Abstract
Recovery of belowground microbial biodiversity is important for soil restoration after mining exploitation. We aimed to compare microbial communities of a mine rehabilitation site to those with native vegetation. Community structure and metabolic potential of soil microbial communities were analyzed in an inactive open-pit coal mine located in northeastern Colombia using GeoChip and high-throughput sequencing techniques. Sites included (1) a spoil dump closed in 2010 with a mix of mesquite trees and native vegetation, and (2) a native dry forest next to the mining area. Most samples had an alkaline pH, high sulfur, zinc, and magnesium contents, and high cation exchange capacity as well as low calcium content. A total of 61,384 bacterial amplicon sequence variants (ASVs) and 402 Eukarya ASVs were obtained. Overall, the most abundant bacterial/archaeal phyla (0.1%) were Acidobacteria, Actinobacteria, Bacteroidetes, and Chlorolexi. The fungal genus Cladosporium dominated all treatments, while Volutella was observed only in the inactive dump. Bacterial alpha diversity was surprisingly higher in the inactive spoil dump than in the forest, while fungal alpha diversity was similar between that treatment and the native dry forest. Fungal genera might be more sensitive to the different treatments, as their abundances were highly influenced by their location compared to bacterial genera. There were no significant differences regarding the metabolic potential of bacterial communities. The GeoChip 5.0S analysis showed that the native dry forest had a higher number of genes related to C, N, P, and Z cycles, metal homeostasis, and organic remediation.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Open-pit coal mining drastically affects terrestrial ecosystem functioning worldwide (Singh and Singh 2006; Maiti 2013). During open-cast coal mining, large amounts of spoil material overlying coal layers are excavated and deposited aboveground. This process adversely affects existing ecosystems either by destroying or burying them under spoil material deposits. Restoration of soil biological activity requires some human intervention for returning disturbed soil to “nearly-natural” ecosystems; however, this is a long and expensive process (Doong and Lei 2003; Vázquez et al. 2013). To implement good soil restoration practices, an initial approach will require comparing complex biotic and abiotic soil interactions between undisturbed and disturbed ecosystems.
Coal mining companies in the dry Colombian Caribbean region produce about 90% of the coal extracted in the country (Agencia Nacional de Mineria 2017; UPME 2016). Díaz et al. (2013) determined that coal mines significantly decreased soil quality, causing its physical degradation and low fertility. Plant populations and the agricultural value of these soils are also negatively affected due to soil degradation and contamination (Betancur and Escobar 2013; Holguin 2011). For example, soil pollution by heavy metals represents an environmental risk because metals are not biodegradable and bioaccumulate, ultimately deteriorating soil quality (Bhattacharyya et al. 2003; Kahn et al. 2010; Ali et al. 2013). Minimizing environmental damages and reconstructing soils after coal mining are of utmost importance, but an initial soil biotic and abiotic characterization is always required. This study provides the baseline for sustainable soil management after coal mining in the dry Colombian Caribbean region. We used physical, microbial, and chemical characterizations of two contrasting soil ecosystems, native dry vegetation, and spoil material deposits under recovery.
Soil microbial communities are essential, as they are responsible for physiological and metabolic processes of great importance for soil quality (Bundy et al. 2009; Lauber et al. 2009; Ranjard et al. 2003; Rutgers et al. 2016), while supporting key ecosystem functions such as nutrient cycling, water infiltration, decomposition, soil respiration, and plant growth, among others (Guerra et al. 2020). When soil degradation happens, it is often irreversible, causing a substantial loss of plant and microbial biodiversity, and their ecosystem functions. After mining, soil’s surface layers are rich in pyrite (FeS2) that later oxidizes to sulfuric acid when exposed to oxygen and water. This process results in high levels of soil acidification (Fanning and Fanning 1989), loss of soil structure, a decrease in soil organic matter (SOM) content, and a low rate of water infiltration—ultimately leading to an increased nutrient leaching and soil erosion (Guebert and Gardner 2001; Kampf et al. 1997). Also, the level of stability, functional compatibility, and/or redundancy of microbial communities will affect the availability of soil nutrients, SOM (soil organic matter), and soil physical stability (Griffiths and Philippot 2013; McGuire and Treseder 2010; Shade et al. 2012; Tardy et al. 2014). SOM is critical for microbial communities; at the same time, these communities aid to increase SOM content, usually deficient in post-mined soils. Assessing the ecosystem functions of soil microorganisms at the site of mining helps to understand soil rehabilitation processes and informs the management of these areas (Gomez et al. 2004; Mishra and Nautiyal 2009; Peng et al. 2015).
Although at a global scale the effects of post-mining rehabilitation treatments in soil biodiversity are not well understood, regional- and local-scale studies show some patterns. Andrés and Mateos (2006) found that even after 12 years, four different post-mining treatments did not lead to the recovery of soil mesofauna biodiversity. In contrast, Madej et al. (2011) found that a soil quality index, based on the biodiversity of soil microarthropods, significantly increased on a post-mining chrono-sequence after abandonment. Kneller et al. (2018) found organic amendments either of plant material or topsoil increased important ecosystem functions of soil microorganisms and their microbial activity. For example, N and C contents increased resulting in plant growth and survivorship after soil was treated with these amendments. Co-introductions of native soil biota and vegetation seem to have the best results when restoring diverse native vegetation on post-mining landscapes (Vahter et al. 2020). However, responses also seem dependent upon the restoration strategy used, time that elapsed after mining, and intensity and type of mining, among other local factors. More baseline studies on the effects of different post-mining restoration strategies on soil biodiversity and ecosystem functions are needed.
Here we ask: are there any differences between the microbiomes in a coal mine and adjacent natural areas? And can we evaluate the effects of post-mining treatments on soil biodiversity and ecosystem functionality? The aim of this study was to identify changes in soil microbial community composition, and taxonomic and functional diversity resulting from land rehabilitation after coal mining and compared them to predisturbance levels from a reference site covered by dry forest vegetation. We sampled two study sites 7 years after rehabilitation efforts, and a reference dry forest site associated with a coal mine located in La Guajira, northern Colombia. We predict the dry forest site will have a higher diversity of fungi and bacteria related to soil formation, organic matter decomposition, and symbiosis. While the rehabilitation site will have stress-tolerant microbiomes. Environmental rehabilitation comprises topographic reformulation after the removal of the coal and revegetation with mesquite trees.
2 Materials and Methods
2.1 Study Site
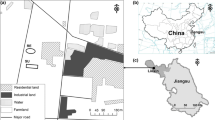
The study site lies within an open-pit coal mine (300 ha) located at Palmito, Barrancas municipality in La Guajira Department, northeastern Colombia (11°02′N, 72°40′W). The average altitude of spoil heaps is 240 m.a.s.l., and mean annual precipitation varies between 500 and 1000 mm, with bimodal seasons of 50–100 rainy days per year. The mean annual temperature is 26 °C. The coal mine is divided into seven main zones: administrative (1), active extracting (2), backfill area (3), southern spoil dump (active) (4), northern spoil dump top (inactive, revegetated) (5), and northern spoil dump terraces with a slope (inactive, revegetated) (6), and a native dry forest (7) next to the mine (Fig. 1).
Main study areas delimited in red.1. Administrative zone, 2. Active extraction zone, 3. Backfill area, 4. Southern spoil dump (active), 5. BiT, northern spoil dump tops 6; Bi, northern spoil dump terraces; Bi represents a different study treatment to BiT in our study because of the slopes, 7. B, native forest. Red lines indicate inactive dumps undergoing recovery and native forest
This study was performed on three of these seven sites: the top northern inactive spoil dump (zone 5, hereafter BiT, initials refer to Spanish translation “Botadero inactivo Top”), its revegetated terraces (zone 6, hereafter Bi, initials refer to Spanish translation “Botadero inactivo”), and a control site of native dry forest (zone 7, hereafter B, initials refer to Spanish translation “Bosque”) (Fig. 1).
This region is part of th Perijá mountain range, an area with high anthropogenic intervention due to coal mining. A total of 16 tree species have been identified around coal mines and in native dry forests such as Spondias mombin, Ficus glabrata, Casearia corimbosa, and Guasuma ulmifolia (Vargas 2011). The Bi dump was closed in 2010 and its six terraces were revegetated with mesquite trees (Prosopis juliflora), but some other vegetation has been naturally colonizing the area ever since.
2.2 Experimental Design
A total of 18 soil samples were collected in February 2018 from three sites: northern spoil dump (BiT), terraces of the northern spoil dump (Bi), and a control site of native dry forest (B) (Fig. 1). BiT and Bi were considered as different treatments since Bi had a 45–65° slope that potentially affects physicochemical and microbiome analysis due to water erosion unlike in BiT. Six composed samples were collected on each site, choosing six random points, and trying to cover as much area as possible. Each of the 18 individual soil samples (3 sites × 6 samples each) included four pulled soil subsamples. These samples were collected in a zigzag random walk, always removing top organic debris, and sampling up to 10 cm soil depth; this depth concentrates most of the roots of the revegetated plant species. The four soil subsamples were combined and sieved through a 2-mm mesh, thoroughly homogenized, and placed until full in sterile 50-mL conical centrifuge tubes, and immediately stored in ice. The remaining soil for each was used for soil physicochemical analyses. Later in the day, samples were flash-frozen in liquid nitrogen for transportation to the laboratory. Once in the laboratory, tubes were stored at − 20 °C until DNA extraction and sequencing were performed.
2.3 Soil Physicochemical Analyses
For physicochemical analysis, three replicates per treatment were analyzed for a total of nine samples. Physicochemical characterization of soil samples was carried out in accordance with the Colombian technical standards (NTC 5526:2007, NTC 5596:2008, NTC 5349:2016, NTC 5350:2020; Instituto Colombiano de Normas Técnicas y Certificación – ICONTEC 2007, 2008, 2016, 2020) at the Laboratorio de Química de Suelos, Aguas y Plantas, of Agrosavia, Tibaitata, Colombia. The following physicochemical parameters were determined on each soil sample: texture (Bouyoucos 1936), pH in a 1:5 ratio of soil:water using a Corning pH-meter 340, electrical conductivity with a conductivimeter (720 WTW), soil organic carbon (Walkley–Black chromic acid wet oxidation method; Nelson and Summers 1982), available P (Bray II method: extraction with HCl + NH4F and ascorbic acid, measured in a spectrophotometer at 800 nm), Fe, Cu, Zn, Al, and Mn (quantification with atomic absorption spectroscopy—AAS 240/280, Agilent, USA); S and B (extraction with calcium phosphate monobasic, quantified in a spectrophotometer UV–vis) and Na, K, Ca, and Mg (extraction with 1 M ammonium acetate at pH 7, quantification with AAS). Cation exchange capacity was calculated as the sum of cations (Al, Na, K, Ca, Mg). For metal concentration in the soil, only two replicates per treatment could be analyzed. Cd determination was carried out by the acid digestion method (Díaz et al. 2013) and flame atomic absorption spectroscopy (FAAS, AA-7000, SHIMADZU, Japan) and Pb by the closed digestion method (with nitric:hydrochloric acid; Dolgopolova et al. 2004) and inductively coupled plasma emission spectrometry (ICP-OES, iCAP 6500, Thermo Scientific, USA).
Due to sampling and analysis limitations, soil chemical analyses were done for samples shown in Tables S1 and S2. Thus, these analyses should be taken as a general characterization of the sites but cannot be used for further analyses (i.e., to relate them with microbial community parameters).
2.4 DNA Extraction, PCR Amplification, and Sequencing
DNA was extracted for each sample from 0.25 g of soil using the Power Soil DNA isolation kit (MoBio Laboratories, Inc., Carlsbad, CA), following the manufacturer’s instructions. All genomic DNA concentrations were quantified in a Qubit 2.0 Fluorometer (Invitrogen, NY, USA). Ribosomal RNA (rRNA) sequenced markers were amplified from the total DNA in each sample using PCR with barcoded primers unique to each sample. The 16S rRNA gene V4 region was amplified from all 18 samples, while the ITS1 sequence was amplified from 9 out of the 18 samples (three samples by site). To obtain PCR products, jagged ends of DNA fragments were converted into blunt ends using T4 DNA polymerase and Klenow Fragment, and T4 Polynucleotide Kinase. Then, an “A” base was added to each 3′ end to make it easier to add adapters. Fragments that were too short were removed by AMPure beads (Thermo Fisher Scientific Inc. Madrid, Spain). Fused primers with dual indexes and adapters were used for PCR, and fragments too short were also removed by AMPure beads. In both cases, qualified libraries were used for sequencing. Fragments were sequenced with Illumina Hiseq 2500 technology 250 bp long pair-ended reads. Amplification and sequence analysis was completed by the BGI Life Tech company, Hong Kong.
2.5 Bioinformatic Analyses
Raw data was preprocessed to get clean reads. All sequenced reads with an average quality Phred score ≤ 20 over a 25 bp sliding window were truncated. Trimmed reads having less than 75% of their original length, reads contaminated by adapters, reads with ambiguous bases (N base), and reads with 10 consecutive equal bases were removed.
Clean sequences were processed using DADA2 (Callahan et al. 2016) designed to resolve exact biological sequences from Illumina sequence data and not involve sequence clustering. All pipelines used in this study are available at: https://github.com/TheAriasLab. Clean paired sequences were processed using the following DADA2 parameters: filterAndTrim based on quality plots; the forward and reverse sequences were trimmed to lengths between 200 and 240 bp. Filtering of sequences was set to maxN = 0, maxEE = 2 (for both reads), truncQ = 11; where maxN was the maximum number of “N” bases, and maxEE corresponded to the maximum expected error calculated from the quality score (EE = sum [10ˆ(-Q/10)]. The truncQ parameter truncated reads at the first instance of a quality score ≤ 11. Other parameters were set to default. Error rates were estimated by learnErrors, where the nbases parameter was set to 10ˆ8.
Sequences were de-replicated using the DADA2 command derepFastq with default parameters. Exact sequence variants were resolved using the DADA2 command dada. Next, chimeras were removed with the DADA2 (Callahan et al. 2016) command removeBimeraDenovo. By applying a consensus method, taxonomy was assigned against version 132 of the Silva database for bacteria released in July 2017 (Glöckner et al. 2017), and against the UNITE database for fungi released in February 2019 (Nilsson et al. 2019) using the command assignTaxonomy from DADA2. Taxa abundances were transformed into percentages.
2.6 Alpha and Beta Diversity Analyses
Species accumulation curves (Ugland et al. 2003) were produced using Vegan (Oksanen et al. 2010) in R Studio (RStudio Team 2021). Summaries of taxonomic composition were produced with Phyloseq v 1.16.2 (McMurdie and Holmes 2012) and plotted in R Studio. Alpha diversity was estimated for each sample with the Phyloseqestimate_richness command. Beta diversity was evaluated using Phyloseq v 1.16.2 as unweighted UniFrac distance (Lozupone et al. 2007), using plot_ordination commands. Weighted UniFrac distance measures were also calculated: the software weights phylogenetic lineages that are not shared between pairs of samples by their abundance. Additionally, correlations to test if the structure of microbial communities was influenced by environmental variables using Adonis in Vegan were also implemented.
Normality for each biodiversity index (Chao1, Shannon, and Simpson; calculated using Vegan) was calculated using the Shapiro–Wilk test with the R base command shapiro.test(x), while homoscedasticity for each index was examined with the Levene test using the R Studio CAR package (Fox and Weisberg 2019) v 3.0–11 (using the command LeveneTest).
2.7 GeoChip 5.0S Hybridization and Data Preprocessing
Two samples, B (native dry forest) and Bi (northern spoil dump terraces), were analyzed using GeoChip 5.0S (Tu et al. 2014). Each sample was the product of combining all six replicates for each treatment. GeoChip 5.0S gene micro-arrays contain 167,044 probes targeting functional genes assigned to several gene categories: carbon (degradation, fixation, methane), nitrogen, sulfur, and phosphorus cycling, energy metabolism, metal homeostasis, organic remediation, secondary metabolism (e.g., antibiotic metabolism, pigments), stress responses, viruses (both bacteriophages and eukaryotic viruses), virulence, and others (phylogenetic genes and CRISPR system). All hybridizations were performed at 42 °C with 40% formamide for 16 h on a MAUI hybridization station (BioMicro, Salt Lake City, UT, USA). After hybridization, the arrays were scanned (NimbleGen MS 200, Madison, WI USA) at 100% laser power. The numbers were normalized by signal intensity, and all showed positive probes detected on each sample. Probes were removed as negative if the SNR < 2 or the signal < 200 or < 1.3 times the background. No further data transformation was performed (He et al. 2010; Lu et al. 2012). Singletons or positive probes detected only in one of the samples were discarded prior to statistical analyses to remove noise from the dataset. All procedures were performed at Glomics Inc. (Norman, OK, USA).
2.8 Differential Abundance Analysis
EdgeR version 4.1 (Robinson et al. 2010) was used to find taxa and genes with differential abundance among pairs of locations. Either (BiT or Bi: treatments) vs. the native dry forest (B: control). Only, significantly abundant taxa with a logFC > 3 or a log FC < -3 and with FDR < = 0.01 were considered.
2.9 Association Analysis
Association analyses were performed by filtering ASVs not assigned to a genus, and those with less than 10 counts in all samples. All ASVs with a prevalence of less than 15% in samples were also filtered. Counts were normalized using a center log ratio (CLR) transformation. Spearman’s rank correlation was performed using distances, and only significant associations with coefficient > 0.6 (FDR < = 0.01) were considered.
2.10 Metabolic Potential Analysis for Bacterial Microbiomes
Metabolic potential for microbial communities was calculated using the software Picrust v2.0 (Douglas et al. 2020). This analysis shows the abundance of predicted genes in each sample. Genes with less than 10 counts across samples and genes present in less than 10% of the samples were filtered, and also gene abundance with low variance among samples (less than 10%; based on inter-quantile range) was filtered. All other data were normalized using a centered log ratio (CLR) normalization. Grouping analyses were also performed using the Bray–Curtis distance. Predicted genes were identified with KEGG release 93.0 (Kanehisa et al. 2016) and grouped according to metabolic function and relative abundance on each site. To search for key genes of interest, an abundance differential analysis between Bi vs B and B vs BiT was completed using EdgeR version 4.1 (Robinson et al. 2010). Correlation analyses were implemented using Spearman’s correlation ranks. The top significant genes were reported here: the first 10 are positive and the last 10 are negative.
3 Results
3.1 Soil Physicochemical Analyses
Soil texture class includes loam-clay for treatments BiT3 and Bi3, loam in Bi1, to loam-silty in B (native dry forest) (Table S1). The amount of Pb in the soil for different treatments ranged between 23.31 and 23.81 mg/kg, and Cd between 1.32 and 3.01 mg/kg, finding the lowest amount for both metals in the native dry forest (B) (Table S2). Most sites had an alkaline pH (7.26–8.15), high values of S, Zn, and Mg content, and high cation exchange capacity (27.93–48.55 cmol(+)/kg), as well as low Ca content (21.66–42.92 cmol(+)/kg) (Table S1). Soil chemical properties in Bi replicates were similar, while those of BiT were heterogeneous. Electric conductivity was five times greater in disturbed soil samples BiT1, Bi1, and Bi3 than in native forest samples B3 and B6 (Table S1). For heavy metals, boron had high values in all sampled sites, while zinc had high values only in BiT. Potassium was low in all samples (Table S2).
3.2 Soil Microbial Diversity and Community Structure
3.2.1 Sequencing
Four million paired-end reads were obtained with read lengths around 250 bp for the 16S rRNA V4 region of Bacteria and Archaea (six replicates per treatment), and the ITS1 region of fungi (three replicates per treatment for Bi and B). However, in fungi, one of the replicates in BiT (top dump) could not be sequenced; this treatment had only two replicates to work with. After filtering singletons, the total number of 16S reads ranged between 1,202,013 in sample B1 and 1,770,139 in sample B5. While for ITS1 ranged between 1,618,254 in Bi6 and 2,158,687 in BiT5. A total of 62,756 16S amplicon sequence variants (ASVs) were found, and taxonomic assignments to kingdom include Archaea 516; Bacteria 61,384; Eukaryote 402; and non-assigned 454. For ITS, a total of 6084 fungal ASVs were identified.
3.2.2 Rarefaction Curves
Species saturation reached for both fungi and bacteria a sequencing depth of 500,000 sequences (Fig. S1). Species number at saturation was lower in Bi than in BiT and B for both 16S- and ITS-based curves. For the 16S, the species number at saturation in BiT is lower than in B (Fig. S1A).
3.2.3 Taxa Relative Abundance
The relative abundance of fungal and bacterial taxa represented more than 0.1% of the total reads. For fungi, the most abundant phyla included Ascomycota and Basidiomycota. Dominant fungal genera included Cladosporium, Nothophoma, Mycocentrospora, Ceratobasidium, Fusarium, and Volutella (Fig. 2A). Cladosporium was well represented in all sampled sites, while Volutella and Strelitziana were quite abundant in B1 (Fig. 2A). For bacteria, most represented phyla include Acidobacteria, Actinobacteria, Bacteroidetes, Chlorolexi, Firmicutes, Gematimonadetes, Planctomycetes, Proteobacteria, and Thaumarchaeota. The genus Microvirga was the most abundant, and Actinomadura, Actinoplanes, Azohydromonas, Byssovorax, Cardidatus, Coochioplanes, Criptosporangium, and Microvirga were also well represented (Fig. 2B).
Relative abundance for genera (relative abundance > 0.1%) expressed as the percentage of recovered sequences in each sampled site. Tropical dry forest (B) sequenced replicates (B1, B3, B5); coal mine dump terraces (Bi) sequenced replicates, for this treatment only two replicates were recovered (Bi5, Bi6); top dump (BiT) sequenced replicates (BiT2, BiT3, BiT5). Panel B shows results for bacteria, and their six replicates in each location (subindexes 1–6)
Analysis of relative abundance for dominant taxa gave a general and qualitative description of the microbial profile in all sampled sites. Next, a differential abundance analysis was performed to test if the qualitative differences observed were statistically significant. Fungal and bacterial ASVs abundance was compared among BiT or Bi, to those from B (Table 1). Bacterial ASVs with differential abundance among BiT or Bi and B did not represent dominant genera and included Ureibacillus, Paraburkholderia, Pelagibius, and Kallotenue among others. The top fungi ASVs displaying differential abundance between Bi and B was Fussariella decreasing in Bi compared to B (FC = 8.5, FDR = 0.008). ASV from non-dominant genera included Podospora, Candida, Alternaria, and Zygosoporium among others (Table S3).
3.2.4 Alpha Diversity Indexes
For bacterial communities, there was a significant difference in indexes between treatments (Chao1 p-value 0.0001, Simpson p-value 0.004, and Shannon p-value 0.001) and data between samples grouped by sampling site. There were significant differences in the Chao1, Simpson, and Shannon indexes between samples BiT and Bi (p-value 0.0004, p-value 0.009, p-value 0.004, respectively), and between BiT and B (p-value 0.005, p-value 0.007, p-value 0.01) (Fig. 3A, B). Fungal communities showed significant differences with the Chao1 index (p-value 0.025) when samples were grouped by sample sites, but not between pairs of comparisons.
Alpha diversity calculated for fungi and bacteria communities. Shannon, Simpson, and Chao1 indexes were calculated for each treatment: tropical dry forest (B), coal mine dump terraces (Bi), and top dump (BiT), and their replicates (black dots). Panel A: alpha diversity distribution in bacterial communities for six replicates. Panel B: alpha diversity values in fungal communities for three replicates in B and BiT, but only two in Bi, since for this treatment data for only two replicates was recovered during sequencing. Values of the Shannon index for fungal communities in the Bi replicates overlap. **Significantly different values (p < 0,01); ns, non-significant
3.2.5 Beta Diversity for Bacterial and Fungal Communities
Weighted UniFrac distance shows bacterial (Adonis R2 = 0.5, p-value = 0.001) and fungal (Adonis R2 = 0.4, p-value = 0.006) communities at each site were different when compared to other sampled sites (Fig. 4). Overall, most samples were grouped within their site, except for BiT6, grouped among Bi samples. BiT and Bi microbial communities are more similar than when compared to B.
Beta diversity calculated for fungal and bacterial communities using weighted UniFrac distance for tropical dry forest (B), coal mine dump terraces (Bi), and top dump (BiT). Panel A: beta diversity values in fungal communities for three replicates in B and BiT, but only two in Bi, since for this treatment data for only two replicates was recovered during sequencing. Panel B: beta diversity for bacterial communities and their six replicates in each location
3.2.6 Association Analysis
Comparing bacteria from B and BiT samples, 285 ASVs from Romboutsia, Pseudenhygromyxa, and Parasegetibacter among other genera had a negative association (FDR < = 0.01) with BiT. Additionally, 280 ASVs from bacterial genera Marisediminicola, Spirosoma, and Methyllocella, among others, had a positive association (FDR < = 0.01) with BiT (Fig. 5A). When analyzing the correlation of bacteria between Bi and B samples, 111 ASVs were negatively associated with Bi, like Romboutsia and Acidicaldus, and 125 ASVs were positively associated with Bi, including Tetrasphaera, Sulfurirhabdus, and Methyllocella (Fig. 5B) (Table S4). No significant associations were found for fungi when BiT, Bi, and B samples were compared with an FDR < = 0.01. However, when the FDR was relaxed to FDR < = 0.05, 24 ASVs from Zygosporium, Podospora, and Candida were negatively associated with BiT, while Alternaria was the only fungal genus positively associated (Fig. 5C).
Abundance association analysis of amplicon sequence variants (ASVs) using Spearman’s rank correlation. The 25 more significant values (with lower p-value) are shown for bacteria. Panel A: comparing the top dump (BiT). vs tropical dry forest (B) sites; and panel B: the coal mine dump terraces (Bi) vs tropical dry forest (B). For fungi, panel C compares top dump (BiT) vs tropical dry forest (B)
4 Functional Diversity of Soils Assessed by GeoChip 5.0S Analysis
The total number of genes detected in the native dry forest (B) was higher than in revegetated dump terraces (Bi) for all gene categories (Table 2). The number of bacterial and fungal-related genes in B was higher than in Bi. In contrast, Archaeal-related genes were more abundant in the dumping site (Table S5). Most abundant genes for forest and dump were related to carbon cycling, followed by organic remediation, while less abundant genes in both forest and dump were related to secondary metabolism and phylogenetic informative genes (Table 2).
4.1 Metabolic Potential Analysis for Bacterial Microbiomes
Using Bray–Curtis distance for gene abundance, samples were grouped by sampled sites (Bi and BiT: dumps, and B: forest). Bi and BiT were clustered but each treatment was separated into distinct groups, different from samples collected in the forest (B) (Fig. 6A). There was no qualitative difference in the relative abundance of metabolic categories, indicating differences could be related to individual genes rather than to broad categories (Fig. 6B). Therefore, we performed a statistical differential abundance analysis and compared individual gene abundance among BiT or Bi, and B. Top genes with significant differential abundance (p < 0.01) among BiT or Bi and B included K12673 N2-(2-carboxyethyl) arginine synthase and K16133 microcystin synthetase protein. McyI increased in treatments and K10842 CDK-activating kinase assembly factor MAT1 and K19658 peroxisomal enoyl-CoA hydratase 2 decreased in treatments (Table S6). Top genes that had a statistically significant positive association (p < 0.01 and correlation > 0.6) with BiT or B included K19428 sugar transferase EpsL, K19576 MFS transporter, DHA1 family, and quinolone resistance protein. Those with a negative association with treatments included K06292 spore germination protein BB and K15076 elongin-A among others (Table S7).
Metabolic potential of bacterial communities calculated using the software Picrust v2.0 (Douglas et al. 2020) for tropical dry forest (B), coal mine dump terraces (Bi), and top dump (BiT) and their replicates. Panel A: dissimilarity of gene abundances using hierarchical clustering of Bray–Curtis. B KEEG metabolic term relative abundance
5 Discussion
Here we provide an analysis of soil microbiomes using metabarcoding in the biggest coal mine exploitation in Colombia. We used comparisons between two contrasting sites, native dry forest vegetation and disturbed coal mining ecosystems undergoing recovery. This work uses alfa and beta diversity of microbial communities and relates diversity with the potential functional roles of microbial communities.
One of the most surprising findings in our results was how alpha bacterial and fungal diversity were differentially affected. Bacterial alpha diversity was higher in the inactive spoil dump (BiT), while was similar between the revegetated dump terraces (Bi) and the native dry forest (B), despite soil organic matter and available phosphorous being the lowest at the spoil dump (BiT) (Table S1). Although this might sound contra-intuitive (Li et al. 2014), previous studies have shown that in areas most contaminated by metals, bacterial diversity was also not affected (Benidire et al. 2020). It has been reported that there is not a linear relationship between the level of metal contamination and bacterial biodiversity (Radeva et al. 2013). Also, it is possible that bacterial taxa inhabiting this system thrive under low organic matter conditions, or that cations, which were similar across treatments, had a higher effect on bacterial alpha diversity than macronutrients. Thus, this is an aspect for future exploration in the specific context of our study. After 7–8 years of ceasing activities, it could mean that levels of metal pollution are not so high, or different from revegetated and native dry forest treatments, as reflected in our results. However, replicates for this study only allowed a general characterization of soil chemistry, and no statistical analyses were possible as sampling for soil chemistry was unbalanced (Table S1, S2). Also, it could mean that the native forest is highly disturbed by mining activities taking place nearby.
Fungal alpha biodiversity did not differ between the native dry forest and the inactive spoil dump. Some studies have shown heavily metal–polluted mine soils reduce fungal biodiversity while increasing the tolerance of some fungal taxa, like Aspergillus spp., to some of those metals (Iram et al., 2009). Also, phytoremediation of metal-polluted soils seems to increase fungal biodiversity, especially ectomycorrhizal and saprophytic fungi (Gil-Martínez et al. 2021). While also showing some fungal tolerance to metals, other studies did show a relatively high fungal biodiversity in mine soils (Taleski et al. 2020). For example, Vahter et al. (2020) showed arbuscular mycorrhizal fungi (AMF) were resilient to very polluted mining conditions in Estonia. In contrast to our results, Guo et al. (2020) have shown that for AMF, alpha biodiversity is higher in undisturbed areas compared to mining dumps. It is also possible that, as the cation contents were similar across treatments, and some fungal guilds are more influenced by cations than C and N contents (Marín and Kohout 2021), this might result in no differences in fungal alpha diversity.
Overall, the most abundant bacterial phyla included Acidobacteria, Actinobacteria, Bacteroidetes, Chlorolexi, Firmicutes, Gematimonadetes, Planctomycetes, Proteobacteria, and Thaumarchaeota, which also represent most abundant and ubiquitous bacterial phyla in diverse soils (Fierer and Ladau 2012; Lauber et al. 2009). Proteobacteria, Actinobacteria, and Acidobacteria are the top three most common phyla at a global scale (Delgado-Baquerizo et al. 2018). Bacterial genera distribution across the three sampled sites was generally equal (i.e., major bacterial genera having similar abundance across treatments), with the dominance of two or three phyla across sampled sites. Other studies have shown that Acidobacteria abundance in soils is mainly influenced by pH, with subdivisions 1 and 3 being more abundant in acidic soils and subdivisions 4 and 6 being more abundant in neutral soils (Jones et al. 2009; Kishimoto et al. 1991; Sait et al. 2006). Members of Acidobacteria have been reported to be dominant in neutral soils of semiarid regions, including cyanobacterial crusts (Gundlapally and Garcia-Pichel 2006; Moquin et al. 2012). In our study, Acidobacteria were highly abundant. This pattern could be common across the northeastern Colombian region. Lastly, members of Actinobacteria also are highly abundant bacterial phyla across our results; they have been isolated from abandoned mining areas and shown resistance to metals like Pb, Cr, Zn, and Cu (El Baz et al. 2015).
Although there were no significant differences in fungi alpha diversity between native dry forests and spoil dumps, samples were grouped together within each site, and beta diversity and the distribution of fungal genera were different between treatments. Genera like Cladosporium, Mycocentrospora, Nothophoma, Ceratobasidium, Fusarium, Preussia, and Volutella showed different abundances among sampled sites. Intriguingly, none of these genera appeared in the association analysis for fungi.
The plant-fungal pathogen Cladosporium displayed resistance against drugs (Talbot et al. 1988) and metals (Shao and Sun 2007), and it is currently being used to remove such metals from water bodies (Mota et al. 2020). This genus was more abundant in one replicate of the native dry forest and one of the revegetated dump terraces. Fusarium is also widely known for its ability to degrade pollutants like polycyclic aromatic hydrocarbons and others (Wei et al. 2017; Zhang et al. 2012; Zhao et al. 2017). Ceratobasidium which could be plant pathogens, saprotrophs, and orchid symbionts has been associated with mining soils elsewhere (Ouanphanivanh et al. 2013). From the association analysis for fungi stand out some saprophytic (and often hallucinogenic) genera like Psilocybe and Conocybe, which are often indicative of advanced decomposition processes and/or decomposition associated with feces. Goats are often found in this area and may be responsible for the abundance of these fungal genera. Overall, fungal genera included a plethora of functional guilds: plant pathogens (Zygosporium, Thecaphora), animal pathogens (Podospora, Candida), and coprophilous fungi (Ascobolus, Polytolypa, Psilocybe, and Conocybe), among others.
A surprising result was that metabolic potential did not differ among treatments, but functional diversity did. Environmental constraints (i.e., metal concentration, harsh weather conditions) suppressing the development of metabolic functions could be responsible for this observation. It is worth noting that Picrust v. 2.0 is a purely speculative tool, of the “potential” of soil microbial communities, and does not represent what is happening in them. In addition, important critiques to possible flaws of this technique have emerged recently (Louca et al. 2018; Sevigny et al. 2019). Our Picrust v. 2.0 analyses show no differences in the bacterial metabolic potential among different treatments: untreated spoil dumps, revegetated terrace dumps, and native dry forests. KEGG predictions for more abundant bacterial phyla found in soil (Delgado-Baquerizo et al. 2018), such as Acidobacteria (Lladó et al. 2016) showed a prevalence of amino acid, carbohydrate, energy, cofactors, and vitamin metabolites, which are commonly dominant (Wrighton et al. 2012). Furthermore, these major metabolic types are crucial for all soil microorganisms, so an additional exploration of specific metabolic and physiological processes—related to bacterial resistance to heavy metals—is necessary. Three treatments overall did not affect bacterial genera distribution; this result is not surprising since metabolism and physiology usually are phylogenetically constrained.
In contrast with bacterial metabolic potential results—and with alpha bacterial diversity results—GeoChip 5.0S analysis showed the native dry forest had a higher number of genes related to carbon cycling, metal homeostasis, phosphorus and sulfur cycling, organic remediation, N cycling, among other processes, compared to the revegetated dump terraces. This result shows the importance of analyzing biodiversity by different metrics of taxonomic, phylogenetic, and, as in this case, functional diversity. To properly address nutrient cycling and other ecosystem processes, as well as ecological restoration targets, it is crucial to establish causal paths between those processes and soil microbial community attributes and ecosystem functions/services (Xu et al. 2020). On a global scale, soil biodiversity and ecosystem functions are rarely addressed at the same time (Guerra et al. 2020; but solutions have been proposed, see Guerra et al. 2021). Hall et al. (2018) propose that microbial processes (i.e., nitrogen fixation) are more directly related to a nutrient pool or cycle than microbial community properties (i.e., biomass C:N ratio, functional gene abundance), and microbial membership (i.e., taxonomic and phylogenetic diversity, community structure, co-occurrence networks), the last two having effects on nutrient cycling via their effect on microbial processes. Our study corroborates this view, as differences among treatments were higher when microbial processes/properties were, directly and indirectly, quantified, compared to plain alpha/beta diversity measurements and metabolic prediction.
6 Conclusions
Microbial diversity in degraded soils is usually lower than in fertile, healthy soils, supporting plant growth. Regarding this, we overall found that (i) bacterial alpha diversity was surprisingly higher in the spoil dump, while fungal alpha diversity was similar between that treatment and the native dry forest. ii. Some fungal genera might be more sensitive to the different treatments, as they were more unequally distributed compared to bacterial taxa. (iii) Some bacterial and fungal genera positively and negatively correlated to different treatments could be good bioindicators of the mining effects in soil. (iv) There were no significant differences regarding the metabolic potential of bacterial communities across treatments—but the technique used for this has many flaws. And more importantly, (v) the native dry forest had a higher number of genes related to carbon cycling, metal homeostasis, phosphorous and sulfur cycling, organic remediation, and N cycling, among other processes, compared to the revegetated dump terraces. This shows that post-mining revegetation strategies in our study system still need more time to reach a target situation of native vegetation. These results are intricate and show that a single measure of soil microbial biodiversity and/or community complexity does not tell the whole story of different treatment effects. Measures of alpha taxonomic diversity could tell a very different story than functional diversity measures (as GeoChip 5.0S). This study constitutes a pioneer study regarding the ecosystem functioning effects of revegetation strategies in post-mining scenarios in Colombia.
References
Agencia Nacional de Mineria (2017) Producción de carbón en ascenso. Available via ANM. https://www.anm.gov.co/?q=produccion_de_carbon_en_ascenso_boletin_prensa. Accessed June 2017
Ali H, Khan E, Sajad MA (2013) Phytoremediation of heavy metals—concepts and applications. Chemosphere 91:869–881. https://doi.org/10.1016/j.chemosphere.2013.01.075
Andrés P, Mateos E (2006) Soil mesofaunal responses to post-mining restoration treatments. Appl Soil Ecol 33:67–78. https://doi.org/10.1016/j.apsoil.2005.08.007
Benidire L, Pereira SI, Naylo A, Castro PM, Boularbah A (2020) Do metal contamination and plant species affect microbial abundance and bacterial diversity in the rhizosphere of metallophytes growing in mining areas in a semiarid climate? J Soils Sediments 20:1003–1017. https://doi.org/10.1007/s11368-019-02475-4
Betancur EO, Escobar JMM (2013) Legislación Colombiana de Cierre de Minas. ¿Es realmente necesaria? Boletín De Ciencias De La Tierra 34:51–64
Bhattacharyya P, Chakrabarti K, Chakraborty A (2003) Effect of MSW compost on microbiological and biochemical soil quality indicators. Compost Sci Util 11:220–227. https://doi.org/10.1080/1065657X.2003.10702130
Bouyoucos GJ (1936) Directions for making mechanical analysis of soils by the hydrometer method. Soil Sci 4:225–228. https://doi.org/10.1097/00010694-193609000-00007
Bundy JG, Davey MP, Viant MR (2009) Environmental metabolomics: a critical review and future perspectives. Metabolomics 5:3. https://doi.org/10.1007/s11306-008-0152-0
Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP (2016) DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. https://doi.org/10.1038/nmeth.3869
Delgado-Baquerizo M, Oliverio AM, Brewer TE et al (2018) A global atlas of the dominant bacteria found in soil. Science 359:320–325. https://doi.org/10.1126/science.aap9516
Díaz LC, Arranz JC, Mesa G (2013) Caracterización físico-química y mineralógica de suelos en zona carbonífera del Cesar Colombia. Interciencia 38:42–47
Dolgopolova A, Weiss DJ, Seltmann R, Stanley C, Coles B, Cheburkin AK (2004) Closed-vessel microwave digestion technique for lichens and leaves prior to determination of trace elements (Pb, Zn, Cu) and stable Pb isotope ratios. Int J Environ Anal Chem 84:889–899. https://doi.org/10.1080/03067310410001729006
Doong RA, Lei WG (2003) Solubilization and mineralization of polycyclic aromatic hydrocarbons by Pseudomonas putida in the presence of surfactant. J Hazard Mater 96:15–27. https://doi.org/10.1016/s0304-3894(02)00167-x
Douglas GM, Maffei VJ, Zaneveld JR et al (2020) PICRUSt2 for prediction of metagenome functions. Nat Biotechnol 38:685–688. https://doi.org/10.1038/s41587-020-0548-6
El Baz S, Baz M, Barakate M, Hassani L, El Gharmali A, Imziln B (2015) Resistance to and accumulation of heavy metals by actinobacteria isolated from abandoned mining areas. Sci World J 2015:761834. https://doi.org/10.1155/2015/761834
Fanning DS, Fanning MCB (1989) Soil morphology, genesis, and classification. John Wiley and Sons Inc.
Fierer N, Ladau J (2012) Predicting microbial distributions in space and time. Nat Methods 9:549–551. https://doi.org/10.1038/nmeth.2041
Fox J, Weisberg S (2019) An R companion to applied regression, 3rd edn. Sage, Thousand Oaks CA
Gil-Martínez M, López-García Á, Domínguez MT, Kjøller R, Navarro-Fernández CM, Rosendahl S, Marañón T (2021) Soil fungal diversity and functionality are driven by plant species used in phytoremediation. Soil Biol Biochem 153:108102. https://doi.org/10.1016/j.soilbio.2020.108102
Glöckner FO, Yilmaz P, Quast C, Gerken J, Beccati A, Ciuprina A, Bruns G, Yarza P, Peplies J, Westram R, Ludwig W (2017) 25 years of serving the community with ribosomal RNA gene reference databases and tools. J Biotechnol 261:169–176. https://doi.org/10.1016/j.jbiotec.2017.06.1198
Gomez E, Garland J, Conti M (2004) Reproducibility in the response of soil bacterial community-level physiological profiles from a land use intensification gradient. Appl Soil Ecol 26:21–30. https://doi.org/10.1016/j.apsoil.2003.10.007
Gómez-Sagasti MT, Alkorta I, Becerril JM et al (2012) Microbial monitoring of the recovery of soil quality during heavy metal phytoremediation. Water Air Soil Pollut 223:3249–3262. https://doi.org/10.1007/s11270-012-1106-8
Griffiths BS, Philippot L (2013) Insights into the resistance and resilience of the soil microbial community. FEMS Microbiol Rev 37:112–129. https://doi.org/10.1111/j.1574-6976.2012.00343.x
Guebert MD, Gardner TW (2001) Macropore flow on a reclaimed surface mine: infiltration and hillslope hydrology. Geomorphology 39:151–169. https://doi.org/10.1016/S0169-555X(00)00107-0
Guerra CA, Heintz-Buschart A, Sikorski J, Chatzinotas A et al (2020) Blind spots in global soil biodiversity and ecosystem function research. Nat Commun 11:3870. https://doi.org/10.1038/s41467-020-17688-2
Guerra CA, Bardgett RD, Caon L, Crowther TW et al (2021) Tracking, targeting, and conserving soil biodiversity. Science 371:239–241. https://doi.org/10.1126/science.abd7926
Gundlapally SR, Garcia-Pichel F (2006) The community and phylogenetic diversity of biological soil crusts in the Colorado Plateau studied by molecular fingerprinting and intensive cultivation. Microb Ecol 52:345–357. https://doi.org/10.1007/s00248-006-9011-6
Guo Y, Chen J, Tsolmon B, He A, Guo J, Yang J, Bao Y (2020) Effects of subsidence and transplanted trees on soil arbuscular mycorrhizal fungal diversity in a coal mining area of the Loess Plateau. Glob Ecol Conserv 24:e01308. https://doi.org/10.1016/j.gecco.2020.e01308
Hall EK, Bernhardt ES, Bier RL, Bradford MA et al (2018) Understanding how microbiomes influence the systems they inhabit. Nat Microbiol 3:977–982. https://doi.org/10.1038/s41564-018-0201-z
He ZL, Deng Y, Van Nostrand JD, Tu QC et al (2010) GeoChip 3.0 as a high throughput tool for analyzing microbial community composition, structure and functional activity. ISME J 4:1167–1179. https://doi.org/10.1038/ismej.2010.46
Holguin G (2011) Contexto minero en el departamento del Cesar. Estado actual y proyecciones, Bogotá, p 10
Instituto Colombiano de Normas Técnicas y Certificación – ICONTEC (2007) Norma técnica colombiana NTC 5526. Calidad de suelo. Determinación de micronutrientes disponibles: cobre, zinc, hierro y manganeso. Bogotá. 8
Instituto Colombiano de Normas Técnicas y Certificación - ICONTEC (2008) Norma técnica colombiana NTC 5596. Calidad de suelo. Determinación de la conductividad electrica. Bogotá. 10
Instituto Colombiano de Normas Técnicas y Certificación – ICONTEC (2020) Norma técnica colombiana NTC 5350: Calidad del suelo determinación de fósforo disponible. Segunda actualización. Bogotá. 18
Instituto Colombiano de Normas Técnicas y Certificación - ICONTEC (2016) Norma técnica colombiana NTC 5349. Calidad de suelo. Determinación de las bases cambiables: Método del acetato amonio 1M y pH 7,0. Segunda actualización. Bogotá. 9 p.
Iram S, Ahmad I, Stuben D (2009) Analysis of mines and contaminated agricultural soil samples for fungal diversity and tolerance to heavy metals. Pak J Bot 41:885–895
Jones RT, Robeson MS, Lauber CL, Hamady M, Knight R, Fierer N (2009) A comprehensive survey of soil Acidobacterial diversity using pyrosequencing and clone library analyses. ISME J 3:442–453. https://doi.org/10.1038/ismej.2008.127
Kampf N, Schneider P, Giasson E (1997) Properties: pedogenesis and classification of constructed soils in coal mining areas of the Baixo Jacuí region in Southern Brazil. Braz J Soil Sci 21:9–88
Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M (2016) KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res 44:D457–D462. https://doi.org/10.1093/nar/gkv1070
Kishimoto N, Kosako Y, Tano T (1991) Acidobacterium capsulatum gen. nov., sp. nov.: an acidophilic chemoorganotrophic bacterium containing menaquinone from acidic mineral environment. Curr Microbiol 22:1–7. https://doi.org/10.1007/BF02106205
Kneller T, Harris RJ, Bateman A, Muñoz-Rojas M (2018) Native-plant amendments and topsoil addition enhance soil function in post-mining arid grasslands. Sci Total Environ 621:744–752. https://doi.org/10.1016/j.scitotenv.2017.11.219
Lauber CL, Hamady M, Knight R, Fierer N (2009) Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl Environ Microbiol 75:5111–5120. https://doi.org/10.1128/AEM.00335-09
Li Y, Chen L, Wen H, Zhou T, Zhang T, Gao X (2014) 454 Pyrosequencing analysis of bacterial diversity revealed by a comparative study of soils from mining subsidence and reclamation areas. J Microbiol Biotech 24:313–323. https://doi.org/10.4014/jmb.1309.09001
Lladó S, Žifčáková L, Větrovský T, Eichlerová I, Baldrian P (2016) Functional screening of abundant bacteria from acidic forest soil indicates the metabolic potential of Acidobacteria subdivision 1 for polysaccharide decomposition. Biol Fertil Soils 52:251–260. https://doi.org/10.1007/s00374-015-1072-6
Louca S, Doebeli M, Parfrey LW (2018) Correcting for 16S rRNA gene copy numbers in microbiome surveys remains an unsolved problem. Microbiome 6:41. https://doi.org/10.1186/s40168-018-0420-9
Lozupone CA, Hamady M, Kelley ST, Knight R (2007) Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol 73:1576–1585. https://doi.org/10.1128/AEM.01996-06
Lu ZM, Deng Y, van Nostrand JD et al (2012) Microbial gene functions enriched in the Deepwater Horizon deep-sea oil plume. ISME J 6:451–460. https://doi.org/10.1038/ismej.2011.91
Madej G, Barczyk G, Gdawiec M (2011) Evaluation of soil biological quality index (QBS-ar): its sensitivity and usefulness in the post-mining chronosequence - preliminary research. Pol J Environ Stud 20(5):1367–1372
Maiti SK (2013). Introduction. In: Ecorestoration of the coalmine degraded lands. Springer, India. https://doi.org/10.1007/978-81-322-0851-8_1
Marín C, Kohout P (2021) Response of soil fungal ecological guilds to global changes. New Phytol 229:656–658. https://doi.org/10.1111/nph.17054
McGuire KL, Treseder KK (2010) Microbial communities and their relevance for ecosystem models: decomposition as a case study. Soil Biol Biochem 42:529–535. https://doi.org/10.1016/j.soilbio.2009.11.016
McMurdie PJ, Holmes S (2012) Phyloseq: a bioconductor package for handling and analysis of high-throughput phylogenetic sequence data. In Biocomputing 2012 (235–246). https://doi.org/10.1142/9789814366496_0023
Mishra A, Nautiyal CS (2009) Functional diversity of the microbial community in the rhizosphere of chickpea grown in diesel fuel-spiked soil amended with Trichoderma ressei using sole-carbon-source utilization profiles. World J Microbiol Biotechnol 25:1175–1180. https://doi.org/10.1007/s11274-009-9998-1
Moquin SA, Garcia JR, Brantley SL, Takacs-Vesbach CD, Shepherd UL (2012) Bacterial diversity of bryophyte-dominant biological soil crusts and associated mites. J Arid Environ 87:110–117. https://doi.org/10.1016/j.jaridenv.2012.05.004
Mota EA, Felestrino ÉB, Leão VA, Guerra-Sá R (2020) Manganese (II) removal from aqueous solutions by Cladosporium halotolerans and Hypocrea jecorina. Biotechn Rep 25:e00431. https://doi.org/10.1016/j.btre.2020.e00431
Nelson DW, Sommers LE (1982) Total carbon, organic carbon, and organic matter. p. 539–579. In: Page AL et al (eds) Methods of soil analysis: Part 2. Chemical and microbiological properties. ASA Monograph Number 9. https://doi.org/10.2134/agronmonogr9.2.2ed.c29
Nilsson RH, Larsson K-H, Taylor AFS, Bengtsson-Palme J et al (2019) The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res 47:D259–D264. https://doi.org/10.1093/nar/gky1022
Oksanen J, Blanchet FG, Kindt R, Legendre P et al (2010) Vegan: community ecology package. R package version 1.17–4. http://cran. r-project.org
Ouanphanivanh N, Illyes Z, Merenyi Z, Orczan AK, Szigeti Z, Bratek Z (2013) Mycorrhizal fungi and endophytes of orchids living in three Hungarian abandoned mines. Sydowia 65:245–266
Peng M, Zi X, Wang Q (2015) Bacterial community diversity of oil contaminated soils assessed by high throughput sequencing of 16S rRNA genes. Int J Environ Res Public Health 12:12002–12015. https://doi.org/10.3390/ijerph121012002
Radeva G, Kenarova A, Bachvarova V, Flemming K, Popov I, Vassilev D, Selenska-Pobell S (2013) Bacterial diversity at abandoned uranium mining and milling sites in Bulgaria as revealed by 16S rRNA genetic diversity study. Water Air Soil Pollut 224:1–14. https://doi.org/10.1007/s11270-013-1748-1
Ranjard L, Lejon DPH, Mougel C, Schehrer L, Merdinoglu D, Chaussod R (2003) Sampling strategy in molecular microbial ecology: influence of soil sample size on DNA fingerprinting analysis of fungal and bacterial communities. Environ Microbiol 5:1111–1120. https://doi.org/10.1046/j.1462-2920.2003.00521.x
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. https://doi.org/10.1093/bioinformatics/btp616
RStudio Team (2021) RStudio: integrated development environment for R. RStudio, PBC, Boston. URL: http://www.rstudio.com/. Accessed 2021
Rutgers M, Wouterse M, Drost SM, Breure AM, Mulder C, Stone D et al (2016) Monitoring soil bacteria with community-level physiological profiles using BiologTM ECO-plates in the Netherlands and Europe. Appl Soil Ecol 97:23–35. https://doi.org/10.1016/j.apsoil.2015.06.007
Sait M, Davis KE, Janssen PH (2006) Effect of pH on isolation and distribution of members of subdivision 1 of the phylum Acidobacteria occurring in soil. Appl Environ Microbiol 72:1852–1857. https://doi.org/10.1128/AEM.72.3.1852-1857.2006
Sevigny JL, Rothenheber D, Diaz KS, Zhang Y, Agustsson K, Bergeron RD, Thomas WK (2019) Marker genes as predictors of shared genomic function. BMC Genomics 20:268. https://doi.org/10.1186/s12864-019-5641-1
Shade A, Peter H, Allison SD, Baho DL, Berga M, Bürgmann H et al (2012) Fundamentals of microbial community resistance and resilience. Front Microbiol 3:1–19. https://doi.org/10.3389/fmicb.2012.00417
Shao Z, Sun F (2007) Intracellular sequestration of manganese and phosphorus in a metal-resistant fungus Cladosporium cladosporioides from deep-sea sediment. Extremophiles 11:435–443. https://doi.org/10.1007/s00792-006-0051-0
Singh AN, Singh JS (2006) Experiments on ecological restoration of coal mine spoil using native trees in a dry tropical environment, India: a synthesis. New for 31:25–39. https://doi.org/10.1007/s11056-004-6795-4
Talbot NJ, Coddington A, Roberts IN, Oliver RP (1988) Diploid construction by protoplast fusion in Fulvia fulva (syn. Cladosporium fulvum): genetic analysis of an imperfect fungal plant pathogen. Curr Genet 14:567–572. https://doi.org/10.1007/BF00434082
Taleski V, Dimkić I, Boev B, Boev I, Živković S, Stanković S (2020) Bacterial and fungal diversity in the lorandite (TlAsS2) mine Allcharin the Republic of North Macedonia. FEMS Microbiol Ecol 96(9):fiaa155. https://doi.org/10.1093/femsec/fiaa155
Tardy V, Mathieu O, Lévêque J, Terrat S, Chabbi A, Lemanceau P, Ranjard L, Maron PA (2014) Stability of soil microbial structure and activity depends on microbial diversity. Environ Microbiol Rep 6:173–183. https://doi.org/10.1111/1758-2229.12126
Tu Q, Yu H, He Z, Deng Y, Wu L, Van Nostrand JD, Zhou A, Voordeckers J, Lee Y-J, Qin Y et al (2014) GeoChip 4: a functional gene-array-based high-throughput environmental technology for microbial community analysis. Mol Ecol Resour 14:914–928. https://doi.org/10.1111/1755-0998.12239
Ugland KI, Gray JS, Ellingsen KE (2003) The species–accumulation curve and estimation of species richness. J of Anim Ecol 72(5):888–897. https://doi.org/10.1046/j.1365-2656.2003.00748.x
Unidad de Planeación Minero-Energética - UPME (2016) Boletín Estadístico de Minas y energía 2012 – 2016. Ministerio de Minas y Energía. Available via http://www1.upme.gov.co/simco/Documents/Boletin_Estadistico_2012_2016.pdf. Accessed December 2016
Vahter T, Bueno CG, Davison J, Herodes K, Hiiesalu I, Kasari-Toussaint L et al (2020) Co-introduction of native mycorrhizal fungi and plant seeds accelerates restoration of post-mining landscapes. J of Appl Ecol 57(9):1741–1751. https://doi.org/10.1111/1365-2664.13663
Vargas C (2011) Caracterización florística y fitogeográfica del sector sur de la serranía de Perijá y áreas adyacentes de la cordillera oriental colombiana. Tesis de Maestría. Universidad Nacional de Colombia. Available via http://www.bdigital.unal.edu.co/5239/1/carlosalbertovargasrincon.2011.pdf. Accessed January 2017
Vazquez S, Nogales B, Ruberto L, Mestre C, Christie-Oleza J, Ferrero M et al (2013) Characterization of bacterial consortia from diesel-contaminated Antarctic soils: towards the design of tailored formulas for bioaugmentation. Int Biodeterior & Biodegrad 77:22–30. https://doi.org/10.1016/j.ibiod.2012.11.002
Wei SHI, Xue-na ZHANG, Hai-bin JIA, Sheng-dong FENG, Zhi-xin YANG, Ou-ya ZHAO, Yu-ling LI (2017) Effective remediation of aged HMW-PAHs polluted agricultural soil by the combination of Fusarium sp. and smooth bromegrass (Bromus inermis Leyss.). J of Integr Agric 16.1:199–209 ISSN 2095–3119. https://doi.org/10.1016/S2095-3119(16)61373-4.
Wrighton KC, Thomas BC, Sharon I, Miller CS et al (2012) Fermentation, hydrogen, and sulfur metabolism in multiple uncultivated bacterial phyla. Science 337:1661–1665. https://doi.org/10.1126/science.1224041
Xu X, Wang N, Lipson D, Sinsabaugh R, Schimel J, He L et al (2020) Microbial macroecology: in search of mechanisms governing microbial biogeographic patterns. Global Ecol Biogeogr 29:1870–1886. https://doi.org/10.1111/geb.13162
Zhang X, Lin L, Chen M, Zhu Z, Yang W, Chen B et al (2012) A nonpathogenic Fusarium oxysporum strain enhances phytoextraction of heavy metals by the hyperaccumulator Sedum alfredii Hance. J of Hazard Mater 229:361–370. https://doi.org/10.1016/j.jhazmat.2012.06.013
Zhao OY, Zhang XN, Feng SD, Zhang LX, Shi W, Yang ZX et al (2017) Starch-enhanced degradation of HMW PAHs by Fusarium sp. in an aged, polluted soil from a coal mining area. Chemosphere 174:774–780. https://doi.org/10.1016/j.chemosphere.2016.12.026
Acknowledgements
We thank Luciano Levin, Luciano Merini, Elka Arzuza, Rafael Arango, and Pablo Patiño, and coal mine employees who prefer to be anonymous for their administrative and logistics support during the development of this study.
Funding
This work was possible due to the financial aid for research of “Fondo Regional de Tecnologia Agropecuaria FONTAGRO FTG-5021/16” that supported the work of T. A. C. M. was supported by “Convocatoria Nacional Subvención a Instalación Academia Convocatoria Año 2021 + Folio SA77210019 (ANID—Chile).”
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Arcila-Galvis, J.E., Marín, C., Ortega-Cuadros, M. et al. A Metagenomic Assessment of Soil Microbial Communities in a Coal Mine Spoil Dump Under Reclaimed Vegetation in La Guajira, Colombia. J Soil Sci Plant Nutr 22, 4377–4390 (2022). https://doi.org/10.1007/s42729-022-01036-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42729-022-01036-y