi.org/10.1128/Jb.00777-08
Zhang B, Zhang L, Dai RX, Yu MY, Zhao GP, Ding XM (2013) An efficient procedure for marker-free mutagenesis of S. coelicolor by site-specific recombination for secondary metabolite overproduction. PLoS One 8(2):e55906. https://doi.org/10.1371/journal.pone.0055906
Zhu CX, Jiang XL, Sun DY, Ji HJ, Tian YL, Xie DL, Ni CF (2002) Zhongshengmycin, a new agro-antibiotics. Fine Specialty Chem 16:14–17 (In Chinese)
Funding
This work was financially supported by grants from the National Key R&D Program of China (2016YFA0500600), the Basic Research Project of Science and Technology Commission of Shanghai Municipality (17JC1401800), and the Youth Talent Training Program of Zhejiang Academy of Agricultural Sciences (2017R19R08E01). This work was also supported by grant from the Natural Science Foundation for the Youth (31700007).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Ethical approval
This article does not contain any studies with human participants performed by any of the authors.
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
ESM 1
(PDF 582 kb)
Rights and permissions
About this article
Cite this article
Yu, Y., Tang, B., Dai, R. et al. Identification of the streptothricin and tunicamycin biosynthetic gene clusters by genome mining in Streptomyces sp. strain fd1-xmd. Appl Microbiol Biotechnol 102, 2621–2633 (2018). https://doi.org/10.1007/s00253-018-8748-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-018-8748-4