Abstract
Purpose
Retinopathies are the leading cause of eyesight loss, especially among diabetics. Due to the low contrast of blood vessels in fundus images, the visual inspection is a challenging job even for specialists. In this context, this work aims to implement image processing techniques to support contrast enhancement and segmentation of retinal blood vessels.
Methods
The initial proposal consisted only of green channel separation, contrast limited adaptive histogram equalization, and 2D Gabor wavelet and mathematical morphology. The new proposal includes the edge and mask detection and the vessel enhancement 2D to preserve image’s characteristics. The development and validation of this work, in MatLab® environment, involved 40 images from Digital Retinal Images for Vessel Extraction (DRIVE), 20 images from Structured Analysis of the Retina (STARE), and 45 images from High-Resolution Fundus (HRF) database.
Results
In the unsupervised method context, the proposal presented the best performance regarding sensitivity and second place for balanced-accuracy on all databases. A subjective validation involving eleven ophthalmology professionals showed higher levels of acceptance (above 80%) after contrast limited adaptive histogram equalization (CLAHE) and vessel enhancement 2D steps and 75.5% for overall quality system.
Conclusion
The main contributions refer to the inclusion of techniques for automatic mask detection, image edge removal, and suppression of vessels background to improve the retinal vessels segmentation process. In addition, this work made a computational interface named “Retinal Lab - A Tool for Fundus Image Analysis” available, which permits the users to adjust the contrast and segmentation of blood vessels in retinal images.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Assertive and early diagnosis of diseases has always been a great concern to public health (SBD 2018). The benefits are diverse, such as cost reduction of medical treatments and early detection of symptoms of potentially irreversible damages. The medical examinations present the ongoing evolution dynamics aligned with the development of acquisition techniques, processing, and image analysis. In the case of computerized exams, image processing is a key factor that allows emphasizing structures of interest for the identification of potential anomalies (Singh and Kaur 2015).
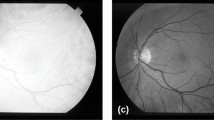
Currently, several researches have been focusing on digital processing of retinal imaging, aiming to identify and analyze various diseases such as diabetic retinopathy, glaucoma, macular degeneration, atherosclerosis, hypertension, and cardiovascular diseases (Singh and Kaur 2015; Zhu et al. 2017). Figure 1 shows images of a healthy retina and a retina with signs of diabetic retinopathy, both from the Structured Analysis of the Retina (STARE) database.
Usually, the blood vessels in the retinal images are manually marked, leading to an increase in the time required for analysis and in financial costs. Supervised or unsupervised image methods intend to obtain automatic vessel segmentation and facilitate the marking process, where one of the main challenges is to improve the low contrast of vessels in relation to the background. Hereafter, the main works on unsupervised methods, which is the same category of this proposal, will be summarized. With few exceptions, all works made use of Digital Retinal Images for Vessel Extraction (DRIVE) and STARE databases.
Nugroho et al. (2018) applied the 2D Gabor wavelet transform (2D-GWT) and morphological operations to fundus images to segment the retinal blood vessels. The pre-processing step consists in extracting the green channel, followed by the complement operation and contrast limited adaptive histogram equalization (CLAHE) stages. In the segmentation step, the 2D-GWT reduces the noise and improves the vascular pattern. The closing operation connects the image points improperly disconnected in the previous processes. Eventually, the morphological reconstruction approaches the object edges through successive operations based on morphological dilation and connectivity.
Neto et al. (2017) segmented the retinal vessels adopting a coarse-to-fine approach, combining Gaussian smoothing, top-hat morphological operator, and contrast enhancement for vessel homogenization and noise reduction. Based on statistics of spatial dependence and probability, the authors present an approximation for the thicker vessels map with a local adaptive threshold. Analyses of curvature and morphological reconstruction refine the segmentation.
Fan et al. (2019) used a hierarchical strategy integrated in the image matting model for blood vessel segmentation based on a trimap created from the characteristics present in the images, separating them in vessels, background, and unknown regions. Then, a hierarchical image matting model defines the pixels of unknown regions as vessel or background.
Sazak et al. (2019) introduced a method called bowler-hat transform to enhance blood vessels in the retina. Based on mathematical morphology, this method combines different structuring elements and minimizes non-uniform illuminating effects, resulting in the preservation of vessel junctions and better detection of fine vessels.
The following works use methods classified as supervised.
Roychowdhury et al. (2015) devised a vessel segmentation method for retinas with abnormalities due to diabetic retinopathy. Initially, a high pass filter and an operation of morphological reconstruction generate two binary images using the green channel. Hence, the pixels classified as vessels in both images will constitute the major vessels (larger vessels). Subsequently, a Gaussian mixture model (GMM) treats the remaining pixels to match them back to the major vessels.
Liskowski and Krawiec (2016) proposed the segmentation of retinal vessels using deep neural networks. The main characteristics in this study are spatial arrangement, local connectivity, parameter sharing, and grouping of hidden units.
In Zhu et al. (2017), a method based on the extreme learning machine (ELM) provides the retinal vascular pattern from a training vector with 39 characteristics for each pixel. These characteristics include the pixel intensity, the result of 2D Gaussian filtering and its derivatives, morphological operations, and top-hat and bottom-hat transformations.
Considering the significance of retinal images in diagnosing various diseases, this work deals with the application of digital processing techniques for contrast enhancement, noise filtering, and automatic retinal vessel segmentation in fundus images. The work was developed in a MatLab®-Version 2017a environment using training and test images obtained from public databases DRIVE (Staal et al. 2004), STARE (Hoover et al. 2000) and HRF (Budai et al. 2013).
Methods
As shown in Fig. 2, this section presents the proposed structure for the contrast enhancement and retinal blood vessels segmentation. Based on Nugroho et al. (2018), this new proposal includes the edge and mask detection using the red channel, retinal edge removal, and suppression of vessels background to remove the retinal edge, preserve vessel junctions, and reduce the noise level. As will be shown, these modifications permitted working better with DRIVE, STARE, and HRF databases.
Flowchart of the proposed method. Pre-processing stage: green and red channels extractions, edge and mask detections, complement operation, retinal edge removal, CLAHE, and suppression of vessels background. Segmentation stage: 2D Gabor wavelet transform and closing operation and morphological reconstruction
Databases
The tests and validation were performed in three public databases, commonly used in most related works. These are DRIVE (digital retinal images for vessel extraction) from Staal et al. (2004), STARE (structured analysis of the retina) from Hoover et al. (2000) and HRF (high-resolution fundus image database), available on www.isi.uu.nl/Research/Databases/DRIVE/, http://cecas.clemson.edu/~ahoover/stare, and https://www5.cs.fau.de/research/data/fundus-images/, respectively.
The DRIVE database provides 40 color fundus images of 584 × 565 pixels size, captured by a Canon CR5 non-mydriatic 3CCD camera with a 45-degree field of view (FOV), in which 7 images present signs of mild early diabetic retinopathy. The 40 images are available in training and test sets, both containing 20 images (Staal et al. 2004).
The STARE database consists of 20 color fundus images of 700 × 605 pixels size, captured by a TopCon TRV-50 fundus camera with a 35-degree FOV. The diagnostics list and the expert annotations of manifestations visible in the images are also available.
The HRF database provides 45 high-resolution color fundus images of 3504 × 2336 pixels size, divided in 3 sets with 15 images of healthy patients, 15 images of patients with diabetic retinopathy, and 15 images presenting with glaucomatous.
All databases supply images with manual medical markings, named ground truth images, which allow performing objective validations.
Pre-processing
The pre-processing techniques, indicated in Fig. 3, aim at image enhancement, suppressing unwanted distortions, and highlighting important characteristics for the segmentation process. Figure 3 shows the fundus image #39 (Fig. 3a), available in the DRIVE database, and the resulting images in the pre-processing stages (Fig. 3b–f).
Green channel extraction
The original images are available in the RGB color space where the pixels intensity can vary from 0 to 255 (8 bits) for each one of these colors. The green channel (Fig. 3b) is the RGB component with the highest contrast between the blood vessels and the background, the reason why this channel is more adequate for the analysis of fundus images.
Complement operation
The complement operation (Fig. 3c) consists of subtracting 255 from the gray level values in the green channel image. The result represents the inversion of image intensity levels, making the blood vessels brighter than the retinal background. The complement of an image A is:
where x and y are pixel coordinates, K = 2l − 1 and l is the number of bits used to represent the intensity z (Gonzalez and Woods 2018).
Edge and mask detections for retinal edge removal
A mask for the region of interest (ROI) is created from the red channel using the Sobel operator and mathematical morphology. As the ROI is not completely circular in some databases, the images must be overlapped on a black rectangle with larger dimensions than the original image to ensure the mask edge detection by the Sobel operator.
As edges are regions characterized for abrupt variations in pixel intensity, they present a high spatial gradient. Sobel operator calculates the 2D spatial gradient in the image and emphasizes the pixels with high spatial gradient (Gupta and Mazumdar 2013). For such, it used a pair of 3 × 3 convolution matrices that slide over the image on the x and y axes, resulting in their respective gradients of magnitude, Gx and Gy. The gradient magnitude Gy is calculated by:
where I is the image. Gx can be obtained in an analogous way, taking the transposed matrix used for calculating Gy. The gradient magnitude |G| is:
Subsequently, the magnitude image is binarized and dilated using a diamond structuring element (SE) with size 2 to eliminate possible gaps in the detected edge. Then, the mask center is filled with binary pixel of true value (1 or white) and smoothed by erosion by a diamond SE with size 7, whose size is adjusted to reduce the white area, keeping only the ROI. The dilation and erosion of an image A by a flat SE B, denoted by A⊕B and A⊖B, respectively, are (Gonzalez and Woods 2018):
Finally, there are the removal of borders and the application of a mask to the resulting image from the complement operation, as shown in Fig. 3d. The mask has the same original image dimensions.
CLAHE
The CLAHE is widely used in medical image processing. The CLAHE algorithm divides the image into rectangular regions, applying some threshold and equalization locally in each region. After setting a threshold for the gray levels, occurrences above this threshold are clipped to minimize saturation, followed by uniform and recursive redistributions along the local histogram. As a result, the background levels become more flattened, increasing the background–vessels contrast (Pizer et al. 1990; Zuiderveld 1994; Nugroho et al. 2018).
Based on the number of regions experimentally determined by Zuiderveld (1994), the images were divided into 32 rows and 32 columns of tiles. The chosen distribution for the histogram was the bell shape, also called Rayleigh, and the value for the clip limit was set to 0.02 within a 0–1 range by successive adjustments, allowing an adequate contrast enhancement with an acceptable noise level for the next steps. Figure 3e shows the enhanced image.
This step offers two different and independent techniques as options to reduce the noise and effects of non-uniform illumination and brightness, preserving vessel junctions and small vessels:
- 1)
Opening operation: according to Gonzalez and Woods (2018), the opening operation suppresses the brighter details that are smaller than the structuring element (SE). For the opening operation, the non-flat SE was defined as a ball, as proposed by Nugroho et al. (2018). The radius (r) and height (h) defined for the SE through the offsetstrel MatLab® function were determined experimentally as 2 and 140, respectively, aiming for the smallest SE and average grayscale values. The increase of h makes the image background uniform, but eliminates thinner blood vessels.
- 2)
Vessel enhancement 2D: Sazak et al. (2019) developed a method based on Zana and Klein (2001) to enhance elongated vessel-like structures in biomedical images. It carries out a series of morphological openings with a line-shaped SE across defined angles. According to Sazak et al. (2019), the SE width is 1 pixel and its length is the biggest vessel expected diameter. For each angle, segments smaller than the SE are removed, whereas the bigger ones remain unchanged. The final enhanced image is a pixel-wise maximum among all images produced in all considered orientations given in the following:
where Iout is the resulting image, I is the input image, the symbols “∘” and bθ indicate the opening operation and the SE respectively, and θ are the angles in the n selected orientations. In this work, following the suggestions of Sazak et al. (2019), the length of SE was set to 10 and the number of orientations n was set to 12.
Aiming to work with high-resolution images such as the ones available at HRF database, this proposal included an automatic adjustment for sizes of SEs used in the pre-processing stage. This adjustment is based on the size ratio between the DRIVE training and the test images, taking the number of columns or rows of those images into account.
Segmentation
The segmentation aims to extract the blood vessels from the retinal background. Figure 4 shows the resulting images for each segmentation step, consisting of 2D-GWT (Fig. 4a), a closing operation (Fig. 4b), and final image, after the morphological reconstruction (Fig. 4e). Figure 4f shows the corresponding ground truth image from DRIVE. Figure 4c and d highlights the same regions from the results of 2D-GWT and closing operation, respectively.
2D Gabor wavelet transform
According to Jain and Farrokhnia (1990), a 2D Gabor function corresponds to a sinusoidal plane wave, with certain frequency and orientation, modulated by a two-dimensional Gaussian envelope, where the image information is extracted by measuring the energy in small windows. Blood vessel extraction by 2D-GWT (Fig. 4a) is due to its ability to provide spatial information, orientation selectivity, and spectral characteristics, which makes it possible to detect directional structures and high frequency regions, such as blood vessel edges. The 2D-GWT is defined as (Soares et al. 2006):
where \( A=\mathit{\operatorname{diag}}\left[{\epsilon}^{-\frac{1}{2}},1\right] \) is a 2 × 2 diagonal matrix and k0 is a vector. ϵ ≥ 1 indicates the filter anisotropy, i.e., its elongation in any direction, and k0 is the complex frequency.
In this work, the orientation starts from 0 to 165o, in steps of 15o, resulting in 12 directions in which the vessel characteristics are extracted. According to Soares et al. (2006), ϵ was set to 4 and the frequency was set to 4 for both databases.
Closing operation
The closing operation (Fig. 4b) suppresses dark details smaller than the SE. This operation allows fulfilling the central parts of larger vessels not fully detected by the 2D-GWT due to its distance from the vessel edges (low frequency region) as well as to link up disconnected vessels especially at junctions. The closing of A by SE B, denoted A ¥ B, is:
which indicates that the closing of A by B is the dilatation (⊕) of A by B followed by the erosion (⊖) of this resulting image by B (Gonzalez and Woods 2018).
The SE was defined as flat diamond (Nugroho et al. 2018) and the distance between the origin and the edge was adjusted to 2, the minimum value available, aiming to fill the larger vessels. Values greater than 2 made the vessels wider and linked different vessels at junctions, increasing the number of pixels wrongly detected as vessels. To illustrate the filling of one vessel after the closing operation, Fig. 4c and d presents pixel details resulting from 2D-GWT and closing operation, respectively.
Morphological reconstruction
The morphological reconstruction involves two images, named marker and mask, and an SE. The marker contains the starting points for the transformation after which the image will suffer successive dilations until it reaches the mask. The mask function is to limit the marker transformation and the SE defines the connectivity. The morphological reconstruction by geodesic dilation of size n, denoted by \( {\mathrm{D}}_{\mathrm{G}}^{\left(\mathrm{n}\right)}\left(\mathrm{F}\right) \), is defined as (Gonzalez and Woods 2018):
where G, F, and B are the mask, marker, and the SE, respectively.
The final image (Fig. 4e) is the result from morphological reconstruction. The resultant image from the closing operation makes up the mask while the 2D-GWT image functions as the marker. Fig. 4f shows the respective ground truth image marked by a specialist, provided by DRIVE database.
The integrated computational interface
The algorithm was implemented in MatLab® environment and a graphical user interface named “Retinal Lab-A Tool for Fundus Image Analysis” was developed as an application for monitoring images through pre-processing, contrast enhancement, and segmentation steps, with parameter adjustments as well as performance indexes. This interface is available at https://github.com/DouglasAbreu/RetinalLab.
On an Intel Core I5 and 4GB RAM computer, the full processing of an image in MatLab® environment took an average time of 2.6 s for DRIVE and STARE databases and 58.67 s for HRF, involving all processing stages. Figure 5 shows the developed graphical user interface, whose operation begins from selecting an image stored in the same computational environment.
Results
The validation included comparative analyses using objective metrics usually applied in medical image processing and content validity tests performed by ophthalmologists and retinal specialists using the “Retinal Lab-A Tool for Fundus Image Analysis.”
Objective validation
For objective evaluation, the tests used the 20 remaining images from the DRIVE and all images available in the STARE and HRF databases. After submitting the selected image to all proposed steps, the segmented image was then compared with the respective ground truth image, available in the databases.
The validation criteria according to the sensitivity, specificity, accuracy, and balanced-accuracy are (Nugroho et al. 2018; Neto et al. 2017):
TP (true positive) and TN (True Negative) are the numbers of pixels correctly detected as vessels and as background, respectively, while FP (false positive) and FN (false negative) indicate the numbers of pixels wrongly detected as vessels and as fundus, respectively. In Eq. 12, the weights α and β were adjusted to 0.5 to equally balance the sensitivity and specificity (Neto et al. 2017).
Aiming to validate this proposal, the obtained results were compared with the main works in available literature, separated into supervised and unsupervised classes, since they involve different methodological approaches. Table 1 shows the average results achieved, based on images from DRIVE, STARE, and HRF, highlighting in bold the highest rates for each category and database.
Subjective validation
The accomplishment of subjective validation involved eleven ophthalmology professionals, including four retinal specialists. Aiming to evaluate the image quality after the 5 main test phases, as well as the system overall quality, the doctors assessed the resultant images of each processing phase using the retinal lab interface and images on DRIVE and STARE databases, randomly selected by the participants. The score tests consisted of discrete grading scales between 1 and 20, encompassing 1–4 (bad), 5–8 (poor), 9–12 (reasonable), 13–16 (good), and 17–20 (excellent), subsequently converted to a 1–100 continuous range. The statistical analysis was performed in R software environment.
Considering the 5 questions in the tests, the answers produced 55 scores while the overall quality was inferred from the scores attributed to the steps previously evaluated. Figure 6 shows typical boxplots for the test results, with the exclusion of one participant (outlier) that did not complete all test steps. The results show the evaluation of image quality after the green channel extraction (median score 82.5), CLAHE (median score 90), suppression of vessels background (median score 80), highlighting of vessels (median score 72.5), removal of details (median score 77.5), and overall quality (median score 75.5).
Discussion
Based on Nugroho et al. (2018), this proposal included steps for automatic mask detection and image edge removal through the red channel, using Sobel operator and mathematical morphology. Furthermore, two additional options for the suppression of vessel background consisted of opening operation and vessel enhancement offer better conditions to the following steps, CLAHE and 2D-GWT, normally used in applications to improve image contrast and segmentation.
From the supervised methods, according to Table 1, on the DRIVE database, Liskowski and Krawiec (2016) and Zhu et al. (2017) presented the best performances regarding sensitivity, specificity, accuracy, and balanced-accuracy. On the STARE database, Liskowski and Krawiec (2016) showed the best results in relation to sensitivity and balanced-accuracy and, Staal et al. (2004) and Roychowdhury et al. (2015) for accuracy and specificity, respectively. A disadvantage of supervised methods refers to the need for prior training, which implies an additional cost in computation and classification time.
Considering the unsupervised methods presented in Table 1, which are the research subject in this study, the proposed method presented the best results regarding sensitivity on all databases. Regarding specificity, Sazak et al. (2019) reached the highest rates on DRIVE (herewith Fan et al. (2019)) and on all the other databases. In relation to accuracy, Fan et al. (2019) obtained the best results on DRIVE and Sazak et al. (2019) on STARE and HRF. With regard to balanced-accuracy, Nugroho et al. (2018), Fan et al. (2019), and Sazak et al. (2019) presented the best results on DRIVE, STARE, and HRF, respectively.
In summary, the proposed method presented the best performance in terms of sensitivity without significant losses of other parameters, and the second place for balanced-accuracy on all the databases. A limitation of this method refers to the presence of lesions with dimensions comparable to biggest blood vessels. These lesions are not fully removed by vessel enhancement 2D and, therefore, are accounted as false–positives.
Based on the subjective validation results, images obtained after green channel extraction, CLAHE, and vessel background suppression stages achieved higher levels of acceptance (above 80%). On the other hand, there was a decrease regarding the results of vessel highlighting and detail removal. From the overall quality, one can infer a positive acceptance from the experts consulted.
Conclusion
The main contributions refer to the inclusion of techniques for automatic mask detection, image edge removal, and suppression of vessels background.
In this proposal, the sensitivity had higher priority due to its importance in the early diagnosis of diabetic retinopathy in the proliferative phase when the disease causes the emergence of new vessels in the retina, which grow towards the vitreous interface and may progress to irreversible loss of eyesight (Bosco et al. 2005).
Another contribution of this work is the computer interface “Retinal Lab-A Tool for Fundus Image Analysis”, developed in MatLab® environment and later made available in an executable version at https://github.com/DouglasAbreu/RetinalLab, which allows users to access retinal blood vessel contrast and segmentation adjustments.
In future works, it is expected the usage of broader databases, which will allow training and testing steps in a wider range of images and to incorporate other techniques to the computational platform aiming to extend the detection to other retinal structures and pathology characterization.
References
Aguirre-Ramos H, Avina-Cervantes JG, Cruz-Aceves I, Ruiz-Pinales J, Ledesma S. Blood vessel segmentation in retinal fundus images using Gabor filters, fractional derivatives, and expectation maximization. Appl Math Comput. 2018;339:568–87. https://doi.org/10.1016/j.amc.2018.07.057.
Bosco A, Lerário AC, Soriano D, dos Santos RF, Massote P, Galvão D, et al. Retinopatia diabética. Arq Brasil Endocrinol Metabol. 2005;49(2):217–27. https://doi.org/10.1590/S0004-27302005000200007.
Budai A, Bock R, Maier A, Hornegger J, Michelson G. Robust vessel segmentation in fundus images. Int J Biomed Imag. 2013;2013. https://doi.org/10.1155/2013/154860.
Fan Z, Lu J, Wei C, Huang H, Cai X, Chen X. A hierarchical image matting model for blood vessel segmentation in fundus images. IEEE Trans Image Process. 2019;28(5):2367–77. https://doi.org/10.1109/TIP.2018.2885495.
Gonzalez RC, Woods RE. Digital image processing. 4th ed. New Jersey: Pearson Education; 2018.
Gupta S, Mazumdar SG. Sobel edge detection algorithm. International Journal of Computer Science and Management Research. 2013;2(2):1578–83.
Hoover A, Kouznetsova V, Goldbaum M. Locating blood vessels in retinal images by piece-wise threhsold probing of a matched filter response. IEEE Trans Med Imaging. 2000;19(3):203–10. https://doi.org/10.1109/TMI.2003.815900.
Jain AK, Farrokhnia F. Unsupervised texture segmentation using Gabor filters. In: 1990 IEEE international conference on systems, man, and cybernetics conference proceedings; 1990 Nov 4–7. Los Angeles: IEEE; 1990. p. 14–9. https://doi.org/10.1109/ICSMC.1990.142050.
Liskowski P, Krawiec K. Segmenting retinal blood vessels with deep neural networks. IEEE Trans Med Imaging. 2016;35(11):2369–80. https://doi.org/10.1109/TMI.2016.2546227.
Neto LC, Ramalho GLB, Neto JFSR, Veras MSR, Medeiros FNS. An unsupervised coarse-to-fine algorithm for blood vessel segmentation in fundus images. Expert Syst Appl. 2017;78:182–92. https://doi.org/10.1016/j.eswa.2017.02.015.
Nugroho HA, Lestari T, Aras RA, Ardiyanto I. Segmentation of retinal blood vessels using Gabor wavelet and morphological reconstruction. In: Proceedings of the 3rd international conference on science in information technology (ICSITech); 2017 Oct 25–26; Bandung, Indonesia. USA: IEEE; 2018. p. 513–6. https://doi.org/10.1109/ICSITech.2017.8257166.
Pizer SM, Johnston RE, Ericksen JP, Yankaskas BC, Muller KE. Contrast-limited adaptive histogram equalization: speed and effectiveness. In: Proceedings of the first conference on visualization in biomedical computing; 1990 may 22–25. Atlanta: IEEE; 1990. p. 1990. https://doi.org/10.1109/VBC.1990.109340.
Roychowdhury S, Koozekanani DD, Parhi KK. Blood vessel segmentation of fundus images by major vessel extraction and subimage classification. IEEE J Biomed Health Inform. 2015;19(3):1118–28. https://doi.org/10.1109/JBHI.2014.2335617.
Sazak Ç, Nelson CJ, Obara B. The multiscale bowler-hat transform for the blood vessel enhancement in retinal images. Pattern Recogn. 2019;88:739–50. https://doi.org/10.1016/j.patcog.2018.10.011.
SBD (Sociedade Brasileira de Diabetes) Oliveira JEP, Junior RMM, Vencio S. Diretrizes da Sociedade Brasileira de Diabetes 2017-2018. Editora Clannad; 2018. ISBN: 978-85-93746-02-4. Available from: https://www.diabetes.org.br/profissionais/images/2017/diretrizes/diretrizes-sbd-2017-2018.pdf. Accessed 03 Nov 2018.
Singh N, Kaur L. A survey on blood vessel segmentation methods in retinal images. In: Proceedings of the 2015 international conference on electronic design, Computer Networks & Automated Verification (EDCAV); 2015 Jan 29–30; Shillong, India. USA: IEEE; 2015. p. 23–8. https://doi.org/10.1109/EDCAV.2015.7060532.
Soares JVB, Leandro JJG, Cesar RM, Jelinek HF, Cree MJ. Retinal vessel segmentation using the 2-D Gabor wavelet and supervised classification. IEEE Trans Med Imaging. 2006;25(9):1214–22. https://doi.org/10.1109/TMI.2006.879967.
Staal J, Abramoff M, Niemeijer M, Viergever M, van Ginneken B. Ridge-based vessel segmentation in color images of the retina. IEEE Trans Med Imaging. 2004;23(4):501–9. https://doi.org/10.1109/TMI.2004.825627.
Zana F, Klein J. Segmentation of vessel-like patterns using mathematical morphology and curvature evaluation. IEEE Trans Image Process. 2001;10(7):1010–9. https://doi.org/10.1109/83.931095.
Zhu C, Zou B, Zhao R, Cui J, Duan X, Chen Z, et al. Retinal vessel segmentation in colour fundus images using extreme learning machine. Comput Med Imaging Graph. 2017;55:68–77. Special Issue on Ophthalmic Medical Image Analysis. https://doi.org/10.1016/j.compmedimag.2016.05.004.
Zuiderveld K. Contrast limited adaptive histograph equalization. Graphic Gems IV. San Diego: Academic Press Professional; 1994. p. 474–85.
Acknowledgments
The authors would like to thank J. J. Staal, A. Hoover, and their colleagues for making their databases publicly available and Dr. Ricardo Guimarães Eye Hospital for the support in taking the subjective tests.
Funding
This study was financially supported in part by CAPES-Finance Code 001.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
da Rocha, D.A., Barbosa, A.B.L., Guimarães, D.S. et al. An unsupervised approach to improve contrast and segmentation of blood vessels in retinal images using CLAHE, 2D Gabor wavelet, and morphological operations. Res. Biomed. Eng. 36, 67–75 (2020). https://doi.org/10.1007/s42600-019-00032-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42600-019-00032-z