Abstract
The aspartate pathway of Streptomyces clavuligerus is an important primary metabolic pathway which provides substrates for β-lactam synthesis. In this study, the hom gene which encodes homoserine dehydrogenase was cloned from the cephamycin C producer S. clavuligerus NRRL 3585 and characterized. The fully sequenced open reading frame encodes 433 amino acids with a deduced M r of 44.9 kDa. The gene was heterologously expressed in the auxotroph mutant Escherichia coli CGSC 5075 and the recombinant protein was purified. The cloned gene was used to construct a plasmid containing a hom disruption cassette which was then transformed into S. clavuligerus. A hom mutant of S. clavuligerus was obtained by insertional inactivation via double crossover, and the effect of hom gene disruption on cephamycin C yield was investigated by comparing antibiotic levels in culture broths of this mutant and in the parental strain. Disruption of hom gene resulted in up to 4.3-fold and twofold increases in intracellular free l-lysine concentration and specific cephamycin C production, respectively, during stationary phase in chemically defined medium.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
β-lactams constitute a very important class of antibiotics, and improvement of β-lactam yields in the producer microorganisms is of great economical importance. For this purpose, there have been many metabolic engineering attempts dealing with the β-lactam biosynthetic genes and gene clusters as reviewed by Thykaer and Nielsen [27].
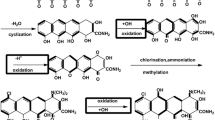
Streptomyces clavuligerus is the producer of the medically important β-lactam antibiotics, including cephamycin C and the potent β-lactamase inhibitor clavulanic acid. The aspartate pathway of S. clavuligerus is an important primary metabolic pathway which provides substrates for β-lactam synthesis (Fig. 1). The carbon flow through the l-lysine-specific branch of aspartate pathway is a rate-limiting step for the formation of cephamycin C [12, 15, 19]. Since several branch points exist in the aspartate pathway, the flow of carbon from aspartate through l-lysine to α-aminoadipyl side chain of cephamycins might be controlled by regulatory mechanisms operating at the initial and branching steps of pathway [11, 19]. Formation of l-homoserine from aspartate-β-semialdehyde is the first step of the other branch of the pathway leading to l-threonine, l-isoleucine and l-methionine syntheses and catalyzed by homoserine dehydrogenase (HSD, EC 1.1.1.3). The gene encoding homoserine dehydrogenase (hom) has been cloned and sequenced from diverse bacteria including Corynebacterium glutamicum [23], Lactobacillus lactis [14], Bacillus subtilis [22], Bacillus sp. ULM1 [16], Pseudomonas aeruginosa [5], Methylobacillus glycogens [21], Methylobacillus flagellatus [18], Esherichia coli and Salmonella typhimurium [6] and Streptomyces sp. NRRL 5331 [7].
Regulation of the activity or biosynthesis of HSD in S. clavuligerus might determine the availability of aspartate-β-semialdehyde for the biosynthesis of diaminopimelate, l-lysine and α-aminoadipic acid [19]. Such a regulation is expected to control the levels of l-threonine which together with l-lysine inhibits aspartokinase, the key enzyme in the control of carbon flow towards α-aminoadipic acid. Hence, the absence of HSD activity in S. clavuligerus is expected to lead to an increase in cephamycin C production levels in two ways. Firstly, all the carbon coming from aspartate would be directed to l-lysine rather than being shared between the two branches; and secondly, concerted feedback inhibition of aspartokinase would be relieved.
In this paper, we describe cloning and characterization of the hom gene from S. clavuligerus NRRL 3585 and some properties of the HSD enzyme expressed in E. coli. The cloned gene was used to disrupt the hom gene in S. clavuligerus via cassette mutagenesis, and the effect of this disruption on cephamycin C yield was investigated.
Materials and methods
Bacterial strains, plasmids, media and culture conditions
Bacterial strains and plasmids used in this study are described in Table 1. E. coli cultures were grown in either Luria Broth (LB) or on Luria agar plates at 37 °C. E. coli CGSC 5075 was grown on M9 medium [25] supplemented with l-methionine and l-threonine (50 μg/ml, each).
Streptomyces clavuligerus was maintained on sporulation agar [10] supplemented with CoCl2 6H2O (20 μg/ml). For isolation of genomic or plasmid DNA and for protoplast preparation, seed culture media containing trypticase soy broth (TSB, Oxoid) supplemented with 0.5% (w/v) maltose were inoculated with either spore or mycelium stocks of S. clavuligerus and incubated at 28 °C on a rotary shaker (220 rpm) in baffled flasks for 24 h. About 5 ml of this seed culture was inoculated into 50 ml of 2:3 (v/v) mixture of TSB:YEME [10] supplemented with 0.3% (v/v) glycine and 3 mM MgCl2 and incubated at 28 °C for 24 h. In the case of plasmid-containing cultures, ampicillin (100 μg/ml, Sigma), kanamycin (25 μg/ml for E. coli and 200 μg/ml for S. clavuligerus, Sigma) or thiostrepton (50 μg/ml, Sigma) was added into the medium. For antibiotic selection, Streptomyces colonies were replica plated on TSA (3 g TSB; 1.5 g agar per 100 ml) supplemented with 8 μg/ml thiostrepton or 200 μg/ml kanamycin. MM [10] containing glycerol and (NH4)2SO4 was used to test auxotrophy in S. clavuligerus. For HSD and cephamycin C assay, 100 ml of chemically defined medium (CDM) [15] was inoculated with a mid-log phase seed culture to give an initial OD595 of 0.03–0.04, and the cells were grown for 96 h. MM and CDM were supplemented with l-methionine (50 μg/ml) and l-threonine (50 μg/ml) for growth of S. clavuligerus AK39.
DNA isolation and manipulations
Genomic DNA was isolated from S. clavuligerus as described by Keiser et al. [10]. Plasmid DNA was isolated using Plasmid Midi Kits (Qiagen). DNA fragments were isolated from agarose gels using the Quickgel Extraction Kits (Qiagen). Restriction enzyme digestion of DNA was carried out as specified by the manufacturers. PCR products were cloned into pGEM-T vector prior to cloning into the desired plasmid vector. DNA manipulations in E. coli were performed as described by Sambrook et al. [25]. E. coli plasmids that were used in construction of recombinant S. clavuligerus plasmids were propagated in E. coli ET12567 to avoid restriction barriers. Southern blot hybridization analysis was performed by standard procedures using AlkPhos Direct labeling and detection kit (Amersham).
Transformation of S. clavuligerus
PEG-mediated protoplast transformation technique [10] with slight modifications was used to transform S. clavuligerus. Transformants were regenerated on R2YE medium [10] at 26 °C for 48 h, and then each plate was overlaid with soft nutrient agar [10] containing antibiotics.
Cloning of S. clavuligerus hom gene
A 1.3 kb DNA fragment containing the hom gene was amplified by PCR using the nucleotide primers 5′-AGGATCCATGATGCTGACGCGTCCG-3′ and 5′-TAAGCTTTTACTCCCCTTCAACACG-3′ and S. clavuligerus genomic DNA as a template. Primer design was based on homologous sequences from other known hom genes. The PCR amplification condition was as follows: 95 °C (5 min) and 30 cycles of 95 °C (1 min), 63 °C (1 min) and 72 °C (2 min). The amplified DNA was cloned into pGEM-T, digested with BamHI and HindIII enzymes and inserted into BamHI–HindIII digested pQE-30 expression vector. This allowed the expression of the gene under the control of T5 promoter and subsequent purification of 6×His-tagged protein. The resulting recombinant plasmid, designated pEBH1, was used to transform E. coli CGSC 5075 which lacks HSD activity. hom complementation was assessed on M9 medium containing 100 μg/ml ampicillin in the absence of l-methionine and l-threonine.
Insertional inactivation of hom gene via double crossing over
Kanamycin resistance cassette was obtained from the plasmid pK19 by PCR amplification of kan gene with SacII restriction site-tagged primers; 5′-GCCGCGGGAACACGTAGAAAGCCAGT-3′ and 5′-CCCGCGGTCAGAAGAACTCGTCAAGA-3′. The resulting 1.1 kb kan gene was inserted into the 1.3 kb hom gene contained in plasmid pEBH1 at unique SacII site giving rise to plasmid pEBHK. 2.4 kb BamHI–HindIII fragment of pEBHK was ligated to BamHI–HindIII digested plasmid pIJ486 which was then used for the transformation of S. clavuligerus protoplasts. Transformants selected for kanamycin resistance were replica plated on sporulation agar. Following five rounds of sporulation, ThioS–KanR colonies were selected. Such cells were next tested on MM for methionine–threonine auxotrophy.
HSD extraction, purification and enzyme assay
In E. coli cells containing recombinant plasmid pEBH1, hom gene was expressed as follows: The cells were grown in LB at 37 °C until OD600 reached 0.4–0.5. Induction was performed by the addition of 1 mM isopropyl-l-thiogalactopyranoside (IPTG, Sigma), and the cells were grown for three more hours at 37 °C to reach an OD600 of 1.5. Cells were centrifuged at 7,000g for 10 min (Sigma) and washed with cold sodium-buffer (50 mM, pH 7.5). The cell pellet was resuspended in 50 mM NaH2PO4, 300 mM NaCl, and 10 mM imidazole, pH 8.0. The cells were lysed by ultrasonication (Ultrasonic Processor, Cole Parmer) and following centrifugation at 25,000g for 30 min, the pellet was discarded. Native His-tagged protein was purified using Ni-NTA agarose column (Qiagen) according to the manufacturer’s instructions. Purity of the protein was visualized by SDS-PAGE [13] after Coomassie Blue staining of the gels. To test HSD activity in S. clavuligerus, cell-free extracts were prepared as mentioned above.
The HSD activity in both E. coli and S. clavuligerus cells was measured in the reverse reaction by determining the initial rate of increase of A 340 at room temperature. Standard assay mixture contained 0.2 ml of 500 mM Tris–HCl buffer containing 5 mM EDTA, pH 8.4; 0.1 ml of 100 mM l-homoserine, pH 7.0; 0.1 ml of 4 mM NADP solution, 0.5 ml distilled water and 0.1 ml of cell-free extract or purified enzyme. For crude extracts, absorbance at 340 nm was measured against a blank containing all the components except the substrate. Specific activity of HSD was expressed as micromoles of NADPH formed per min per mg of protein. Protein concentration was measured by the method of Bradford [4] with BSA as the standard.
Determination of cultural growth
Cultural growth was monitored by dry cell weight (DCW) determination. For this, a 5 ml of culture was passed through a cellulose nitrate membrane filter (0.2 μm pore size, Whatman) under vacuum. Filters were dried at 80 °C for 24 h and weighed. DCW values tabulated herein represented the mean of three independent measurements.
Bioassay of β-lactam antibiotics
β-lactam antibiotic bioassay was conducted by using agar plate diffusion method with E. coli ESS as the indicator organism [1]. Zones of inhibition were measured, and the amount of cephamycin C in samples was calculated by using the standard curve constructed with cephalosporin C (Sigma) as the standard. One unit of β-lactam produces an inhibition zone equivalent to that formed by 1 g of cephalosporin C.
Determination of intracellular-free l-lysine concentration
Cells were grown in CDM, and samples were collected at intervals during cultivation. Intracellular-free amino acids were extracted as described by Betancort Rodriguez et al. [3]. Following automated pre-column derivatization with o-phthaladehyde (OPA), amino acid analysis was performed by conventional reverse-phase HPLC (Shimadzu VP series) on a Novapac C18 column (Waters) followed by fluorescence detection. l-lysine concentration was expressed as micromoles of l-lysine extracted from 1 g wet weight of bacteria.
DNA sequencing and sequence analysis
DNA sequencing was performed by the dideoxy chain-termination method using an ABI Prism 377 instrument (Perkin–Elmer). The data were analyzed using the DNAstar software package (Madison, WI, USA). Deduced polypeptide sequences were compared with sequences in the GenBank database using BLAST. The GenBank accession no. for the sequence reported is AY802988.
Results and discussion
Cloning of the hom gene from S. clavuligerus
The cloning and sequencing showed that the hom coding region is 1,302 bp in length and encodes 433 amino acids with a predicted M r of about 44.9 kDa. The predicted M r is consistent with the observed M r of most homoserine dehydrogenase enzymes reported in the GenBank. The G + C content of the nucleotide sequence is 72.1 mol%, well within the range of the reference values for Streptomyces DNA [ 29 ] and extremely biased towards codons containing G or C in the third position. This position-specific preference for G and C bases is characteristic of Streptomyces coding regions and thought to reflect the adaptations to available tRNA pools for the regulation of gene expression.
Comparison with the predicted amino acid sequences of the hom genes
The predicted amino acid sequence of S. clavuligerus hom gene was compared with those of other bacteria. The deduced amino acid sequence showed 89.8% identity to HSD from aminoethoxyvinylglycine-producing Streptomyces sp. NRRL 5331[ 7 ]. Comparison of the sequence of S. clavuligerus with those of S. avermitilis, S. coelicolor, C. glutamicum, B. subtilis, P. aeruginosa and M. glycogens revealed identities of 89, 87.9, 54.4, 38.4, 36.6 and 31.1%, respectively.
Several well-conserved domains were found in the amino acid sequences compared. The motif at positions 13 to 18, G-X-G-X-X-G, surrounded by small hydrophobic amino acid residues is the characteristic of the NAD(P)-binding site of HSDs [7, 21–23]. Like other HSDs, NAD(P), the cofactor of the enzyme, should bind this region. The most conserved region among the HSD sequences lies between the positions 199 and 228 in S. clavuligerus HSD, where 21 out of 29 amino acid residues are strictly conserved, suggesting the importance of this region in substrate binding or catalytic activity. Parsot and Cohen [22] showed that the amino acid sequences between positions 200 and 221 of B. subtilis are well conserved and suggested that this region could be involved in substrate binding or catalysis. The regulatory domains involved in feedback inhibition by l-threonine were postulated to reside in the C-terminal regions in B. subtilis [22], C. glutamicum [2], M. flagellatus [17] and M. glycogens [21]. The C-terminal region of the S. clavuligerus HSD show some homology with those of other HSD.
Expression of the S. clavuligerus hom gene in E. coli and the properties of the recombinant enzyme
When introduced into E. coli CGSC 5075 (hom−), pEBH1 restored the ability of the mutant to grow in the absence of l-methionine and l-threonine, suggesting that the cloned gene was functionally expressed in E. coli. Specific activities of the recombinant (in the presence of IPTG) and parental HSD were 0.019 and 0.013 μmol min−1 mg-1 of proteins in the cell-free extracts, respectively. Recombinant HSD was next purified, and its specific activity was shown to be about 70-fold higher than that of the crude extract of the recombinant organism. By performing SDS-PAGE, the molecular mass of the purified HSD expressed by E. coli CGSC 5075 was estimated as 45 kDa (data not shown), almost equal to the value deduced from the nucleotide sequence.
Disruption of the hom gene in S. clavuligerus
Streptomyces clavuligerus was transformed with the plasmid construct pAEHK containing a hom disruption cassette. After plasmid curing, kanamycin-resistant and thiostrepton-sensitive colonies were selected. One putative mutant that did not grow on MM, but grew when supplemented with l-methionine and l-threonine was selected and named AK39 (Fig. 2a). HSD activities of wild type and mutant S. clavuligerus were determined in CDM at time intervals, and no activity was detected in the hom mutant (data not shown). Insertional inactivation of hom via double crossing over (Fig. 2b) was verified by Southern blot hybridization by using the 1,300 bp hom (Fig. 2c) and 1,100 bp kan (Fig. 2d) gene fragments as the probes.
a hom-Disrupted mutant AK39 exhibiting methionine and threonine auxothrophy. Identical volumes of the exponentially growing wild and mutant strains were streaked onto MM plates and incubated at 28 °C for 72 h. b Diagram of the hom gene disruption via double crossing over. Restriction enzyme sites are shown as B BamHI, S SacII, H HindIII. c hom Gene amplified by PCR from the chromosome of the wild type (lane 1) and mutant (lane 2) strains, blotted and hybridized to hom probe. d SacII-digested chromosomal DNA of the wild type (lane 1) and the mutant (lane 2), blotted and hybridized to kan probe. Size marker (M) in c and d: PstI-digested lambda DNA
Effect of hom gene disruption on cephamycin C production
The effect of hom gene disruption on cephamycin C production was determined by growing the wild type and mutant S. clavuligerus in CDM (Fig. 3). hom gene disruption in AK39 resulted in a poorer growth in CDM supplemented with l-methionine and l-threonine; however, the organism produced 1.7-fold to twofold higher specific yields of cephamycin C between 48 and 72 h. It is to be noted that the entire experiment was repeated three times with similar results. Mendelovitz and Aharonowitz [19] studied the effects of amino acids of the aspartic acid pathway on cephamycin C production by S. clavuligerus NRRL 3585. It was reported that externally added dl-meso-diaminopimelate and l-lysine exerted precursor effect, increasing specific cephamycin C production by 1.9- and 1.7-fold, respectively. In our study, it was demonstrated that the intracellular free l-lysine levels increased by 1.6- to 4.3-fold upon hom-disruption (Fig. 4). Besides the positive impact of elevated l-lysine levels on cephamycin C production, the prevention of concerted feedback inhibition of aspartokinase in our threonine-minus mutant is also thought to make contribution to increased specific production of the antibiotic.
In Streptomyces, threonine biosynthetic genes hom, thrC (threonine synthase) and thrB (homoserine kinase) are clustered; hom and thrC being organized as a single transcriptional unit while thrB is transcribed as a monocistronic transcript under a strong promoter [7]. Hence, hom disruption in S. clavuligerus was not expected to have any polar effects other than that on thrC. It was therefore interesting to observe some pleiotropic effects of hom disruption in our mutant. Besides the aforementioned growth reduction, AK39 was defective in sporulation on MM supplemented with l-methionine and l-threonine, but not on sporulation agar. This finding accorded well with that of Gehring et al. [9] who reported that an l-methionine auxotroph of S. coelicolor had developmental defects which also increased secondary metabolite production. A functional metH (methionine synthase gene) was required for conversion of aerial hyphae into chains of spores in this mutant.
When AK39 was grown in CDM, the color of the culture fluid turned into faint red after 48 h. A comparison of the absorption spectra (200–800 nm) of 60 h culture supernatants of the wild and mutant strains grown in CDM revealed the existence of a major peak at 265 nm in the mutant (data not shown). Red pigment accumulation by the blocked mutant was reminiscent of characteristic red pigment formation in certain adenine auxotrophs of Schizosacharomyces pombe due to the accumulation of two intermediates (phosphoribosyl aminoimidazole and phosphoribosyl aminoimidazole carboxylate) of purine biosynthetic pathway which was relieved by intracellular homocysteine accumulation due to a methionine synthase mutation [8]. Yet, the structure of the colored substance produced by AK39 and its relation to hom mutation remains to be elucidated.
There have been only a limited number of studies focusing on improvement of cephamycin C yields by manipulating aspartate pathway in S. clavuligerus. Malmberg et al. [15] was able to elevate cephamycin C yields twofold to fivefold by inserting an additional copy of l -lysine ɛ-aminotransferase gene into the chromosome of S. clavuligerus NRRL 3585. In another study, aspartokinase deregulated mutants (AEC-resistant strains) produced 2 to 7 times more cephamycin C than the wild type [20]. Very recently, we accomplished the expression of ask (aspartokinase) gene in a multi-copy plasmid in S. clavuligerus with a twofold to threefold increase in specific cephamycin C yields (unpublished), and the studies are under way to express multiple copies of ask gene in AK39. In view of specific cephamycin C titers, the present study demonstrated a beneficial effect of closing l-threonine branch of aspartate biosynthetic pathway. Further work will focus on determination of the impact of hom inactivation on pathway flux distributions and optimization of the nutritional composition of CDM to improve the growth of the disrupted mutant which may lead to a further increase in cephamycin C production.
References
Aharonowitz Y, Demain AL (1978) Carbon catabolite regulation of cephalosporin production in Streptomyces clavuligerus. Antimicrob Agents Chemother 14:159–164
Archer JA, Solow-Cordero DE, Sinskey AJ (1991) A C-terminal deletion in Corynebacterium glutamicum homoserine dehydrogenase abolishes allosteric inhibition by l-threonine. Gene 107:53–59
Betancort Rodriguez JR, Garcia Reina G, Santana Rodriguez JJ (1997) Determination of free amino acids in microalgae by high-performance liquid chromatography using pre-column fluorescence derivatization. Biomed Chromatogr 11:335–336
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Clepet C, Borne F, Baird C, Patte JC, Cami B (1992), Isolation, organization and expression of Pseudomonas aeruginosa threonine genes. Mol Microbiol 6:3109–3119
Cohen GN, Saint-Girons I (1987) Biosynthesis of threonine, lysine and methionine. In: E. coli and S. typhimurium: cellular and molecular biology. ASM, Washington DC, pp 429–444
Fernandez M, Cuadrado Y, Recio E, Aparicio JF, Martin JF (2002) Characterization of the hom-thrC-thrB cluster in aminoethoxyvinylglycine-producing Streptomyces sp. NRRL 5331. Microbiology 148:1413–1420
Fujita Y, Ukena E, Iefuji H, Giga-Hama Y, Takegawa K (2006) Homocysteine accumulation causes a defect in purine biosynthesis: further characterization of Schizosaccharomyces pombe methionine auxotrophs. Microbiology 152:397–404
Gehring AM, Wang ST, Kearns DB, Storer NY, Losick R (2004) Novel genes that influence development in Streptomyces coelicolor. J Bacteriol 86:3570–3577
Keiser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA (2000) Practical Streptomyces genetics, The John Innes Foundation, Norwich
Kern BA, Hendlin D, Inamine E (1980) l-lysine-ɛ-aminotransferase involved in cephamycin C synthesis in Streptomyces lactamdurans. Antimicrob Agents Chemother 17:679–685
Khetan A, Malmberg LH, Kyung YS, Sherman DH, Hu WS (1999) Precursor and cofactor as a check valve for cephamycin biosynthesis in Streptomyces clavuligerus. Biotechnol Prog 15:1020–1027
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Madsen SM, Albrechtsen B, Hansen EB, Israelsen H (1996) Cloning and transcriptional analysis of two threonine biosynthetic genes from Lactococcus lactis MG1614. J Bacteriol 178:3689–3694
Malmberg LH, Hu WS, Sherman DH (1993) Precursor flux control through targeted chromosomal insertion of the lysine epsilon-aminotransferase (lat) gene in cephamycin C biosynthesis. J Bacteriol 175:6916–6924
Malumbres M, Mateos LM, Guerrero C, Martin JF (1995) Molecular cloning of the hom-thrC-thrB cluster from Bacillus sp. ULM1: expression of the thrC gene in Escherichia coli and corynebacteria, and evolutionary relationships of the threonine genes. Folia Microbiol (Praha) 40:595–606
Marchenko G, Tsygankov Y (1992) Genes for threonine biosynthesis in Methylobacillus flagellatum. In: Abstract in proceedings of the 7th international symposium on microbial growth on C1, B79
Marchenko GN, Marchenko DN, Tsygankov YD, Chistoserdov AY (1999) Organization of threonine biosynthesis genes from the obligate methylotroph Methylobacillus flagellatus. Microbiology 145:3273–3282
Mendelovitz S, Aharonowitz Y (1982) Regulation of cephamycin C synthesis, aspartokinase, dihydrodipicolinic acid synthetase and homoserine dehydrogenase by aspartic acid family amino acids in Streptomyces clavuligerus. Antimicrob Agents Chemother 21:74–84
Mendelovitz S, Aharonowitz Y (1983) β-lactam antibiotic production by Streptomyces clavuligerus mutants impaired in regulation of aspartokinase. J Gen Microbiol 129:2063–2069
Motoyama H, Maki K, Anazawa H, Ishino S, Teshiba S (1994) Cloning and nucleotide sequences of the homoserine dehydrogenase genes (hom) and the threonine synthase genes (thrC) of the gram-negative obligate methylotroph Methylobacillus glycogens. Appl Environ Microbiol 60:11–119
Parsot C, Cohen GN (1988) Cloning and nucleotide sequence of the Bacillus subtilis hom gene coding for homoserine dehydrogenase. J Biol Chem 263:14654–14660
Peoples OP, Liebl W, Bodis M, Maeng PJ, Folletie JT, Archer JA, Sinskey AJ (1988) Nucleotide sequence and structural analysis of the Corynebacterium glutamicum hom-thrB operon. Mol Microbiol 2:63–72
Pridmore RD (1987) New and versatile cloning vectors with kanamycin-resistance marker. Gene 56:309–312
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbour Laboratory Press, New York
Shio I, Miyajima R (1969) Concerted inhibition and its reversal by end products of aspartate kinase in Brevibacterium flavum. J Biochem 65:849–859
Thykaer J, Nielsen J (2003) Metabolic engineering of β-lactam production. Metab Eng 5:56–69
Tunca S, Yılmaz EI, Piret J, Liras P, Özcengiz G (2004) Cloning, characterization and heterologous expression of the aspartokinase and aspartate semialdehyde dehydrogenase genes of cephamycin C-producer Streptomyces clavuligerus. Res Microbiol 155:525–534
Wright F, Bibb MJ (1992) Codon usage in the G−C-rich Streptomyces genome. Gene 113:55–65
Acknowledgments
This study was supported by the Scientific and Technical Research Council of Turkey (SBAG 2753). We thank Gulveren Taskin for her technical assistance in amino acid analysis.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yılmaz, E.I., Çaydasi, A.K. & Özcengiz, G. Targeted disruption of homoserine dehydrogenase gene and its effect on cephamycin C production in Streptomyces clavuligerus . J Ind Microbiol Biotechnol 35, 1–7 (2008). https://doi.org/10.1007/s10295-007-0259-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-007-0259-8