Abstract
Medicago sativa L. (alfalfa) is a perennial leguminous forage crop cultivated worldwide. Unique water-soaked brown spots surrounded by yellow haloes were reported extensively on leaves of alfalfa crop from the farmer fields of Hafizabad and Bahawalpur districts in 2018. Olive gray colonies of a fungus, isolated with 95% frequency from the symptomatic tissues, were identified as Curvularia sp. based on morphocultural characteristics. Thus, representative Curvularia isolate “FMB-ALF2-MS” was subjected to molecular analysis to validate its phylogeny. Four genetic regions, ITS, LSU, TEF1-α, and GAPDH, were directly sequenced. BLASTn search showed 100% homology to C. buchloes strain CBS 246.49. Phylogenetic analysis is also identified and congruent with this isolate as C. buchloes with 100% bootstrap support. Four repeated pathogenicity tests were carried out with C. buchloes, and the etiology of the leaf spots was determined. To our best knowledge, this is the first report of C. buchloes causing leaf spots on alfalfa from Pakistan. The new and emerging foliar disease's occurrence and intensity were documented across the Punjab province during 2018–2019. The highest D.I (57%) with an average D.S of 78.5% was recorded from the Bahawalpur district. Eight different (four contact and four systemic) fungicides were tested against the pathogen under in vitro conditions at 50, 100, 150, and 200 ppm concentration. Among all the tested fungicides, maximum inhibition (95%) was recorded through mancozeb (contact fungicide), followed by 92% pathogen growth inhibition through propiconazole (systemic fungicide) at 200 ppm. Characterized pathogenic fungal isolate was deposited to the Culture Collection (FMB-CC-UAF) with accession number FMB0177.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Alfalfa (Medicago sativa L.) is the largest cultivated perennial, high-yielding forage legume worldwide (Abd El-Naby Zeinab et al. 2014). It is grown for haymaking, silage preparation, and grazing purpose, provides more protein, high nutritional quality, and abundant biomass to livestock than other forages (Capstaff and Miller 2018). Its cultivation enhances soil fertility, improves soil structure, and prevents soil erosion (Butler et al. 2012; Sabanci et al. 2013).
Various biotic and abiotic stresses adversely affect the growth and lower the fodder quality and yield of alfalfa; among these, diseases are major hindrance factor (Morsy et al. 2011; Li and Nan 2015). Among the diseases, fungal infections affect the foliar parts, imposing severe restrictions on the persistence and productivity of M. sativa (Beuselinck et al. 1994). Curvularia leaf spots are common and occurred worldwide on many kinds of grass species caused by C. lunata, C. eragrostidis, C. geniculata, C. inaequalis, C. intermedia, C. pallescens, and C. trifolii (Weng et al. 1997; Smith et al. 1989; Huang et al. 2004). Many species of Curvularia have been reported as plant pathogenic, causing necrotic leaf spots of several vegetables and other plant families (Dasgupta et al. 2005).
Curvularia species are the predominant foliar pathogens of M. sativa and are associated with lessening dry matter production (Avila et al. 2017). The Curvularia is a ubiquitous genus; species of this genus could be either endophytes, saprobes, or opportunistic pathogens (Gautam et al. 2013; Madrid et al. 2014; Manamgoda et al. 2015) causing diseases in plants, animals, and humans (Wijayawardene et al. 2018). The genus Curvularia has more than 40 species distinguished by their morphological characters Meng et al. (2004).
While conducting field surveys, initially, water-soaked, circular to oval brown color spots were observed; later on, these spots became irregular, enlarged, and oblong surrounded by yellow haloes on alfalfa leaves. The symptoms were unusual, not reported before by plant pathologists or farmers on alfalfa from Pakistan. This study's objective was to determine the extent of new emerging foliar disease on alfalfa, accurate identification of the putative fungal pathogen employing morphocultural and molecular characterization, testing a scenario of contact and systemic fungicides under laboratory conditions to select the appropriate fungicide(s) for field application to control the disease.
Material and methods
Disease estimation and sampling
An extensive survey of alfalfa fields from twenty districts across the Punjab province, Pakistan, was conducted for disease estimation and sample collection. Disease severity (D.S) was recorded using a scale described by Godoy et al. (2006). In total, 160 samples of symptomatic leaves were collected following method described by Stubbs et al. (1986) from 20 different locations across the province, including (1) Bahawalpur (29.3544° N, 71.6911° E), (2) Bahawalnagar (30.0025° N, 73.2412° E), (3) Multan (30.1575° N, 71.5249° E), (4) Shiekhpura (25.1417° N, 85.8629° E), (5) Hafizabad (32.0712° N, 73.6895° E), (6) Samundri (31.0646° N, 72.9520° E), (7) Toba Tek Singh (30.9709° N, 72.4826° E), (8) Vehari (30.0442° N, 79 2.3441° E), (9) Sargodha (32.0740° N, 72.6861° E), (10) Chiniot (31.7292° N, 72.9822° E), (11) Faisalabad (31.4504° N, 73.1350° E), (12) Nankna Sahib (31.4487° N, 73.7949° E), (13) Okara (30.8138° N, 73.4534° E), (14) Jhang (31.2781° N, 72.3317° E), (15) Layyah (30.9693° N, 70.9428° E), (16) Muzafar Garh (30.0736° N, 71.1805° E), (17) Gujranwala (32.1877° N, 74.1945° E), (18) Narowal (32.1014° N, 74.8800° E), (19) Sialkot (32.4945° N, 74.5229° E) and (20) Sahiwal (30.6682° N, 73.1114° E). The collected samples were processed for further examination in fungal molecular biology (FMB) Laboratory, Department of Plant Pathology University of Agriculture Faisalabad (UAF), Pakistan.
Isolation and identification
Diseased leaves were rinsed with tap water. Symptomatic and asymptomatic leaf tissues were cut into 4–5 mm pieces and surface-disinfested with 1% sodium hypochlorite for 45 s, followed by washing with sterilized distilled water and then dried using blotter paper to absorb excessive moisture. Isolation of putative fungal pathogens was done on potato dextrose agar (PDA) medium. Bits of leaf tissue were placed on PDA and incubated at 25–30 °C in an incubator (Sanyo MIR, Japan). Fungal colonies appeared in 3–4 days of incubation. A small portion of mycelium of the fungi of interest was shifted/ transferred to a new PDA plates through single hyphal tip technique (Hildebrand 1938). Morphocultural characters of seven-day-old putative fungal isolates were done following the study of Manamgoda et al. (2012). The color was determined following the Methuen handbook of color (Kornerup and Wanscher 1967). Twenty (20) arbitrarily selected conidia from a conidial suspension of the pathogenic fungal isolate prepared in sterile distilled water using compound microscope Meji Techno, Japan model HD1600T fitted with Olympus (DP25) digital camera was observed for the measurements for each character.
Molecular characterization
For molecular taxonomy, the genomic scenario of representative fungal isolate, FMB-ALF2-MS, was exploited by characterizing taxonomically informative genetic regions such as internal transcribed spacer (ITS) region of ribosomal DNA, translation elongation factor 1-alpha (TEF1-α), glyceraldehyde 3-phosphate dehydrogenase (GAPDH), and large subunit (LSU) of ribosomal RNA gene. Genomic DNA was extracted using GeneJET Genomic DNA Purification Kit (Thermos scientific, USA) following the manufacturer's protocol. The PCR analysis was performed for the amplification of investigated loci. The PCR conditions (thermal and reagent profiles) were optimized to get the required amplification. The optimized annealing temperatures for ITS, TEF1-α, GAPDH, and LSU were 54, 53, 61, and 54 °C, respectively. Initial denaturation was at 95 °C for 5 min, followed by 35 cycles comprising denaturation at 95 °C for 60 s, annealing at respective temperature for 30 s, and extension at 72 °C 45 s. The 1 × reaction mixture contained 0.5 ul Phusion High-Fidelity PCR Master Mix with HF Buffer, primers (reverse and forward) 10 μM each, and 50 ng template DNA. PCR products were resolved on high-resolution agarose through gel electrophoresis under 80 Voltage. Required amplicons were eluted using FavorPrep Gel Extraction Kit (FAVORGEN, BIOTECH CORP, Taiwan) and sequenced (Eurofins Genomics DNA sequencing services, USA). Sequences were in silico analyzed for homology search through the BLASTn tool. Multiple sequence alignments were made with ClustalW by aligning the DNA sequences of each loci with DNA sequences retrieved from public databases (Table 1). Concatenated data set of all investigated loci was generated using Geneious software, and phylogram using concatenated DNA sequence dataset was generated in the MEGA7 software package. We sequenced the fungal isolate FMB-ALF2-MS to investigate its molecular taxonomy. The PCR analysis was performed using primer pairs, ITS1-F/ITS4 (White et al. 1990), TEF 983/2218R (Schoch et al. 2009), gpd1/gpd2 (Berbee et al. 1999) and LR7/LROR (Vilgalys and Hester 1990).
Pathogenicity test
Pathogenicity test was done by fulfilling Koch's postulates under greenhouse conditions in the FMB greenhouse research area department of plant pathology UAF. Viable seeds of alfalfa were surface-sterilized with 1% sodium hypochlorite solution (5 min) and washed thrice with distilled water. Disinfested seeds were sown in a 1 × 1 m bed of sterilized soil. Spore suspension of the investigated pathogen with a concentration of 1 × 106 was prepared, and the foliar application was made on four-week-old healthy seedlings. Control plants were sprayed with sterilized distilled water. Inoculated seedlings were covered with polythene sheath for 12 h to attain the high relative humidity required for the establishment of pathogen and disease development. Observations in terms of appearance and development of leaf spot symptoms were made at regular intervals. Re-isolation of the pathogen was done from the infected leaves for confirmation.
In vitro evaluation of fungicides against C. buchloes
Efficacy of mancozeb, propineb, thiram, copper oxychloride, iprodione (contact) and metalaxyl, propiconazole, and Trifloxystrobin (systemic) fungicides was tested in vitro against the C. buchloes using PDA medium. Each fungicide's stock solution was prepared by dissolving in sterile distilled water and then added to the PDA medium to obtain four concentrations, 50, 100, 150, and 200 ppm fungicide. PDA medium amended with fungicides (20 ml) at different concentrations was poured in 09 mm sterilized Petri plate individually. Mycelial disk of 5 mm in diameter taken from the periphery of seven-day-old culture of the pathogen was placed in the center of PDA medium amended with fungicides and incubated at 30ºC until the fungal growth in control plate (PDA medium without fungicide) touched the periphery. Six replications were maintained for each treatment. Colony diameter was measured, and the percentage of inhibition was recorded (Vincent 1947).
Results
The data regarding the disease estimation (D.I and D.S) revealed that the disease prevailed across the surveyed locations with varying intensities. D.I was ranging between 57% (higher) and 15% (lower). As far as district-wise estimation of disease incidence is concerned, Bahawalpur, Bahawalnagar, and Multan districts were found maximum D.I 57, 48, and 43% respectively, with average 78.5% D.S. Alfalfa fields of Shiekhpura, Hafizabad, Samundri, Toba Tek Singh, and Vehari districts showed D.I 32, 35, 29, 33, and 39%, respectively, with an average 42% D.S. From Sargodha, Chiniot, Faisalabad, Nankna Sahib, Okara, Jhang, Layyah, and Muzafar Garh, D.I recorded 20, 18, 24, 27, 22, 23, 29, and 25%, respectively, with 18% D.S. From Gujranwala, Narowal, Sialkot, and Sahiwal, D.I was recorded as 19, 15, 17, and 17%, respectively, with 7% D.S (Fig. 1).
Morphology
The fungal colonies appeared as Olive gray; conidiophore was sympodial, erect, geniculate branched with an average of 5.3 μm in width. Conidia were oblong, straight, 2–3 transverse septations with average 9.47–19.8 × 6.13–8.3 μm in size. Based on morphological characterization, the fungal isolate was identified as C. buchloes.
Molecular characterization
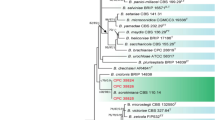
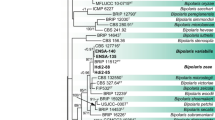
The PCR analysis yielded specific amplicons with their approximate sizes, i.e., ~ 684 (ITS), ~ 1000 bp (TEF1-α), ~ 550 bp (GPDH), and ~ 1200 bp (LSU). The DNA sequences of these loci (Table 1) were trimmed to get high-quality sequences by using BioEdit version 7.2.6.1 and were subjected to the Basic Local Alignment searching (BLAST) tool. The BLASTn results indicated this isolate as C. buchloes because these isolates showed 100% homology to C. buchloes strain CBS 246.49. Generated sequences were deposited to GenBank with accession numbers given in Table 1. For phylogenetic study, sequences of each dataset were then used by aligning them with other supplemented DNA sequences retrieved from NCBI database separately ClustalW under set conditions of Gap Open Penalty (15), Gap Extension Penalty (6.66), 0.5 transition weight, and 30% delay divergence cutoff value to generate phylograms. The phylogenetic status of this isolate was inferred through neighbor joining (NJ) approach in the MEGA7 program. The neighbor joining analysis was used under the p-distance method to infer the evolutionary hierarchy. Then generated alignments of each dataset were concatenated to make concatenated dataset with Geneious software v. 4.8.5. Test of phylogeny was bootstrap with 1000 bootstrap replication with uniform rate following homogenous pattern among lineage. Missing data and gaps were excluded. In the phylogenetic tree based on concatenated DNA sequence dataset of ITS, TEF1-α, GAPDH, and LSU region, fungal isolate (FMB0177) was revealed nested with Curvularia buchloes strain CBS 246.49 with 100% bootstrap support (Fig. 2). The characterized culture of FMB-ALF2-MS was deposited to Fungal Molecular Biology Culture Collection (FMB-CC-UAF) University of Agriculture Faisalabad, Pakistan, with accession number FMB0177.
Pathogenicity test
After 15 days of post-incubation of inoculated alfalfa plants, initially, water-soaked, circular to oval brown color spots on the leaves were appeared, which turned irregular, enlarged, and oblong lesions surrounded by yellow haloes at later stages, very similar to, as were observed on alfalfa leaves during field surveys. Re-isolation of the pathogen was done from infected leaves, and morphological characters of the isolated fungus were observed and identified as C. buchloes. Pathogenicity test revealed C. buchloes, the causative agent for leaf spot disease on alfalfa. To our best knowledge, this is the first report of C. buchloes causing leaf spots of alfalfa in Pakistan.
In vitro evaluation of fungicides against C. buchloes
The effect of fungicides on inhibition of C. buchloes is presented in Fig. 3. Data revealed that all the tested fungicides significantly inhibited the mycelial growth of C. buchloes up to varying extent depending upon the type and concentration of fungicide tested. The highest inhibition, 95%, was recorded at 200 ppm concentration, whereas the least was 26% at 50 ppm. The highest percent inhibition at 200 ppm was 95% with mancozeb among contact fungicides, while the least inhibition, 84% with Iprodion was recorded. Among systemic fungicides, the highest percent inhibition at 200 ppm was 92% with propiconazole, while the least inhibition, 42% with Trifloxystrobin, was observed. The results depicted that mancozeb and propiconazole could provide promising results in controlling the disease under field conditions.
Discussion
During 2018–2019, we found that the alfalfa crop seems to be affected with a unique leaf spot disease, never reported before, from Pakistan. However, foliar diseases are common and more damaging to alfalfa and are considered one of the major issues responsible for lower crop productivity worldwide (Sheaffer et al. 1992; Beuselinck et al. 1994; Samac et al. 2014; Ávila et al. 2017). The appearance of the unusual leaf spots on alfalfa was an eye-catcher for the research group of FMB Lab, UAF, while surveying to assess the foliar diseases of Egyptian clover from farmer fields. Water-soaked, circular to oval brown spots on alfalfa leaves were noticed extensively in Hafizabad and Bahawalpur districts, while irregular, enlarged, and oblong lesions surrounded by yellow haloes on leaves of alfalfa were observed where the infection was severe. Similar kinds of leaf spots are attributed to curvularia leaf spots, mainly producing necrotic leaf spots on several plant species, including forage grasses (Dasgupta et al. 2005; Santos et al. 2014; Silva et al. 2014; Sunpapao et al. 2014; Kusai et al. 2016). Olivaceous gray conidiophore was sympodial, erect, geniculate branched, and Conidia were oblong, straight, which were found similar to Ellis (1971), and Sivanesan (1987) description for the Curvularia genus. As far as the morphology of the genus Curvularia is concerned, overlapping in the morphological features is present (Manamgoda et al. 2012). The characterization of Curvularia species based on morphological characters is very subtle attributes that are varied by varying the host, growth conditions, and life stages (da Cunha et al. 2013). Therefore, the identification of species based on phenotype is ambiguous. The molecular characterization by primary and secondary DNA barcodes has supported accurate species recognition (Manamgoda et al. 2011, 2012), because the literature supports that the morphologically identified Curvularia species are different upon molecular identification (Yanagihara et al. 2010). Therefore, fungal isolate, FMB-ALF2-MS was characterized using ITS, TEF1-α, LSU, GAPDH genetic regions, which was revealed to be nested with Curvularia buchloes strain CBS 246.49 with 100% bootstrap support. This finding was similar to the finding of Manamgoda et al. (2012), where they resolved Curvularia species' boundaries. Chemical management of plant diseases is unavoidable because of rapid action and efficacy. Thus, keeping in view the importance of fungicides, in the present study, the efficacy of all the tested fungicides was found to be effective at all concentrations. Among different concentrations of both contact and systemic fungicides, the highest inhibition, 95%, was revealed at 200 ppm concentration, whereas the least was at 26% at 50 ppm concentration. Adaangadi et al. (2018) reported similar results in their studies to manage the Curvularia leaf spot of maize. Among the different contacts, fungicides tested the highest percent inhibition at 200 ppm was 95 with Mancozeband. Among systemic fungicides, the highest percent inhibition at 200 ppm was 92 with propiconazole. Sumangala et al. (2008) also studied different contacts and systemic fungicides for managing leaf spot of rice and described that contact fungicide mancozeb and systemic fungicide propiconazole were the most effective fungicides. Bisht et al. (2018) also gave similar results on the efficacy of different systemic and non-systemic fungicides against Curvularia lunata in in vitro conditions and described that carboxin at 25 ppm and mancozeb at 200 ppm gave significant inhibition.
References
Abd El-Naby Zeinab M, Azzam CR, Abd El-Rahman SS (2014) Evaluation of ten alfalfa populations for forage yield, protein content, susceptibility to seedling damping-off disease and associated biochemical markers with levels of resistance. J Am Sci 10(3s):73–85
Adaangadi KC, Harlapur SI, Savita C, Hadimani BR (2018) Studies on evaluation of fungicides against Curvularia lunata in maize. Int J Agric Sci 14(1):225–228
Avila MR, Dall’agnol MI, Martinelli JA, Silva GB, Bremm C, Nunes T (2017) Selection of alfalfa genotypes for resistance to the foliar pathogen Curvularia geniculata. Anais da Academia Brasileira de Ciências. 89(3):1801–1813
Berbee ML, Pirseyedi M, Hubbard S (1999) Cochliobolus phylogenetics and the origin of known, highly virulent pathogens, inferred from ITS and glyceraldehyde-3-phosphate dehydrogenase gene sequences. Mycologia 91(6):964–977
Beuselinck PR, Bouton JH, Lamp WO, Matches AG, McCaslin MH, Nelson CJ, Rhodes LH, Sheaffer CC, Volenec JJ (1994) Improving legume persistence in forage crop systems. J Prod Agric 7(3):311–322
Bisht S, Balodi R, Ghatak A, Kumar P (2018) Determination of susceptible growth stage and efficacy of fungicidal management of Curvularia leaf spot of maize caused by Curvularia lunata (Wakker) Boedijn. Maydica 61(3):5
Butler TJ, Biermacher JT, Interrante SM, Sledge MK, Hopkins AA, Bouton JH (2012) Production and economics of grazing alfalfa in the southern great plains. Crop Sci 52(3):1424–1429
Capstaff NM, Miller AJ (2018) Improving the yield and nutritional quality of forage crops. Front Plant Sci 24(9):535
da Cunha KC, Sutton DA, Fothergill AW, Gené J, Cano J, Madrid H, de Hoog S, Crous PW, Guarro J (2013) In vitro antifungal susceptibility and molecular identity of 99 clinical isolates of the opportunistic fungal genus Curvularia. Diagn Microbiol Infect Dis 76(2):168–174
Dasgupta S, Saha D, Saha A (2005) Levels of common antigens in determining pathogenicity of Curvularia eragrostidisin different tea varieties. J Appl Microbiol 98:1084–1092
Ellis MB (1971) Dematiaceous hyphomycetes. Dematiaceous hyphomycetes 86
Gautam AK, Kant M, Thakur Y (2013) Isolation of endophytic fungi from Cannabis sativa and study their antifungal potential. Arch Phytopathol Plant Protect 46:627–635
Godoy CV, Koga LJ, Canteri MG (2006) Diagramatic scale for assessment of soybean rust severity. FitopatologiaBrasileira 31:63–68
Hildebrand EM (1938) Techniques for the isolation of single microorganisms. Botan Rev 4(12):627–664
Huang J, Zheng L, Hsiang T (2004) First report of leaf spot caused by Curvularia verruculosa on Cynodon sp. in Hubei, China. New Dis Rep 10:5
Kornerup A, Wanscher JH (1967) Methuen handbook of colour. Methuen & Co. Ltd., London, p 243
Kusai NA, Azmi MMZ, Zulkifly S, Yusof MT, Zainudin NAIM (2016) Morphological and molecular characterization of Curvularia and related species associated with leaf spot disease of rice in Peninsular Malaysia. Rend Lincei: Sci Fis Nat 27:205–214
Li YZ, Nan ZB (2015) The methods of diagnose, investigation and loss evaluation for forage diseases. Phoenix Science Press, Nanjing
Madrid H, Da Cunha KC, Gené J, Dijksterhuis J, Cano J, Sutton DA, Guarro J, Crous P (2014) Novel Curvularia species from clinical specimens. Persoonia 33:48–60
Manamgoda DS, Cai L, Bahkali AH, Chukeatirote E, Hyde KD (2011) Cochliobolus: an overview and current status of species. Fungal Divers 51:3–42
Manamgoda DS, Cai L, McKenzie EH, Crous PW, Madrid H, Chukeatirote E, Shivas RG, Tan YP, Hyde KD (2012) A phylogenetic and taxonomic re-evaluation of the Bipolaris–Cochliobolus–Curvularia complex. Fungal Divers 56(1):131–144
Manamgoda DS, Rossman AY, Castlebury LA, Chukeatirote E, Hyde KD (2015) Taxonomic and phylogenetic re-appraisal of the genus Curvularia (Pleosporaceae): human and plant pathogens. Phytotaxa 212:175–198
Meng Z, Tianyu Z, Yueming W (2004) A new name and a new variety in Curvularia. Mycosystema 23:177–178
Morsy KM, Abdel-Monaim MF, Mazen MM (2011) Use of abiotic and biotic inducers for controlling fungal diseases and improving growth of Alfalfa. World J Agric Sci 7(5):566–576
Sabanci CO, Ertus MM, Celebi SZ (2013) Collection, conservation and evaluation for forage yield of alfalfa landraces grown in East Anatolia. Turk J Field Crops 18(1):46–51
Samac DA, Rhodes LH, Lamp WO (2014) Compendium of alfalfa diseases and pests, 3rd edn. APS Press, St. Paul
Santos GR, Tschoeke PH, Silva LD, Silveira MC, Reis HB, Brito DR, Carlos DD (2014) Sanitary analysis, transmission and pathogenicity of fungi associated with forage plant seeds in tropical regions of Brazil. J Seed Sci 36:54–62
Schoch CL, Wang Z, Townsend JP, Spatafora JW (2009) Geoglossomycetes cl. nov., Geoglossales ord. nov. and taxa above class rank in the Ascomycota Tree of Life. Persoonia 22:129
Sheaffer CC, Marten GC, Jordan RM, Ristau EA (1992) Forage potential of kura clover and birdsfoot trefoil when grazed by sheep. Agron J 84(2):176–180
Silva MSBS, Rodrigues AAC, Oliveira LJMG, Silva EKC, Pereira TS (2014) Sanidade de sementes de arroz, biocontrole, caracterização e transmissão de Curvularia lunataem semente-plântula de arroz. Rev Ceres 61:511–517
Sivanesan A (1987) Graminicolous species of Bipolaris, Curvularia, Drechslera, Exserohilum and their teleomorphs. CAB International, Kew
Smith JD, Jackson N, Woolhouse AR (1989) Fungal diseases of amenity turf grasses, 3rd edn. E. & FN Spon, London
Stubbs RI, Prescott JM, Saari EE, Dubin HJ (1986) Cereal disease methodology manual
Sumangala K, Patil MB, Nargund VB, Ramegowda G (2008) Evaluation of fungicides, botanicals and bioagents against Curvularia lunata, a causal agent of grain discolouration in rice. J Pl Dis Sci 3(2):159–164
Sunpapao A, Kittimorakul J, Pornsuriya C (2014) Disease note: identification of Curvularia oryzae as cause of leaf spot disease on oil palm seedlings in nurseries of Thailand. Phytoparasitica 42(4):529–533
Vilgalys R, Hester M (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J bac 172(8):4238–4246
Vincent JM (1947) Distortion of fungal hyphae in presence of certain inhibitors. Nature 159(4051):850
Weng Q, Wang Q, He Y, Liu M, Yu D (1997) The occurrence of turf diseases in Fujian Province. Pratacult Trans 6:70–73
White TJ, Bruns T, Lee SJ, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc: Guide Methods Appl 18(1):315–322
Wijayawardene NN, Hyde KD, Lumbsch HT, Liu JK, Maharachchikumbura SS, Ekanayaka AH, Tian Q, Phookamsak R (2018) Outline of ascomycota: 2017. Fungal Divers 88(1):167–263
Yanagihara M, Kawasaki M, Ishizaki H, Anzawa K, Udagawa SI, Mochizuki T, Sato Y, Tachikawa N, Hanakawa H (2010) Tiny keratotic brown lesions on the interdigital web between the toes of a healthy man caused by Curvularia species infection and a review of cutaneous Curvularia infections. Mycoscience 51(3):224–233
Acknowledgments
We acknowledge Fungal Molecular Biology Laboratory (FMB Lab.) for providing research facilities, supplies, and FMB Culture Collection (FMB-CC-UAF) for fungal culture identification and preservation services.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Haq, I.U., Ijaz, S., Faraz, A. et al. Characterization of Curvularia buchloes causing leaf spots on Medicago sativa L. (alfalfa) and its management through fungicides. J Plant Dis Prot 128, 493–500 (2021). https://doi.org/10.1007/s41348-020-00414-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s41348-020-00414-x