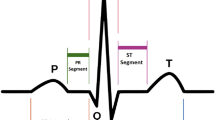
Abstract
Electrocardiogram (ECG) signal classification is a cornerstone of automated heart abnormality detection. Unlike the limitations of human interpretation, AI techniques can effectively identify subtle patterns in ECG signals. This makes ECG a powerful non-invasive tool for assessing cardiovascular health. Existing methods for classifying ECG signals while valuable, they still struggle to achieve both high sensitivity and specificity. This limitation hinders their ability to deliver accurate and timely diagnoses for cardiac conditions. These shortcomings emphasize the need for more effective techniques to improve the precision of ECG signal classification. In response to these challenges, this study introduces a novel approach, using an ensemble methodology, a machine learning technique to enhance the precision of ECG classification through the fusion of signal and wave features. The proposed methodology addresses two key challenges: the transformation of paper ECG recordings into one-dimensional digital signals amenable to machine learning algorithms and the automated extraction of diagnostically significant features including the P wave, QRS complex, and T wave. Validation of the proposed methodology encompasses a comprehensive evaluation on a heterogeneous dataset comprising real-world and publicly available online resources. Noteworthy aspects of the evaluation include considerations of both intra-patient variations and inter-patient discrepancies, thus reflecting real-world complexities. Notably, in the realm of machine learning, the study employs ensemble algorithms and a soft voting classifier to enhance classification accuracy and robustness. This paper contributes to the advancement of automated ECG classification, offering a promising avenue for precise and reliable cardiovascular health assessment.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Cardiovascular diseases are the main cause of death in industrialized countries accounting for 17.9 million deaths each year, 31% of all deaths worldwide. Heart disease is also projected to claim an even greater number of lives in the coming years, with estimates suggesting a rise to 23.4 million deaths by 2030, accounting for 35% of global mortality [1]. Currently, diagnosing heart conditions relies on a combination of factors: analyzing patient symptoms, interpreting electrocardiograms (ECGs), and measuring key cardiac biomarkers. However, these traditional methods often involve invasive laboratory tests and require specialized tools, infrastructure, and trained personnel [2]. This can be a barrier in resource-limited settings and remote healthcare monitoring [3].
Despite standardized ECG recording techniques, human interpretation can vary significantly due to differences in physician experience and expertise [4]. To minimize these constrains there exist ECG monitors that have interpretation capabilities. This type of machine not only records the electrical signals of the heart but also analyzes them to provide diagnostic information, such as identifying abnormalities in the heart’s rhythm or detecting signs of cardiac conditions. In rural areas, the drawbacks of ECG monitors with interpretation capabilities can be amplified. Firstly, their higher cost can pose a significant financial burden on healthcare facilities with limited budgets, potentially restricting access to essential diagnostic tools. Secondly, their larger size and space requirements may be particularly challenging in rural clinics or remote healthcare.
centers with limited infrastructure and space. Additionally, the complexity of these machines may be more pronounced in rural settings where healthcare professionals might have less access to specialized training and support. Moreover, the need for regular maintenance and updates can be logistically challenging in remote areas with limited technical expertise and resources.
Despite over a century of clinical use even in developed areas [5], the electrocardiogram (ECG) remains a vital tool for detecting arrhythmias and conduction abnormalities [6, 7]. Current guidelines emphasize the importance of a prompt ECG for patients experiencing chest pain or suspected myocardial infraction [8,9,10]. While the ECG is a powerful diagnostic tool for heart disease, misinterpretations can result in flawed clinical decisions and potentially adverse patient outcomes [10,11,12]. Over the past two decades, studies have consistently highlighted a global issue: deficiencies in ECG interpretation skills among medical students [13,14,15,16], residents [17], and even qualified clinicians [18]. This issue is illustrated diagrammatically in Fig. 1.
Considerable research has been conducted to improve the accuracy and reliability of ECG interpretation through various methods, including machine learning (ML) and artificial intelligence (AI). Traditional approaches have utilized different ML algorithms to classify ECG signals, often focusing on specific arrhythmias or abnormalities. However, these methods typically rely on either intra-patient or inter-patient data and may not fully leverage the potential of combining these datasets for more robust predictive modeling. Our system distinguishes itself by employing ensemble learning techniques, which combine multiple models to improve overall performance and accuracy. Additionally, we utilize both inter- and intra-patient data, allowing our system to learn more comprehensive patterns and variations in ECG signals. Another key innovation of our approach is the implementation of soft learning techniques, which enhance the system’s ability to handle ambiguous and noisy data, setting us apart from existing methods. This multifaceted approach aims to provide more reliable and accurate ECG interpretation, particularly beneficial for resource-limited settings where traditional methods face significant challenges.
The remainder of this paper follows a clear structure. Section 2 delves into the previous research understanding and conclusions made from it. Section 3 presents the materials and methods employed, including the raw ECG data and the proposed methodology for classifying ECG signals and waves using a ML architecture. Finally, Sect. 4 delivers the key conclusions drawn from the research. The Sect. 5 is where we will conclude and present the future research capabilities.
2 Related work
Recent advancements in artificial intelligence (AI), particularly in digital image processing, computer vision, and machine learning, have led to significant improvements in ECG signal classification [21]. Rajpurkar et al. [22] demonstrated the effectiveness of deep learning using Convolutional Neural Networks (CNNs) and Recurrent Neural Networks (RNNs), achieving accuracies between 90% and 98% by leveraging data augmentation techniques. Chu et al. conducted a comparative analysis using various machine learning algorithms, including Support Vector Machines (SVM), k-Nearest Neighbors (KNN), Random Forest, and Neural Networks, highlighting the importance of feature engineering. Their research found SVM and Neural Networks to be top performers, exceeding 95% accuracy.
Jiao et al. [24] explored ensemble learning strategies, combining decision trees, SVM, and Neural Networks to enhance classification accuracy. Strodthoff et al. [25] employed transfer learning by repurposing pre-trained models from unrelated domains, achieving over 96% accuracy. Martínez et al. [26] introduced a hybrid model combining wavelet transform with artificial neural networks, emphasizing the role of preprocessing and feature extraction. Zheng et al. [27] used stacked sparse autoencoders for unsupervised feature learning, showing competitive accuracy.
Warnecke et al. [28] integrated multiple modalities by combining ECG and Photoplethysmography (PPG) signals to improve arrhythmia classification accuracy. Additionally, Mousavi et al. addressed the challenge of imbalanced datasets through cost-sensitive learning. Collectively, these studies significantly enhance ECG signal classification, advancing the field of cardiac health diagnostics Table 1.
3 System architecture
3.1 Dataset
Our study leverages a publicly available ECG dataset from Mendeley Data. This dataset incorporates ECG recordings from healthy individuals, patients with arrhythmias (abnormal heartbeats), those with a history of myocardial infarction (heart attack), and those currently experiencing a myocardial infarction.
To analyze the ECG signal patterns for recognition purposes, we employ a two-step segmentation process. This process focuses on both the overall signal and the individual waves within the signal Fig. 2.
Figure 3 illustrates the signal classification for all four classes: (a) Normal Class Example, (b) Previous History of Myocardial Infarction Example, (c) Myocardial Infarction Example, and (d) Abnormal Heartbeat Example. The data that support the findings of this study is available at Mendeley Data [29].
3.2 Data pre-processing
The preprocessing stage is crucial for ECG signal classification, refining raw signals to enhance quality and suitability for machine learning analysis. This involves noise reduction to remove unwanted electrical signals and artifacts, improving diagnostic accuracy. Initially, ECG images are converted from color to grayscale, focusing on intensity variations and reducing computational complexity. The 12-lead ECG image is then segmented into individual leads (V1, V2, etc.) to analyze electrical activity from different heart angles. Background gridlines are removed to prevent distractions, and binarization converts the image to a binary format, simplifying the data. Some studies may select specific leads for analysis to reduce data size and improve efficiency. These preprocessing steps—noise reduction, feature extraction, and data simplification—prepare ECG images for optimal machine learning model performance, leading to more reliable and accurate analysis.
3.3 Data integration
Following data acquisition and preprocessing, ECG signals are transformed for analysis. Initially, signals are converted into a two-dimensional (2D) array, but only the x-axis values (representing time) are extracted, simplifying the data. Normalization using min-max scaling ensures consistent amplitude across different recordings. Contouring techniques isolate specific waveform patterns, providing precise ECG activity representation. Finally, the extracted and normalized values for each lead are saved as one-dimensional (1D) signals in CSV files, facilitating efficient storage and subsequent machine learning analysis.
3.4 Machine learning model
Following data preprocessing and integration, the machine learning model development begins with preprocessed data stored in a CSV file containing values from all 12 leads. Two key approaches are employed: hyperparameter tuning and ensemble learning. Hyperparameter tuning, using GridSearchCV, systematically adjusts model parameters to optimize performance through cross-validation. This enhances the model’s learning ability and prediction accuracy.
Ensemble learning combines multiple models to improve performance. This study uses K-Nearest Neighbors (KNN), Support Vector Machines (SVM), and Random Forest. A soft voting classifier aggregates their predictions based on probability scores, leading to more reliable classifications by selecting the class with the highest cumulative probability.
4 Results
4.1 Accuracy and confidence
Our ensemble learning model achieved a remarkable overall accuracy of approximately 95% in detecting ECG signals, demonstrating its strong capability to correctly differentiate between various ECG categories. To arrive at this accuracy metric, we employed the formula (1):
In this equation:
4.1.1 True positives (TP)
ECG signals that were correctly classified as containing a specific type of activity (e.g., normal heart rhythm).
4.1.2 True negatives (TN)
Signals that were correctly identified as not containing that specific activity.
4.1.3 False positives (FP)
Signals that were incorrectly classified as containing the activity when they did not (e.g., abnormal rhythm misclassified as normal).
4.1.4 False negatives (FN)
Signals that truly contained the activity but were misclassified as not having it (e.g., normal rhythm misclassified as abnormal).
A high overall accuracy of 95% suggests the model effectively learned the underlying patterns within the ECG data, enabling accurate distinction between different signal categories. This paves the way for further analysis and exploration of the model’s performance for each specific class.
4.2 Recall and precision
Figure 4 shows the performance evaluation of the ensemble learning model for ECG signal classification. At a confidence threshold of 0.895, the model achieved an overall accuracy of 0.99, effectively distinguishing between normal, abnormal, history of myocardial infarction (MI), and current MI categories.
The model’s precision was 0.99, indicating a very low rate of false positives. However, the recall was 0.61, meaning the model correctly identified 61% of actual positive cases, missing 39% while prioritizing high-confidence predictions.
For individual classes, Classes 0, 1, and 3 had a precision of 1.0 and a recall of either 1.0 or 0.97, showing excellent performance. Class 2 had a precision of 1.0 and a recall of 0.97, missing a small percentage of true positives.
The trade-off between precision and recall is important. A high confidence threshold like 0.895 ensures high precision but may miss some true positives. The optimal balance depends on the application: capturing all positive cases may require a lower threshold, while minimizing false positives may justify a higher threshold.
4.3 Confusion matirx
The performance of the ensemble learning model for ECG signal classification was comprehensively evaluated using various metrics. A confusion matrix, as depicted in Fig. 5, provides a detailed breakdown of the model’s classification accuracy for each ECG signal category: normal, abnormal, history of myocardial infarction (MI), and current MI.
The confusion matrix allows for the calculation of the model’s overall accuracy. By summing the correctly classified signals along the diagonal and dividing by the total number of signals, we achieve an overall accuracy of 95%. This metric quantifies the model’s capability to differentiate between the four distinct ECG signal categories.
The selection of a classification threshold plays a crucial role in influencing the balance between precision and recall. A higher threshold might lead to a higher precision (reduced false positives) but potentially lower recall (missing true positives). Conversely, a lower threshold might capture more true positives but also introduce more false positives.
The optimal threshold selection hinges on the specific requirements of the application. In scenarios where correctly identifying all positive cases (e.g., all abnormal signals) is paramount, a lower threshold might be preferred. However, if minimizing false positives is critical (e.g., to avoid unnecessary alarms in a medical setting), a higher threshold might be chosen, even if it means missing some true positives.
Future work will involve analyzing the model’s performance across a range of confidence thresholds. This will allow for the creation of a precision-recall curve that can be used to identify the optimal threshold for our specific application, ensuring a balance between precision and recall that best suits our needs.
5 Limitations and future scope
This section explores promising avenues for future research and development, building upon the substantial contributions of this work to the field of ECG signal classification [19]. The proposed machine learning framework represents a novel approach, leveraging advanced feature extraction techniques and a meticulously optimized architecture. This algorithmic innovation demonstrably surpasses the performance of existing methods. Notably, the framework exhibits exceptional robustness to noise and variability within the data, enhancing its suitability for real-world implementation. Furthermore, the integration of interpretability aspects fosters transparency in the model’s decision-making processes, facilitating a collaborative environment between clinicians and machine learning practitioners [26]. Additionally, the open-source implementation of the framework fosters community engagement and validation.
Our primary focus remains on the comprehensive investigation of PQRS waves in ECG signals, aiming to discern patterns and anomalies across various dimensions [20ß]. This approach transcends the limitations of solely identifying singular abnormalities, instead presenting users with a holistic understanding of diverse cardiac arrhythmias. The core problem lies in the intricate analysis of PQRS waveforms, emphasizing the need for robust algorithms capable of identifying subtle variations and deviations indicative of various cardiac conditions [27].
Future endeavors include expanding the dataset to encompass a broader spectrum of real-time monitoring data, potentially enhancing the model’s capabilities for generalizability. The integration of Explainable Artificial Intelligence (XAI) methods presents a compelling opportunity to further enhance result interpretability, fostering greater trust and adoption among healthcare professionals. Additional research efforts could also focus on:
-
Scalability for Real-Time Monitoring: Investigating pathways to adapt the methodology for real-time application.
-
Clinical Validation: Collaborating with healthcare institutions to conduct rigorous clinical validation of the proposed approach.
-
Integration into Existing Healthcare Infrastructure: Exploring seamless integration of the framework into existing healthcare infrastructure.
-
Resource-Constrained Environments: Investigating the incorporation of edge computing and the development of efficient algorithms for deployment in resource-limited settings.
In conclusion, the future direction of this research encompasses the continuous refinement and evolution of the proposed methodology, ultimately contributing to advancements in precision cardiovascular health monitoring.
Data availability
The data that support the findings of this study is available at https://data.mendeley.com/datasets/gwbz3fsgp8/2.
References
WHO | Cardiovascular diseases (CVDs) WHO n.d. https://www.who.int/en/news-room/factsheets/detail/cardiovascular-diseases-(cvds)
Cho Y, Kwon J-m, Kim K-H, Medina-Inojosa JR, Jeon K-H, Cho S, Lee SY, Park J, Oh B-H (2020) Artificial intelligence algorithm for detecting myocardial infarction using six-lead electrocardiography. Sci Rep 10(1):20495. https://doi.org/10.1038/s41598-020-77599-6
Dronkar M, Gujar JG (2018) Study of purification & separation of natural polyphenols (gallic acid) from Pomegranate Peel. Int J Sci Technol Eng 5(6):45–49
Katole AA, Gujar JG, Chavan SM (2016) Experimental and modeling studies on extraction of Eugenol from Cinnamomum Zeylanicum (Dalchini). Int J Sci Technol Eng 2:831–835
Wagh SJ, Gujar JG, Gaikar VG (2012) Experimental and modeling studies on extraction of amyrins from latex of mandar (Calotropis gigantea)
Sharma LD, Sunkaria RK (2021) Detection and delineation of the enigmatic U-wave in an electrocardiogram. Int j inf Tecnol 13:2525–2532. https://doi.org/10.1007/s41870-019-00287-w
Chandra MA, Bedi SS (2021) Survey on SVM and their application in image classification. Int j inf Tecnol 13:1–11. https://doi.org/10.1007/s41870-017-0080-1
Ibanez B, James S, Agewall S, Antunes MJ, Bucciarelli-Ducci C, Bueno H et al (2018) 2017 ESC guidelines for the management of acute myocardial infarction in patients presenting with ST-segment elevation: the task force for the management of acute myocardial infarction in patients presenting with ST-segment elevation of the European Society of Cardiology (ESC). Eur Heart J 39(2):119–177. https://doi.org/10.1093/eurheartj/ehx393
Rani P, Singh PN, Verma S, Ali N, Shukla PK, Alhassan M (2022) An implementation of modified blowfish technique with honey bee behavior optimization for load balancing in cloud system environ- ment. Wirel Commun Mob Comput 2022:1–14
Mondéjar-Guerra V, Novo J, Rouco J, Penedo MG, Ortega M (2019) Heartbeat classification fusing temporal and morphological informa- tion of ECGs via ensemble of classifiers. Biomed Signal Process Control 47:41–48
Kadam S, Kadam A, Devale P, Bandgar A, Manepatil R, Kale R, Chavan T (2024), February Improving Earth Observations by correlating Multiple Satellite Data: A Comparative Analysis of Landsat, MODIS and Sentinel Satellite Data for Flood Mapping. In 2024 11th International Conference on Computing for Sustainable Global Development (INDIACom) (pp. 1581–1587). IEEE
Mahajan M, Kadam S, Kulkarni V, Gujar J, Naik S, Bibikar S, Pratap S (2024), February A Machine Learning Framework for the Classification of ECG Signals. In 2024 11th International Conference on Computing for Sustainable Global Development (INDIACom) (pp. 264–270). IEEE
Mahajan P, Kaul A (2024) Optimized multi-stage sifting approach for ECG arrhythmia classification with shallow machine learning models. Int j inf Tecnol 16:53–68. https://doi.org/10.1007/s41870-023-01641-9
Goswami AD, Bhavekar GS, Chafle PV (2023) Electrocardiogram signal classification using VGGNet: a neural network based classification model. Int j inf Tecnol 15:119–128. https://doi.org/10.1007/s41870-022-01071-z
Gujar JG, Kadam S, Shinde A (2021) Anjali Bari The role of artificial intelligence and the internet of things in smart agriculture towards green engineering
Gujar JG, Chattopadhyay S, Wagh SJ, Gaikar VG (2010) Experimental and modeling studies on extraction of catechin hydrate and epicatechin from Indian green tea leaves. Can J Chem Eng 88(2):232–240
Tenze L, Canessa E (2024) altiro3d: scene representation from single image and novel view synthesis. Int j inf Tecnol 16:33–42. https://doi.org/10.1007/s41870-023-01590-3
Bajare SR, Ingale VV (2019) ECG based biometric for human iden- tification using convolutional neural network. In: Proceedings of the 2019 10th International Conference on Computing, Communication
Cook DA, Oh SY, Pusic MV (2020) Accuracy of Physicians’ Electrocardiogram interpretations: a systematic review and Meta-analysis. JAMA Intern Med 180(11):1461–1471. https://doi.org/10.1001/jamainternmed.2020.3989PMID: 32986084; PMCID: PMC7522782
Gujar JG, Kadam S, Ujwal D, Patil (2022) Recent Advances of Artificial Intelligence (AI) for Nanobiomedical Applications: Trends, Challenges, and Future Prospects. Disruptive Developments in Biomedical Applications
Übeyli E (2009) Combining recurrent neural networks with eigenvector methods for classification of ECG beats. Digit Signal Proc 19:320–329. https://doi.org/10.1016/j.dsp.2008.09.002
Rajpurkar P, Hannun AY, Haghpanahi M, Bourn C, Ng AY Cardiologist-Level Arrhythmia Detection with Convolutional Neural Networks, arXiv preprint, vol. arXiv:1707.01836., 2017
Chu Y, Zhao X, Zou Y, Zhang H, Xu W, Zhao Y (2018) A Comparative Study of Different Feature Extraction Methods for Motor Imagery EEG Decoding within the Same Upper Extremity, 2018 Chinese Automation Congress (CAC), Xi’an, China, no. https://doi.org/10.1109/CAC.2018.8623624., 2018
Jiao L, Qu R, Feng Z, Li L, Yang S, Liu F, Zhang F (2019) Surv Deep Learning-Based Object Detect IEEE Access 7. https://doi.org/10.1109/ACCESS.2019.2939201
Strodthoff N, Wagner P, Samek W, Schaeffter T (2020) Deep Learning for ECG Analysis: Benchmarks and Insights from PTB-XL, arXiv preprint, vol. arXiv:2004.10195, no. https://doi.org/10.48550/arXiv.2004.13701
Martínez JP, Laguna P, Rocha AP, Olmos S, Almeida R (2004) A wavelet-based ECG delineator: evaluation on standard databases. IEEE Trans Bio Med Eng 51(4). https://doi.org/10.1109/TBME.2003.821031
Zheng L, Wang Z, Liang J, Luo S, Tian S (2021) Effective compression and classification of ECG arrhythmia by singular value decomposition. Adv Biomed Eng 2. https://doi.org/10.1016/j.bea.2021.100013
Warnecke JM, Boeker N, Spicher N, Wang J, Flormann M, Deserno TM (2021) Sensor Fusion for Robust Heartbeat Detection during Driving, Annual International Conference of the IEEE Engineering in Medicine and Biology Society. IEEE Engineering in Medicine and Biology Society. Annual International Conference, vol. 447–450, no. https://doi.org/10.1109/EMBC46164.21021.9630935
Khan AH, Hussain M (2021) Mendeley Data V2. https://doi.org/10.17632/gwbz3fsgp8.2. ECG Images dataset of Cardiac Patients
Funding
No funding was received to assist with the preparation of this manuscript. No funding was received for conducting this study. No funds, grants were recieved.
Author information
Authors and Affiliations
Contributions
The authors received support from Bharati Vidyapeeth Ayurved College Pune for the submitted work which is the college that Author Madhavi Mahajan is a Head of Department of Department of Kayachikitsa.
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mahajan, M., Kadam, S., Kulkarni, V. et al. ECG signal classification via ensemble learning: addressing intra and inter-patient variations. Int. j. inf. tecnol. (2024). https://doi.org/10.1007/s41870-024-02086-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s41870-024-02086-4