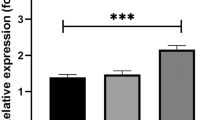
Abstract
Osteoarthritis (OA) is one of the most common musculoskeletal diseases of adults worldwide. Inflammation plays a crucial part in OA pathology, but role of cytokines is still inconclusive. Purpose of this study was to assess the function of tumor necrosis factor α (TNF-α -308 and -238) and interleukin-6 (IL6-572) polymorphisms in OA disease susceptibility in a Pakistani population. 280 OA patients and 308 ethnically matched healthy controls were enrolled in the current study. Demographic data were collected from all participants via a questionnaire. Genomic DNA was isolated from all subjects. TNF-α (-308 G>A, -238 G>A) and IL6 (-572 G>C) polymorphisms in both groups were identified by PCR-coupled restriction fragment length polymorphism (PCR–RFLP) technique. GraphPad Prism software was used to perform statistical analysis. Genotypic and allelic frequencies were determined in both groups. Basic characteristics were mentioned as SD ± mean. p value above 0.05 was considered nonsignificant statistically. Age and body mass index (BMI) differences were not significant (> 0.05) between patients and control groups. Genotype frequencies were in agreement with Hardy–Weinberg equilibrium for all single nucleotide polymorphisms (SNPs) in control and patient group. TNF-α (-308, -238) GA+AA genotypes and IL6 (-572) GC+CC genotypes were considerably associated with higher risk of OA compared to homozygous wild-type genotypes (p < 0.01). Variant alleles were more expressed in knee OA patients as compared to healthy controls for all loci (p < 0.05). Our finding suggests there is an association between TNF-α -308G/A, -238G/A and IL6-572G/C polymorphisms and OA disease susceptibility in a Pakistani population. Further studies with large sample size and in diverse ethnic groups are vital to evaluate and confirm the function of these SNPs in OA disease pathology.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Knee osteoarthritis (OA), identified by the degeneration of articular cartilage, is a common chronic disorder (Orita et al. 2011). It is the most common reason of disability among elders globally. A study, conducted in Pakistan, has reported that 25% inhabitant of countryside areas and 28% population living in cities were affected with knee OA (Haq 2011).
The complete mechanism of action of knee OA is not known yet. Contributions of genetic components in etiology of OA disease have been shown by few recent studies. Although, classically OA is regarded as a noninflammatory disorder due to the absence of certain proinflammatory cells like neutrophils (Valdes et al. 2010), inflammatory reaction seems to have an important role in commencement and progression of OA. There are evidences that suggest the crucial involvement of various soluble inflammatory mediators in orthopedic and rheumatic disorders (Lieberthal et al. 2015). Cartilage degradation, a hallmark of OA, is an expression of inflammatory reaction (Helmy et al. 2001). OA is now suggested as a chronic inflammatory disease (Chen et al. 2017).
Cytokines contribution has been evident in the initiation and expansion of OA over the years (Han et al. 2012). Many OA patients, with degenerated cartilage, are found to have an elevated level of cytokines like tumor necrosis factor alpha (TNF-α), interleukin-6 (IL6) and other acute phase proteins (Fernandes et al. 2002).
TNF-α seems to have a central role in degeneration of cartilage associated with OA. TNF-α not only stimulates its own production but also induces the release of other cytokines like IL6 by activating the chondrocytes and synoviocytes (Sezgin et al. 2008). Additionally, TNF-α has been observed to stimulate osteoclastic bone resorption in vitro (Pelletier et al. 2001). It may also affect the process of remodeling of subchondral bone in OA patients (Pelletier et al. 2001). High level of TNF-α has been reported in synovial fluid of OA patients and, it has crucial involvement in inflammatory process and joint degradation (Özler et al. 2016). TNF-α encoding region is located on chromosome 6 (p21.3). Numerous single nucleotide polymorphisms (SNPs) are present in TNF-α gene regulatory region, two of which are -308G>A (rs1800629) and -238 G>A (rs361525). These polymorphisms are shown to be linked with an altered secretary response of this cytokine (Jamil et al. 2017). Theses SNPs have also been related to high TNF-α level in rheumatoid arthritis (RA) patients (Munoz et al. 2014). Furthermore, TNF-α -308 genetic variant is reported to be associated with variety of arthritis such as rheumatoid arthritis, psoriatic arthritis and juvenile rheumatoid arthritis (Sezgin et al. 2008). So far, many studies have been published to elucidate the association between TNF-α -308 and -238 polymorphisms and knee OA risk, but results are not consistent, and variable findings are reported among different ethnicities. So in view of the uncertain relationship between these SNPs and OA susceptibility, the current study was aimed to clarify the role of theses polymorphisms in a Pakistani population.
IL6 is another important regulator of inflammation, and it induces localized inflammatory response in the joints. Both TNF-α and IL6 are well known for their vital role in bone remodeling process (Manolagas and Jilka 1995). An increased IL6 level has been observed in synovial fluid and serum of OA patients (Kaneko et al. 2000). IL6 coding gene is located on chromosome 7p21–24. Polymorphisms at position -174 (rs1800795), -597 (rs1800797) and -572 (rs1800796) are shown to be associated with various illnesses such as the radiographic severity of RA (Marinou et al. 2007). Published data have reported that -174 and -572 promoter polymorphisms influence transcriptional level of IL6 and may also alter the IL6 serum level in RA patients (-572 IL6 polymorphism and hand RA 2014, Wang et al. 2013). Alterations in cytokine concentration in different individuals are possibly the reason for difference in disease susceptibility and mainly attributed to SNPs in cytokine genes (genetic polymorphism in IL16 and OA 2015#7). Several reports have been published about the association of IL6 -174 polymorphism and OA, but role of -572 SNP is still not clear (Fernandes et al. 2015).
The objective of this study was to investigate the connection of TNF-α (-308G/A and -238G/A) and IL6 (-572 G/C) SNPs with knee OA in a Pakistani population. To the extent of our knowledge, this is the pioneer report to estimate the effects of these genetic variations on knee OA in a Pakistani population.
2 Patients and Material
2.1 Study Subjects
This research project was carried out at the Quaid-i-Azam University (QAU), Department of Biochemistry, Islamabad. It was approved by the ethics committee of QAU. Two hundred and eighty unrelated patients with primary knee osteoarthritis were enrolled in this study from the orthopedic outpatient department in hospitals of Islamabad/Rawalpindi, Pakistan, from March 2014–Jan 2015. Disease was confirmed by experienced orthopedists using American College of Rheumatology (ACR) radiographic criteria (Altman et al. 1986). Patients with any other type of arthritis, post traumatic OA, any inflammatory, systematic and chronic diseases, depression and neoplasm were excluded. In addition, participants who were using intra-articular injections and nonsteroidal antiinflammatory drugs (NSAID) were also excluded. Informed and written consent was also taken from all subjects. A questionnaire containing personal details was filled from all participants through personal interview (Table 1). Three hundred and eight (308) healthy people were recruited as controls. The control subjects were clinically healthy with no personal or family history of OA and any other chronic and inflammatory disease (Table 1).
2.2 DNA Extraction
5 ml blood was taken from all study subjects in EDTA containing tubes to prevent blood coagulation. Jena Biosciences® Blood DNA isolation kit (Jena Biosciences, Germany) was used to extract gDNA from all samples as per manufacturer’s instructions.
2.3 PCR–RFLP Analysis
The polymorphisms in TNF-α and IL-6 genes were assessed by PCR–RFLP technique using three sets of forward and reverse primers (Table 2). PCR and genotyping analysis were performed using the protocol already established in laboratory by Liaquat et al. (2014, 2015). Briefly, each 50-μl PCR mix contained 1 unit of Taq DNA polymerase (MBI Fermentas, England), 10× PCR buffer, 25mMMgCl2 (MBI Fermentas, England), 10 pmol of each forward and reverse primer, 10 mM dNTPs and 100 ng of genomic DNA. GeneAmp PCR cycler 9700 (Applied Biosystems Inc., USA) was used for amplification using following thermal profile: 5 min of initial denaturation at 95 °C followed by 40 cycles of 1 min of denaturation at 95 °C, 1 min of annealing (60 °C for TNF-α and 55 °C for IL-6) and 1 min of elongation at 72 °C. A final extension of 10 min was given at 72 °C. The PCR products were verified on 2% agarose gel along with positive and negative control samples (Table 2) and 100-bp DNA ladder (MBI Fermentas, England). NcoI, MspI and MbiI restriction enzymes were used for digestion of TNF-α -308, TNF-α -238 and IL6 -572, respectively. Each digestion mixture consisted of 12-μl PCR aliquot, 1 μl of the respective enzyme (10 U/μl) (MBI Fermentas, England), 2 μl of 10× Buffer G (MBI Fermentas, England) and 5 μl of PCR water. The reaction mixture was incubated for 16 h at 37 °C. The digested products (TNF-α -308 114 bp and 20 bp, TNF-α -238 133 bp and 19 bp, IL6 -572 146 bp and 105 bp) were electrophoresed on 4% agarose gel, stained with ethidium bromide, and viewed using UV transilluminator. Around 25% samples were randomly picked, and assay was repeated to confirm the results. Results were 100% in accordance.
2.4 Statistics
GraphPad Prism for windows (v6.0; GraphPad Software Inc., USA) was used for all statistical analyses. Basic population characteristics were presented as mean ± SD. The statistical significances between patient and control groups were analyzed by Student’s t test and Chi-square test, where indicated. The genotypic and allelic frequencies were measured by a Fisher’s exact test. Deviation from Hardy–Weinberg equilibrium (HWE) was assessed by Chi-square test for both case and control groups separately. Haplotype analysis for TNF-α polymorphisms was done by Fisher’s exact test. Probability value ≤ 0.05 was taken as significant.
3 Results
Two hundred and eighty patients (mean age; 54.5 ± 10 years) and three hundred and eight healthy controls (mean age; 53.5 ± 10 years) were recruited for this case control study. The patient group contained 43% and 57%, whereas control group contained 49% and 51% males and females, respectively. No considerable difference was observed in gender, age and BMI of OA patients and controls (Table 1). Alleles and genotypes frequencies are given in Table 3. The genotypic frequencies were in equilibrium with HW principle for all three SNPs in both study groups (p > 0.05). The GA+AA genotypes of TNF-α -308 and -238 showed significant association with risk of OA as compared to GG genotype (p = 0.0011 and p = 0.0001, respectively). Similarly, IL6 -572 mutant GC+CC genotype was notably higher in OA patients in comparison with controls (p = 0.0001). Additionally, A alleles of TNF-α (-308 and -238) and C alleles of IL-6 (-572) were linked with increased susceptibility of OA compared to G allele. Haplotype analysis of TNF-α polymorphisms (Table 4) showed that mutant haplotypes (G–A and A–A) were significantly associated with higher risk of disease susceptibility compared to wild-type haplotype (G–G). The G–A haplotype (p = 0.0001, OR .12, CI .1–.2) and A–A haplotype (p = 0.0001, OR .3, CI .2–.4) were revealed as high-risk haplotypes. The A–G haplotype was not correlated with increased vulnerability of knee OA (p = 0.46; Table 4).
4 Discussion
In the current study, we intended to find out the involvement of selected genetic variations in TNF-α (-308G/A and -238 G/A) and IL6 (-572G/C) genes in knee OA. We observed these SNPs are correlated with knee OA in a Pakistani population.
Knee OA is a whole joint disorder involves tearing of cartilage, sclerosis of underlying bone and formation of osteophytes. It is the major source of disability and socioeconomic burden globally. Because of complex disease pathology and involvement of multiple genes, it becomes a challenge to find a universal biomarker for early diagnosis and effective treatment of OA. It is suggested that inflammation plays an important role, but its molecular mechanism with involvement of gene polymorphism is still under discussion. There are several SNPs in a range of cytokine genes which show association with various diseases. Identification and association of these SNPs would be helpful in understanding of OA disease pathology.
Taking into consideration the involvement of the genetic component in OA pathogenesis, the study of TNF-α and IL6 gene polymorphisms might be helpful in identification of the individual’s susceptibility to disease.
TNF-α SNPs have established associations with many disorders like RA, ankylosing spondylitis and systemic lupus erythematosus (Lee et al. 1998). It is observed that allele A is strongly linked with increased level of TNF and disease vulnerability in human (Shlopov et al. 2000).
Our study reveals that A allele of TNF-α -308 and -238 polymorphism is related to enhanced risk of knee OA. Our results are in agreement with Han et al. (2012). He found strong correlation with -308 TNF-α and OA in Korean population. Different previous researches have shown diverse results. Sezgin et al. (2008) did not observe any relation between TNF-α -308 SNP and OA in a Turkish population. This is, perhaps, due to a small sample size of the study or potential involvement may be weakened by the environmental factor. Actually, strong linkage disequilibrium also makes it difficult to study the function of SNP separately (Brinkman et al. 1996). Bin et al. (2013) on the contrary, described a positive relationship between TNF-α -308 polymorphism and OA in Han population; however, they did not find any involvement of TNF-α -238 polymorphism in OA. Another research conducted on Mexican population, with relevant small sample size (n = 150), demonstrated that there is no significant difference in TNF-α -308 and -238 genotype frequencies between patient and control groups (Munoz et al. 2014). Due to small sample size, they could not identify any subject with AA homozygous genotype for both studied polymorphisms and this may have reduced the statistical power of the analysis. A meta-analysis by Kou and Wu (2014) based on published data on TNF -308 polymorphisms, suggested that GA and AA genotypes are coupled with an enhanced risk of knee OA. This might be explained by an increased level of TNF-α in -308A allele carrier compared to -308G carriers. Abdel et al. (2017) in a recently published study have reported that TNF-α -308 mutation is linked with an early onset of primary knee OA in Egyptian female population. Many earlier studies have shown the connection between TNF-α -238 polymorphisms and some rheumatic diseases (Lee et al. 2010; Sousa et al. 2009), but there are only few studies, including current study, which describe its link with OA disease susceptibility, and its role in OA is not completely understood yet. A recent case–control study and meta-analysis by Sobhan et al. (2018) revealed TNF-α -238 polymorphism is not linked with genetic susceptibility to knee OA. They also reported, contrary to previous meta-analysis, that no significant difference existed between TNF-α -308 genotypes in patient and control groups; however, -308 A allele was prominent in patient group compared to G allele (p < 0.05). They have suggested that previous meta-analysis by Kou and Wu (2014) was not accurately conducted due to the selection of inappropriate studies with overlapping findings which distorted the final results. Though, current meta-analysis also have certain limitations as researchers were not able to perform meta-analysis for relationship of the combined TNF-α -308 and -238 polymorphisms and knee OA due to limited number of studies. Their findings may also be influenced by gene-environment interaction factors and heterogeneity of control group.
IL6 also considered as a key player in OA pathology. A direct relationship between an advanced serum level of IL-6 and the radiographic progression of knee OA is also observed (Livshits et al. 2009). Several SNPs in IL6 gene promoter region, particularly -174, -597 and -572, are important and have been established to be linked with numerous diseases like radiography severity in RA (Marinou et al. 2007). Previously published data have proposed that genotypic variants of IL6 gene are associated with cartilage degradation (Osiri et al. 1999; Jordanies et al. 2000).
For current study, IL6 -572 polymorphism was selected as it’s a TAG SNP and showed perfect linkage disequilibrium with other polymorphisms in the promoter region (-174 and -597) (Fernandes et al. 2015). The IL6 -572 SNP exhibited a high frequency of the -572C allele from OA cases versus controls implying a positive relation of this polymorphism with the disease in subjects of current study. Our findings are contradictory to previously published studies which reported no such association between -572C variant and risk of OA.
Kämäräinen et al. (2008) did not find any connection between -572 C variant allele and risk of distal interphalangeal (DIP) OA in a Finish population. They also found that G allele at -572 position is associated with an increased IL6 level in serum. Another recent report by Fernandes et al. also documented similar results for Brazilian population 2015). They found that G allele was more common in OA patients in comparison with healthy subjects. They reported that individual with C allele showed less susceptibility to knee OA. We reported a strong positive association between IL6 -572 C allele and risk of knee OA in a Pakistani population. Our results demonstrate the influence of diverse ethnicity in our genetic background that may explicate the genotype and allele differences compared with the European and American population. As not much data are available on IL6 -572 SNP regarding its association with OA, impact of this polymorphism on knee OA is still ambiguous.
Limitations of the present study may include small sample size. Secondly, we were not able to relate these polymorphisms to their respective serum level in our study subjects. Furthermore, combination of hospital and population based controls for the current study may not fully represent the general population. Although there are studies on other population like European and American on these loci, very limited data are available from Asian region. Despite of aforementioned limitation, to the best of our understanding, this is the primary report to demonstrate the associations of TNF-α (-308 G/A and -238 G/A) and IL6 (-572 G/C) polymorphisms with knee OA in a Pakistani population. Furthermore, no previous data are available on the relationship of IL-6 -572 G/C SNP with OA in any Asian population yet. This study contributes to the ongoing research on OA pathogenesis. Additional studies are considered necessary to validate these findings.
In conclusion, our findings demonstrated that the A allele of TNF-α (-308G>A and -238G>A) and C allele of IL6 (-572G/C) are linked with elevated risk of knee OA is a Pakistani population. To fully understand the role of these polymorphisms, further studies with larger and ethnically diverse groups are needed. The effects of these SNPs on their respective serum level in OA patients and their functional impact on disease severity should also be analyzed.
References
Abdel GSM, Ezzeldin N, Fawzy F, El-Boshym M (2017) The single-nucleotide polymorphism (SNP) of tumor necrosis factor α -308G/A gene is associated with early-onset primary knee osteoarthritis in an Egyptian female population. Clin Rheumatol 36(11):2525–2530
Altman R, Asch E, Bloch D, Bole G, Borenstein D, Brandt K et al (1986) Development of criteria for the classification and reporting of osteoarthritis, classification of osteoarthritis of the knee. Diagnostic and Therapeutic Criteria Committee of the American Rheumatism Association. Arthritis Rheumatol 29:1039–1049
Bin J, Shi J, Cheng X, Zhou J, Zhou Q, Cao C (2013) Association analysis of two candidate polymorphisms in the Tumor Necrosis Factor-α gene with osteoarthritis in a Chinese population. Int Orthop 37(10):2061–2063
Brinkman BM, Zuijdeest D, Kaijzel EL, Breedveld FC, Verweij CL (1996) Relevance of the tumor necrosis factor alpha (TNF alpha) -308 promoter polymorphism in TNF alpha gene regulation. J Inflamm 46(1):32–41
Chen D, Shen J, Zhao W, Wang T, Han L, Hamilton JL et al (2017) Osteoarthritis: toward a comprehensive understanding of pathological mechanism. Bone Res 5:16044
Fernandes JC, Martel-Pelletier J, Pelletier JP (2002) The role of cytokines in osteoarthritis pathophysiology. Biorheology 39:237–246
Fernandes MTP, Fernandes KBP, Marquez AS, Cólus MS, Souza MF, Santos JPM et al (2015) Association of interleukin-6 gene polymorphism (rs1800796) with severity and functional status of osteoarthritis in elderly individuals. Cytokine 75(2):316–320
Han L, Song JH, Yoon JH, Paer YG, Lee SW, Choi YJ et al (2012) TNF-a and TNF-β polymorphisms are associated with susceptibility to osteoarthritis in a Korean population. Korean J Pathol 46(1):30–37
Haq SA (2011) Osteoarthritis of the knees in the COPCORD world. Int J Rheum Dis 14:122–129
Helmy N, Maly FE, Bestmann L et al (2001) Detection of the single-base substitution -174 G > C in the interleukin-6 gene by real-time polymerase chain reaction: comment on the article by Moos et al. Arthritis Rheumatol 44:2213–2214
Jamil K, Archana J, Javeed A, Sindhu J, Sk Yerra (2017) TNF-alpha -308G/A and -238G/A polymorphisms and its protein network associated with type 2 diabetes mellitus. Saudi J Biol Sci 24(6):1195–1203
Jordanies N, Eskdale J, Stuart R, Gallagher G (2000) Allele associations reveal four prominent haplotypes at the human interleukin-6 (IL-6) locus. Genes Immun 1:451–455
Kämäräinen OP, Solovieva S, Vehmas T, Luoma K, Riihimäki H, Ala-Kokko L et al (2008) Common interleukin-6 promoter variants associate with the more severe forms of distal interphalangeal osteoarthritis. Arthritis Res Ther 10(1):R21. https://doi.org/10.1186/ar2374
Kaneko S, Satoh T, Chiba J, Ju C, Inoue K, Kagawa J (2000) Interleukin-6 and interleukin-8 levels in serum and synovial fluid of patients with osteoarthritis. Cytokines Cell Mol Ther 6(2):71–79
Kou S, Wu Y (2014) Meta-analysis of tumor necrosis factor alpha -308 polymorphism and knee osteoarthritis risk. BMC Musculoskeletal Disord 15:373
Lee JY, Dong SM, Kim SY, Yoo NJ, Lee SH, Park WS (1998) A simple, precise and economical micro dissection technique for analysis of genomic DNA from archival tissue sections. Virchows Arch 433(4):305–309
Lee YH, Ji JD, Bae SC, Song GG (2010) Associations between tumor necrosis factor-alpha (TNF-alpha) -308 and -238 G/A polymorphisms and shared epitope status and responsiveness to TNF-alpha blockers in rheumatoid arthritis: a meta-analysis update. J Rheumatol 37(4):740–746
Liaquat A, Asifa GZ, Zeenat A, Javed Q (2014) Polymorphisms of tumor necrosis factor-alpha and interleukin-6 gene and C-reactive protein profiles in patients with idiopathic dilated cardiomyopathy. Ann Saudi Med 34(5):407–414
Liaquat A, Shauket U, Ahmad W, Javed Q (2015) The tumor necrosis factor-α-238G/A and IL-6 -572G/C gene polymorphisms and the risk of idiopathic dilated cardiomyopathy: a meta-analysis of 25 studies including 9493 cases and 13,971 controls. Clin Chem Lab Med 53:307–318
Lieberthal J, Sambamurthy N, Scanzello CR (2015) Inflammation in joint injury and post-traumatic osteoarthritis. Osteoarthr Cartil 23(11):1825–1834
Livshits G, Zhai G, Hart DJ, Kato BS, Wang H, Williams FM et al (2009) Interleukin-6 is a significant predictor of radiographic knee osteoarthritis: the Chingford study. Arthritis Rheumatol 60(7):2037–2045
Manolagas SC, Jilka RL (1995) Bone Marrow, Cytokines, and Bone Remodeling — Emerging Insights into the Pathophysiology of Osteoporosis. N Engl J Med 332(5):305–311
Marinou I, Healy J, Mewar D, Moore DJ, Dickson MC, Binks MH et al (2007) Association of interleukin-6 and interleukin-10 genotypes with radiographic damage in rheumatoid arthritis is dependent on autoantibody status. Arthritis Rheumatol 56(8):2549–2556
Munoz VJF, Oregon RE, Rangel VH, Martinez BGE, Castaneda SE, Salgado GL et al (2014) High expression of TNF alpha is associated with -308 and -238 TNF alpha polymorphisms in knee osteoarthritis. Clin Exp Med 14:61–67
Orita S, Koshi T, Mitsuka T, Miyagi M, Inoue G, Arai G et al (2011) Associations between proinflammatory cytokines in the synovial fluid and radiographic grading and pain-related scores in 47 consecutive patients with osteoarthritis of the knee. BMC Musculoskelet Disord 12(1):144
Osiri M, McNicholl J, Moreland LW, Bridges SL (1999) A novel single nucleotide polymorphism and five probable haplotypes in the 5′ flanking region of the IL-6 gene in African-Americans. Genes Immun 1(2):166–167
Özler K, Aktaş E, Atay Ç, Yılmaz B, Arıkan M, Güngör Ş (2016) Serum and knee synovial fluid matrixmetalloproteinase-13 and tumor necrosis factor-alpha levels in patients with late stage osteoarthritis. Acta Orthop Traumatol Turc 50(6):670–673
Pelletier JP, Martel-Pelletier J, Abramson SB (2001) Osteoarthritis, an inflammatory disease: potential implication for the selection of the new therapeutic targets. Arthritis Rheumatol 44(6):1237–1247
Sezgin M, Barlas IO, Ankarali HC, Altintas ZM, Türkmen E, Gökdoğan (2008) Tumor necrosis factor alpha -308G/A gene polymorphism: lack of association with knee osteoarthritis in a Turkish population. Clin Exp Rheumatol 5:763–768
Shlopov BV, Gumanovskaya ML, Hasty KA (2000) Autocrine regulation of collagenase 3 (matrix metalloproteinase 13) during osteoarthritis. Arthritis Rheumatol 43:195–205
Sobhan MR, Mahdinezhad YM, Aghili K, Zare SM et al (2018) Association of TNF-α-308 G > A and -238G > A polymorphisms with knee osteoarthritis risk: a case-control study and meta-analysis. J Orthop 15(3):747–753
Sousa E, Caetano LJ, Pint P, Pimentel F, Teles J, Canhã H et al (2009) Ankylosing spondylitis susceptibility and severity–contribution of TNF gene promoter polymorphisms at positions -238 and -308. Ann N Y Acad Sci 1173:581–588
Valdes AM, Arden NK, Tamm A, Kisand K, Doherty S, Pola E et al (2010) A meta-analysis of interleukin-6 promoter polymorphisms on risk of hip and knee osteoarthritis. Osteoarthr Cartil 18(5):699–704
Wang J, Platt A, Upmanyu R, Germer S et al (2013) IL-6 pathway-driven investigation of response to IL-6 receptor inhibition in rheumatoid arthritis. BMJ Open 3:e003199
Acknowledgements
The authors are thankful to all participants of this study for their cooperation. We are especially grateful to Dr. Afrose Liaqat for providing valuable help in statistical analysis.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest:
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Naqvi, S.K.B., Bibi, M., Murtaza, I. et al. Interleukin-6 (-572) and Tumor Necrosis Factor α (-308 and -238) Polymorphisms and Risk of Knee Osteoarthritis in a Pakistani Population: A Case–Control Study. Iran J Sci Technol Trans Sci 43, 1485–1490 (2019). https://doi.org/10.1007/s40995-019-00678-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40995-019-00678-5