Abstract
A promising approach to solve the problem of tolerance of plants in hostile environments is to focus of stress tolerance mechanisms of extremophilic plants, in particular mosses. Along with the universal stress mechanisms, bryophytes exhibit a unique spectrum of secondary metabolites such as carotenoids, a lipophilic metabolite derived from the isoprenoid pathway. The main representatives of carotenoids in mosses are α- and β-carotene, lutein, neo-, viola- and zeaxanthins. Hylocomium splendens is one of the most common and widespread mosses of Northern Hemisphere. The genome of this moss has not been sequenced, and the carotenoid biosynthesis pathway (CBP) genes of this species have not been reported to date. This is the first report to of an attempt to identify and characterize the CBP genes in H. splendens. As a result of cloning, sequencing, and in silico analysis, we identified and characterized ten CBP genes in H. splendens with a full ORF, and prediction of subcellular localization suggests chloroplast localization of CBP proteins. Using multiple alignments and phylogenetic and homology analyses, we demonstrated that the CBP genes of H. splendens share high similarity with the sequences in other bryophytes. Differential expression of CBP transcripts during abiotic stresses was more evident for genes in the middle and downstream steps of CBP. This work provides information on the molecular genetics of CBP in extremophilic bryophytes. Analysis of CBP genes can help to unravel the genetic evolution of carotenoid biosynthesis in plants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
The ability of plants to adapt to unfavorable living conditions is a prerequisite for plant survival, evolution of the genotype, and maintenance of flora diversity. Detailed knowledge about the mechanisms of sensitivity and/or resistance of plant species to changing environmental conditions is required. A promising approach to solve the problem of plant resistance to stress is to study the mechanisms of stress resistance of “extremophiles”, in particular mosses, higher non-vascular plants. The relative simplicity of the anatomical structure and the ability to survive in adverse environmental conditions make these evolutionary ancient plants ideal models for studying adaptation mechanisms.
Plants experience numerous stresses during their life cycles, such as drought, and low and high temperatures. Extreme temperature has an adverse impact on photosynthesis (Fan et al. 2017), hormone homeostasis (Maestri et al. 2002), and often results in changes in secondary metabolite concentration (Wahid et al. 2007). Among secondary metabolites, carotenoids are organic pigments, which are widespread in algae, mosses, ferns, gymnosperms and angiosperms (Park et al. 2021). In addition, they are found in the membranes of photosynthetic bacteria such as phototropic bacteria and cyanobacteria (Moeller et al. 2005; Sathasivam et al. 2021). Although classed as secondary metabolites, carotenoids have been implicated in a large number of biological functions of plant growth and development. In photosystem II (PS II), carotenoids contribute to the transfer of energy to the reaction center (Amarnath et al. 2016) and protect chlorophyll from excessive energy by sequestering single oxygen (Ramel et al. 2012; Ruban 2016; Leuenberger et al. 2017). Furthermore, carotenoids play roles in protecting plants from oxidative damages as effective antioxidants (Truscott 1990). They are also the precursors in the production of apocarotenoid hormones such as abscisic acid and strigolactones (Ruiz-Sola and Rodríguez-Concepción 2012).
Several studies have shown that carotenoids contribute to the increased survival of plants under abiotic stress (Davidson et al. 2002; Li et al. 2008). It is well-known that abiotic stresses, such as drought, waterlogging, elevated or freezing temperatures, are accompanied by an increased production of reactive oxygen species (ROS) (Choudhury et al. 2013). Carotenoids as antioxidants are able to detoxify various ROS and also directly quench triplet chlorophylls that are sources of 1O2 in the leaves (Ramel et al. 2012).
High temperatures adversely impact photosynthesis (Fan et al. 2017), hormone homeostasis (Maestri et al. 2002), and metabolism of secondary compounds (Wahid et al. 2007). Plants display a variety of responses to heat stress, including accumulation of carotenoids; heat stress initiates abscisic acid (ABA) and salicylic acid mediated signaling (Wahid et al. 2007; Du et al. 2010). However, understanding how these processes are involved in plant defenses against heat stress remains unclear. Jackson (2015) reported tissue-specific changes in gene expression and carotenoid accumulation in strawberry at elevated temperature. Changes in carotenoid concentration under elevated temperature have also been observed in some green algae, e.g. in Haematococcus pluvialis astaxanthin formation increased three fold, while in a Chlorococcum sp. a two-fold increase in total carotenoid concentration occurred (Juneja et al. 2013). High temperatures increase lutein accumulation in microalgae such as Muriellopsis sp. and Scenedesmus almeriensis (Del Campo et al. 2000; Sanchez et al. 2008). In contrast, low temperature slows lutein accumulation in microalgal species (Bhosale 2004). In Angiosperms, water deficit and prolonged waterlogging cause changes in carotenoid concentration, which are highly depend on plant species, and the duration and intensity of the water restriction or flooding period. A decrease in the concentration of carotenoids during drought have been reported in cherry tomatoes, wheat, sorghum, sunflower, and some plants of the family Asteraceae (Hammad and Ali 2014; Manivannan et al. 2014; Cicevan et al. 2016). It is likely that carotenoid concentration tend to decrease under drought and moderate flooding but slightly increase in response to severe drought stress (Sudrajat et al. 2015). Thus, the concentration of carotenoids in plants can change following abiotic stresses such as temperature variations, drought, and high salinity.
Reviews have summarized information about genes involved in the transcriptional regulation of the carotenoid biosynthesis pathway (CBP) in plants (Stanley and Yuan 2019; Sathasivam et al. 2021). The first important event in carotenogenesis is the production of phytoene from condensation of two geranylgeranyl diphosphate (GGPP) molecules, a reaction catalyzed by phytoene synthase (PSY). In several crop species, PSY is one of the most important and rate-limiting enzymes in the CBP (López Emparán et al. 2014). Four sequential dehydrogenation and two-isomerization increase the number of conjugated double bonds and transform 15-cis-phytoene into all-trans-lycopene (or simply lycopene), a pink colored carotenoid. In summary, during desaturation, phytoene is successively transformed to phytofluene, ζ-carotene, neurosporene and lycopene, with the enzymes phytoene desaturase (PDS), 15-cis-ζ-carotene isomerase (Z-ISO), ζ-carotene desaturase (ZDS) and carotenoid isomerase (CRTISO) being involved in these reactions (Ruiz-Sola and Rodríguez-Concepción 2012).
The regulation of the CBP genes during transcription is critically important for the syntheses of photosynthetic pigments and plant hormones (Stanley and Yuan 2019). The CBP genes have been identified and characterized in several higher plants such as Arabidopsis, Chinese cabbage, citrus, Ixeris dentate, papaya, Scutellaria baicalensis, strawberry, and wolfberry (Tan et al. 2003; Kato et al. 2004; Devitt et al. 2010; Tuan et al. 2012; 2014; Zhu et al. 2015; Li et al. 2020; Reddy et al. 2017). The pathway for the biosynthesis of carotenoids in bryophytes has yet to be clarified. To date, there have only been few studies analysing CBP genes in the bryophytes, e.g. in Physcomitrium patens and Marchantia polymorpha (Takemura et al. 2014; He et al. 2019). Our preliminary analysis of the antioxidant activity and concentration of carotenoids in Dicranum scoparium, Pleurozium schreberi, Hylocomium splendens, and Sphagnum magellanicum indicated that the greatest antioxidant activity and highest concentration of carotenoids was observed in the moss H. splendens (unpublished data). This moss species was therefore chosen to study CBP genes.
Hylocomium splendens Hedw. is one of the most common and widespread mosses of the circumboreal forest and Arctic tundra, covering large areas of Canada, Alaska, northern Europe, and Siberia. To date, the full genome of H. splendens has not been sequenced yet. Moreover, CBP genes in H. splendens were not identified and characterized and their expression has not been studied. Therefore, here we aimed for identifying and characterizing the CBP genes (HsPSY_1, HsPSY_2, HsPDS, HsZ-ISO, HsZDS, HsLCYb, HsCHYb_1, HsCHYb_2, HsLUT1_1, HsLUT1_2) in the moss H. splendens. Gene expression following treatment of H. splendens with low negative (-20 °C) and elevated (+ 30 °C) temperatures, dehydration/rehydration was analyzed using quantitative reverse transcription-polymerase chain reaction (RT-qPCR). We hypothesized that differential expression of certain CBP genes in response to abiotic stresses can contribute to the targeted and timely defense of H. splendens against a particular unfavorable environmental factor.
2 Material and methods
2.1 Plant material
Hylocomium splendens Hedw. was collected in the Aisha Forest in Tatarstan, Russia (55°53 21.3 N 48°38 14.3 E). Plant material was placed between sheets of paper and left to dry slowly in the open air for 2 d before being stored in the refrigerator at + 4 °C in the dark until use (Onele et al. 2022).
2.2 In silico identification of CBP genes
Metatranscriptome data of the moss H. splendens found in the Sequence Read Archive (SRA) NCBI GenBank database under the accession number SRR2518082 (Johnson et al. 2016) were used to in silico identify CBP genes. The de novo transcriptome assembly was constructed as reported in Onele et al. (2022). Sequences of CBP genes from P. patens were used as queries to search against H. splendens transcript/CDS databases using the BLAST + program (Camacho et al. 2009). To confirm the CBP genes members the predicted sequences were submitted against BLAST NCBI, BLAST Phytozome, NCBI Conserved Domains Da tabase (CDD), NCBI Conserved Protein Domain Family (Marchler-Bauer et al. 2017). The specific genes were identified and subjected to NCBI’s open reading frame (ORF) finder program to identify whether the gene possesses the full ORF with a maximum nucleotide length.
2.3 Cloning of CBP genes
Total H. splendens RNA was extracted using the RNeasy Plant Mini Kit (Qiagen, Venlo, Netherlands). First strand and double strand cDNA were synthesized using Evrogen Mint 2 synthesis kit according to manufacturer’s protocols (Evrogen, Moscow, Russia). cDNA was obtained in standard PCR with Taq polymerase (Evrogen) and specific primers listed in Table S1. The optimal annealing temperature was selected experimentally. The size of the PCR products was confirmed by agarose gel electrophoresis, cDNA of the expected sizes were eluted from gel and cloned into the pAL2-T vector (Evrogen).
2.4 Sequence analysis
Plasmid DNA containing cDNA insert (200 ng) of each gene was used for sequencing. The Sanger reaction was performed with the Big-Dye Terminator v3.1 Cycle Sequencing kit (Thermo Scinetific, Waltham, USA). The reaction was carried out using M13-For and M13-Rev primers. BLASTN software available online at (BLAST: Basic Local Alignment Search Tool (nih.gov)) was used to perform a homology search to compare the sequenced gene with other genes in the database. Files in Fasta format were downloaded from the NCBI database after BLAST search and then subjected to sequence alignments.
2.5 Alignment and phylogenetic analysis
EMBOSS web service (EMBOSS Transeq < Sequence Translation Sites < EMBL-EBI) was used to translates nucleic acid sequences of the H. splendens CBP cDNA to their corresponding peptide sequences. BLASTP software available online at (BLAST: Basic Local Alignment Search Tool (nih.gov) was used to perform a homology search in the database of predicted protein sequences with proteins from other Bryophyta species. The homologous sequences of CBP proteins obtained after BLASTX and other known CBP proteins from the NCBI database were aligned by ClustalW (Thompson et al. 1994) in MEGA X (Kumar et al. 2018). A phylogenetic tree was constructed in MEGA X using the neighbor joining method for 1500 bootstraps (Saitou and Nei 1987).
2.6 Stress treatments
Before the experiment, 2 cm apical stem segments of dry mosses were prehydrated at + 4 °C for 24 h on wet filter paper in the dark. For stress treatments, we followed the protocol developed in our early studies (Onele et al. 2022). Hydrated apical stem segments were thermally stressed by their exposure to − 20 °C or + 30 °C during 12 h in a dark temperature-controlled chamber (Thermostat LOIP, St. Petersburg, Russia). For gene expression analysis, moss samples were taken after 1, 3, 6, and 12 h of temperature treatments. For each time point of temperature treatments, hydrated moss, which was kept at room temperature (+ 22 °C), served as a control.
For desiccation stress, three biological replicates per treatment were used, each containing 0.1 g dry mass from 2 cm apical stem segments. Initially, air-dry mosses were fully hydrated by immersing them in a 20 mL volume of distilled water for 1 h while slowly shaking them on an orbital shaker. Then, the hydrated moss was gently blotted with filter paper and placed in the desiccator above silica gel. After 12 h of desiccation, moss samples were rehydrated for 1 h. For gene expression analysis, moss samples were taken after 12 h of hydration (control), 6 and 12 h of desiccation (D), and after rehydration (R). The change in relative water content (RWC) was monitored according to the protocol previously described in Onele et al. (2022).
2.7 RNA extraction, cDNA synthesis and RT-qPCR
Samples exposed to stresses were immersed in liquid nitrogen, then, each sample was ground into a fine powder. For RT-qPCR, 0.1 g of material from each replicate was immediately frozen in liquid nitrogen and stored at − 80 °C until use. Extraction of total RNA from H. splendens thalli was performed using the RNeasy Plant Mini kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. RNA concentration and purity were measured with NanoDrop® ND-1000 spectrophotometer (Thermo Scientific, Waltham, MA, USA), and the integrity was further evaluated by gel electrophoresis in a 1% (w/v) agarose gel. First strand cDNA was synthesized using protocols from the Evrogen Mint 2 synthesis kit (Evrogen, Moscow, Russia).
For RT-qPCR, ribosomal RNA (18 S), glyceraldehyde-3-phosphate dehydrogenase (GAPDH2) and a-tubulin (a-TUB1 and a-TUB2) were used as internal controls. The vector NTI Suite 9 software was used to design RT-qPCR primers with the following parameters: amplicon length from 60 to 300 bp and a Tm range of 55 to 65 °C. Primers used in this study are shown in Table S2. RT-qPCR conditions used in our study followed the protocol described by Onele et al. (2022). Gene expression at each time point was calculated relative to the corresponding control using ΔCt method (Livak and Schmittgen 2001).
The visualization of expression analysis of CBP genes in the heatmap was conducted using an online heat mapper software ggplot R package.
2.8 Data analysis
Three biological and six analytical replicates were used to run all reactions. Gene expression differences were assessed using normalized expression (Cq) in the Bio-Rad CFX Maestro Software version 2.3 and were found to be significant for p ≤ 0.05 (a), p ≤ 0.01 (b), p ≤ 0.001 (c) after ANOVA and Shapiro-Wilk Normality tests. The standard errors of the mean are shown as vertical bars (n = 6).
3 Results
3.1 In silico identification and sequence analysis of CBP genes
As the full genome of H. splendens is not sequenced, we found the coding sequence (CDS) of H. splendens, predicted based on transcriptome assemble (Johnson et al. 2016). ORFs corresponding to seven known CBP genes were determined using this cDNA library by the bioinformatic approach. One gene was identified for each of the transcripts encoding 15-cis-phytoene desaturase (PDS), zeta-carotene isomerase (Z-ISO), zeta-carotene desaturase (ZDS), lycopene beta-cyclase (LCYb). Two genes were identified for transcripts encoding phytoene synthase (PSY), HsPSY_1 and HsPSY_2, β-carotene-3-hydroxylase (CHYb), HsCHYb_1 and HsCHYb_2, as well as carotene epsilon monooxygenase (LUT1), HsLUT1_1 and HsLUT1_2.
3.2 Sequence analysis of CBP genes
For each ORFs found, cDNA was obtained, cloned, and sequenced. The sizes of the obtained cDNA products for HsPSY_2, HsZ-ISO, HsPDS, HsCHYb_1, HsLCYb, HsLUT1_1 and HsLUT1_2 corresponded to the sizes of transcripts predicted in silico, while the sizes of HsPSY_1, HsCHYb_2 and HsZDS cDNA differed from those of the predicted transcripts (Table 1). The HsPSY1 cDNA sequence had a fragment corresponding to the P. patens PSY_2 mRNA (LOC112276781) that was missing from the bioinformatic transcript sequence, causing the cDNA to be larger than expected (Table 1).
Homology between the cDNA sequences of HsPSY_1 and HsPSY_2 was not found, however, their respective protein sequences obtained by bioinformatics coincided by 71%. Both HsPSY_1 and HsPSY_2 showed homology to P. patens phytoene synthase 2, chloroplastic-like mRNA, encoded by two different genes (LOC112276781 and LOC112276694, respectively) (Table 2). The sequences of PpPSY2 (LOC112276781) and PpPSY2 (LOC112276694) did not share any homology, but their respective protein sequences were 74% identical.
The HsCHYb_1 and HsCHYb_2 cDNA nucleotide sequences and their respective protein sequences were 75% and 67% identical, with both predicted proteins showing homology to the same P. patens beta-carotene-3-hydroxylase 2, chloroplast-like protein (XP_024403077.1). HsCHYb_1 was homologous to PpCHY mRNA (LOC112295550), while HsCHYb_2 was homologous to PpCHY mRNA (LOC112273933) (Table 2). Both PpCHY transcripts sequences were 80% identical and their respective protein sequences were 100% identical. Thus, these two HsCHYb genes may be duplicates encoding the same protein.
No homology was found between the cDNA sequences of HsLUT1_1 and HsLUT1_2, while their respective protein sequences showed low (42%) homology. HsLUT1_1 cDNA and predicted protein were homologous to P. patens carotene epsilon-monooxygenase (CYP97C1), chloroplastic-like, mRNA (LOC112283348) and protein (XP_024377702.1) by 83 and 87% respectively (Table 2). HsLUT1_2 cDNA and predicted protein were homologous to P. patens cytochrome P450 97B2, chloroplastic-like mRNA (LOC112281394) and protein (XM_024517858.1) by 85% and 87%, respectively. P. patens CYP97C1 enzyme and cytochrome P450 97B2 had 43% homology. Thus, HsLUT1_1 is a gene involved in CBP, while the HsLUT1_2 gene most likely performs a different function.
Blast analysis of the cloned CBP cDNA from H. splendens revealed the sequence homology of all these transcripts with the corresponding transcripts of two mosses whose genomes have been completely sequenced, P. patens and Ceratodon purpureus, ranging from 81 to 87% (Table 2). All cloned H. splendens cDNAs showed also strong homology ranging from 90 to 98% to the genomic DNA of two mosses, namely, Rhytidiadelphus loreus and Thuidium tamariscinum, although this was limited by the scores of matches identified.
BLASTp alignment of the predicted peptide sequences also confirmed the extended homology of the H. splendens CBP proteins with the corresponding proteins of P. patens ranging from 68% (HsCHYb_2) to 88% (HsPDS) and of C. purpureus ranging from 68% (HsCHYb_2) to 92% (HsPSY_1) (Table 2). Interestingly, one of the predicted proteins, HsLCYb, showed no homology with the lycopene beta-cyclase protein sequences of these two mosses. The protein sequences of R. loreus and T. tamariscinum protein sequences are not available in the database.
We performed analyses of the protein sequence using online services. Physico-chemical properties such as protein length (aa), molecular mass (MW, kDa) and isoelectric point (pI) were predicted (Table 3). All CBP proteins of H. splendens were predicted to be targeted to the chloroplast (CP).
3.3 Phylogenetic and homology analyses
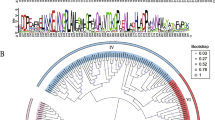
To investigate molecular evolutionary relationships between CBP proteins in H. splendens and those in other bryophytes, neighbor-joining phylogenetic trees were constructed using all ten CBP sequences and a set consisting of each specific CBP sequence. Fig. S1 and Table S3 describe the phylogenetic relationships of the retrieved homologous peptide sequences. Phylogenetic analysis showed that the CBP proteins are highly conserved within bryophytes. HsPSY_1/HsPSY_2, HsCHYb_1/HsCHYb_2 and HsLUT1_1/HsLUT1_2 isoforms form clades with Ceratodon purpureus hypothetical proteins and P. patens respective isoforms. HsPDS or HsZDS belong to a clade with two homologous C. purpureus hypothetical proteins, each of which has common ancestor with respective P. patens 15-cis-phytoene desaturase or zeta-carotene desaturases. HsZ-ISO belongs to P. patens 15-cis-zeta-carotene isomerase clade with common C. purpureus ancestor. S. magellanicum / S. fallax and Marchantia paleacea / M. polymorpha hypothetical proteins are descendants of more remote ancestors for these groups of the proteins. HsLCYb is in a clade with S. magellanicum and S. fallax hypothetical proteins but this clade has also common ancestors with P. patens lycopene beta cyclase and C. purpureus hypothetical proteins.
3.4 Expression of CBP genes of H. splendens under different stress treatment
RT-qPCR analysis was used to analyze the expression patterns of CBP genes in response of H. splendens to different abiotic stress treatments such as exposure to elevated (+ 30 °C) and freezing (− 20 °C) temperatures and desiccation/rehydration cycles. PCR analysis revealed the presence of CBP transcripts in control plants. The results showed that all CBP genes were differentially expressed in H. splendens depending on the type of stress (Figs. 1, 2, 3 and 4). Expression of the CBP genes in moss at low negative temperature remained practically unchanged (Fig. 1), except for HsLUT1_2, which expression increased almost five times after 1 h treatment (Fig. 1).
qPCR analysis of the time-course of CBP gene expression in H. splendens exposed to − 20 °C. For each time point of temperature treatments, hydrated moss, which was kept at room temperature (+ 22 °C), served as a control. The transcripts concentration in the control was taken as one. p ≤ 0.05 (a), p ≤ 0.01 (b), p ≤ 0.001 (c) denote significant differences between the control and treatments according to ANOVA (n = 6)
Expression of CBP genes of H. splendens exposed to high (+ 30 °C) temperature differed from expression of these genes exposed to freezing temperature. Among these, the highest expression was observed for HsPSY_2, both isoforms of HsCHYb and HsLUT1_2 under high temperature treatment (Fig. 2), whereas the lowest expression was found for HsZ-ISO. Transcription of HsPSY_2 increased significantly by 3 h and gradually decreased during 6–12 h high temperature condition. Transcription of HsZDS also demonstrated the same trend as of HsPSY_2. Transcription of HsCHYb_1 and HsCHYb_2 gradually increased during 1–3–6 h and declined sharply after 12 h high temperature treatment. Interestingly, HsLUT1_2 showed the highest expression after short-term (1 h) exposure to both low negative and high temperatures (Fig. 2).
qPCR analysis of the time-course of CBP gene expression in H. splendens exposed to + 30 °C. For each time point of temperature treatments, hydrated moss, which was kept at room temperature (+ 22 °C), served as a control. The transcripts concentration in the control was taken as one. p ≤ 0.05 (a), p ≤ 0.01 (b), p ≤ 0.001 (c) denote significant differences between the control and treatments according to ANOVA (n = 6)
Desiccation of the hydrated moss over silica gel resulted in a considerable drop in water content in the moss samples, down to 43.4% and 5.6% water content after 6 and 12 h of treatments, respectively. It was found that the downstream CBP genes are more sensitive to desiccation and rehydration cycles. Indeed, desiccation of the moss showed a gradual increase in HsCHYb_1 expression to 3.5 times after 6 h and 4.5 times after 12 h. Rehydration of the moss during 1 h after 12 h desiccation showed a slight decrease in HsCHYb_1 expression (Fig. 3). In addition, a slight increase in expressions was observed for HsZDS, HsLCYb and HsLUT1_1. Interestingly that HsLUT1_1 was only sensitive to this stress treatment.
qPCR analysis of the time-course of CBP gene expression in H. splendens during desiccation over silica gel and rehydration. For each time point of temperature treatments, hydrated moss, which was kept at room temperature (+ 22 °C), served as a control. The transcripts concentration in the control was taken as one. p ≤ 0.05 (a), p ≤ 0.01 (b), p ≤ 0.001 (c) denote significant differences between the control and treatments according to ANOVA (n = 6)
4 Discussion
H. splendens is one of the most common and widespread mosses of Northern Hemisphere. The genome of this moss is not sequenced, and there have been no reports on the CBP genes of this species. The present study is the first report to identify and characterize the CBP genes in the moss H. splendens. As a result of cloning, sequencing and in silico analysis, we identified and characterized ten CBP genes in H. splendens with a full ORF. Scheme illustrates CBP enzymes whose genes were identified in present work in H. splendens (Fig. 4).
Using multiple alignments and phylogenetic and homology analyses, we demonstrated that CBP gene sequences in H. splendens share high similarity with such sequences in other bryophytes. Prediction of subcellular localization suggests chloroplast localization of most CBP proteins of H. splendens. Differential expression of CBP genes demonstrates a stress-responsive patterns at the transcriptional with most CBP genes being highly upregulated during early stress response. A heatmap provides graphical representations of the expression profiles of CBP genes of H. splendens across the samples (Fig. 5).
Heatmap of the expression profiles of CBP genes of H. splendens exposed to low (− 20 °C) and high temperature (+ 30 °C), desiccation (D) and rehydration (R). The heatmap was constructed using fold-change values gained from RT-qPCR. Heatmap was plotted using ggplot R package. Downregulated gene expression is indicated by green, while upregulated gene expression is indicated by red
The ten genes encoding seven enzymes of CBP were characterized, because three genes occur with two isoforms (HsPSY_1 and HsPSY_2; HsCHYb_1 and HsCHYb_2; HsLUT1_1 and HsLUT1_2). Most genes generate multiple mRNA isoforms as a result of alternative splicing, intron retention, and alternative transcription start/stop sites. The result is a diversity of mRNA sequences, yielding isoforms that often differ in their protein-coding capacity. Isoforms with very similar sequences can have substantially different morphological outcomes that will reflect a host of gene expression changes in different organs of the plant (Syed et al. 2012). Analysis of the sequences of isoforms of CBP genes in H. splendens indicates similar sequences of ORFs and differ in 5′ and 3′ untranslated regions, which is characteristic of isoforms; in addition, they have differential expression patterns under abiotic stresses (Fig. 5).
Sequence homology analysis of the CBP genes of H. splendens exhibits high similarity with those of other bryophytes, more than 80% with sequences of P. patens and C. purpureus, as well as significant homology with the DNA genomes of R. loreus and T. tamariscinum (Table 2). The genera Hylocomium and Rhytidiadelphus are the parts of the family Hylocomiacea, and together with the genus Thuidium they belong to the order Hypnales, a group of feather mosses that is abundant and young compared to others (Merget and Wolf 2010). We demonstrate that the CBP genes of bryophytes are highly conserved and share higher sequence identities with vascular plants. T. tamariscinum exhibits an almost cosmopolitan distribution, as does C. purpureus, which grows in a wide range of habitats, mainly in urban areas and next to roads on dry sand soils. In contrast, R. loreus grows on decaying logs, the forest floor and as an epiphyte on living trees, and is considered to be a key component of a healthy, thriving forest ecosystem, serving as a water-retaining and thermal insulator. Genera Ceratodon, Marchantia and Sphagnum all belong to the other than H. splendens orders (respectively, Dicranales, Marchantiales and Sphagnales), and inhabit very different ecosystems. C. purpureus grows in a very wide variety of habitats, mainly in urban areas and next to roads on dry sand soils, Sphagnum mosses occur mainly in the Northern Hemisphere in peat bogs, conifer forests, and moist tundra areas, M. polymorpha is a widespread plant often found beside rivers. Thus, despite the wide distribution of these bryophytes in very different ecosystems, homology analysis of CBP gene suggests that these genes are universal and conserved.
Analyses of the physico-chemical properties and the subcellular localization indicate that all CBP proteins of H. splendens share a high similarity with those of vascular plants. In A. thaliana, sweet potato, and in some other vascular plants, most CBP enzymes are localized within the chloroplasts (Han et al. 2015; Kang et al. 2018). Prediction of subcellular location of all CBP proteins of H. splendens also suggests a chloroplast localization (Table 3).
To understand the evolutionary relationships between CBP proteins of H. splendens and well-known CBP proteins from bryophytes, neighbor-joining trees were constructed using the deduced amino acid residues of these CBP proteins (Fig. S1 and Table S3). Analyses of these trees demonstrate that the CBP proteins of H. splendens are highly homologous to other CBP proteins in bryophytes.
Recently, some studies indicated that overexpression of the CBP genes results in an increased concentration of carotenoid and improved the tolerance of plants to abiotic stresses. For example, overexpression of IbZDS (Li et al. 2017) and IbLCYb (Kim et al. 2014) genes enhanced salt stress tolerance of transgenic sweet potato plants, while Lycium chinenses plants overexpressing LcLCYe displayed enhanced tolerance to chilling stress (Chen et al., 2015). In transgenic tobacco, overexpression LcPDS, LcZDS, or LcCRTISO led to an increased carotenoid concentration in leaves and changed the ratio between various carotenoids (Li et al. 2020). This study indicated that LcPDS, LcZDS, and LcCRTISO are able to improve carotenoid concentration and salt tolerance in higher plant breeding.
It is known that isoforms of some genes can adopt new functions, with differential expression patterns. For example, in rice, both OsPSY1 and OsPSY2 containing light responsive cis-acting elements play predominant roles in carotenogenesis in green tissues, whereas OsPSY3 is induced in the roots by high salt and/or drought stresses (Welsch et al. 2008). In the moss H. splendens, the HsPSY_1 and HsPSY_2 transcripts differ when moss experiences abiotic stresses (Fig. 5). Stresses do not change the expression of HsPSY_1, while the HsPSY_2 transcripts change in response to high temperature and desiccation (Fig. 5). The highest expression is observed for HsPSY_2 in response to high temperature treatment (Fig. 2). According to Busch et al. (2002), the over-expression of NtPSY significantly accelerates the biosynthesis of carotenoids and promotes accumulation of extra carotenoids in tobacco leaves. Interestingly, HsPDS and HsZDS transcripts involved in desaturation of phytoene do not change significantly under abiotic stresses (Figs. 1, 2 and 3). It is known that the dysfunction of these genes results in the deficiency of carotenoid accumulation and can lead to the death of the plants (Wang et al. 2009). Thus, among upstream CBP genes, HsPSY is a stress inducible gene, while HsPDS, HsZ-ISO and HsZDS are more stable in stress conditions.
Differential expression of CBP transcripts during abiotic stresses was more evident for genes in the middle and downstream steps of CBP. These genes encode the enzymes responsible for carotenoid diversity, such as β-carotene, violaxanthin, zeaxanthin, lutein, and neoxanthin, as a result of the activities of lycopene β-cyclase (LCYb), ε-cyclase (LCYe), and β-carotene hydroxylase (CHYb) (Bouvier et al. 2000; Kim and DellaPenna 2006; Zhu et al. 2008). As known, zeaxanthin, all-trans-violaxanthin, and neoxanthin are carotenoid precursors of ABA (Du et al. 2010). Another pattern of expression is observed for other pair of isoforms of CBP genes in H. splendens. Indeed, HsCHYb_1 and HsCHYb_2 respond similarly to temperature stress, in particular both are activated by elevated temperature but do not respond to freezing temperature. In contrast, these genes display differential response to desiccation and rehydration, e.g. HsCHYb_1 is not activated, while HsCHYb_2 is activated by this treatment. Overexpression of CHYb gene was shown to increase carotenoid concentration in tobacco (Hasunuma et al. 2008), tomato (Huang et al. 2013), maize (Farre et al. 2016), and potato (Morris et al. 2006). Furthermore, previous studies in Arabidopsis with overexpressed CHYb reported that these plants were more tolerant of high temperature and high light (Davison et al. 2002).
In the next pair of isoforms HsLUT1_1 and HsLUT1_2, the first is activated only during dehydration, while the second responds only to the short-term temperature stress (Fig. 5). It is possible that these gene isoforms contain different stress-sensitive cis-elements in promoter regions that can explain their differential stress induced expression and suggest the performance of various functions during carotenogenesis. Moreover, considering that moss H. splendens was collected in the woods, the potential contribution of endophytic bacteria to the gene expression can not be excluded, although such contribution would be minimal for the expression of genes in the apical stem segments.
Thus, an increase in CBP transcripts of xanthophyll cycle during elevated temperature, desiccation and rehydration may be related to the roles of xanthophylls in plant stress tolerance and involvement in ABA synthesis. Stresses will increase xanthophyll cycle activity because extra energy dissipation is needed when carbon fixation is reduced but light interception continues. High conservation of CBP genes during evolution indicates their essential role in carotenogenesis in photosynthetic organisms.
5 Conclusion
In the present study, ten genes involved in the carotenoid biosynthesis pathway in H. splendens were identified and found to share high similarity with those in other bryophytes. Adverse temperatures and desiccation/rehydration treatment induce the expression of some of these CPB genes, providing insights into the roles of CBP genes in response to abiotic stresses and expanding our understanding of the molecular mechanisms regulating carotenoid biosynthesis in mosses. Future research is needed to enable us to evaluate how differential expression of CBP genes can influence the total carotenoid content and the concentrations of individual carotenoids in H. splendens in response to stress treatment.
References
Amarnath K, Bennett DI, Schneider AR, Fleming GR (2016) Multiscale model of light harvesting by photosystem II in plants. Proc Natl Acad Sci USA 113(5):1156–1161. https://doi.org/10.1073/pnas.1524999113
Bhosale P (2004) Environmental and cultural stimulants in the production of carotenoids from microorganisms. Appl Microbiol Biotechnol 63(4):351–361. https://doi.org/10.1007/s00253-003-1441-1
Bouvier J, Bordes P, Romeo Y, Fourçans A, Bouvier I, Gutierrez C (2000) Characterization of OpuA, a glycine-betaine uptake system of Lactococcus lactis. J Mol Microbiol Biotechnol 2(2):199–205. PMID: 10939245
Busch M, Seuter A, Hain R (2002) Functional analysis of the early steps of carotenoid biosynthesis in tobacco. Plant Physiol 128(2):439–453. https://doi.org/10.1104/pp.010573
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL (2009) BLAST+: architecture and applications. BMC Bioinf 10:421. https://doi.org/10.1186/1471-2105-10-421
Cheng T, Chen J, Ef AA, Wang P, Wang G, Hu X, Shi J (2015) Quantitative proteomics analysis reveals that S-nitrosoglutathione reductase (GSNOR) and nitric oxide signaling enhance poplar defense against chilling stress. Planta 242(6):1361–1390. https://doi.org/10.1007/s00425-015-2374-5
Choudhury S, Panda P, Sahoo L, Panda SK (2013) Reactive oxygen species signaling in plants under abiotic stress. Plant Signal Behav 8(4):e23681. https://doi.org/10.4161/psb.23681
Cicevan R, Al Hassan M, Sestras AF, Prohens J, Vicente O, Sestras RE, Boscaiu M (2016) Screening for drought tolerance in cultivars of the ornamental genus Tagetes (Asteraceae). PeerJ 4:e2133. https://doi.org/10.7717/peerj.2133
Davidson PA, Hunter CN, Horton P (2002) Overexpression of β-carotene hydroxylase enhances stress tolerance in Arabidopsis. Nature 418:203–206. https://doi.org/10.1038/nature00861
Del Campo JA, Moreno J, Rodríguez H, Vargas MA, Rivas J, Guerrero MG (2000) Carotenoid content of chlorophyceanmicroalgae. Factors determining lutein accumulation in Muriellopsis sp. (Chlorophyta). J Biotechnol 76:51–59. https://doi.org/10.1016/S0168-1656(99)00178-9
Devitt LC, Fanning K, Dietzgen RG, Holton TA (2010) Isolation and functional characterization of a lycopene beta-cyclase gene that controls fruit colour of papaya (Carica papaya L). J Exp Bot 61:33–39. https://doi.org/10.1093/jxb/erp284
Du H, Wang N, Cui F, Li X, Xiao J, Xiong L (2010) Characterization of the beta-carotene hydroxylase gene DSM2 conferring drought and oxidative stress resistance by increasing xanthophylls and abscisic acid synthesis in rice. Plant Physiol 154:1304–1318. https://doi.org/10.1104/pp.110.163741
Fan M, Sun X, Xu N, Liao Z, Li Y, Wang J, Fan Y, Cui D, Li P, Miao Z (2017) Integration of deep transcriptome and proteome analyses of salicylic acid regulation high temperature stress in Ulva prolifera. Sci Rep 7:11052. https://doi.org/10.1038/s41598-017-11449-w
Farré G, Perez-Fons L, Decourcelle M, Breitenbach J, Hem S, Zhu C, Capell T, Christou P, Fraser PD, Sandmann G (2016) Metabolic engineering of astaxanthin biosynthesis in maize endosperm and characterization of a prototype high oil hybrid. Transgenic Res 25(4):477–489. https://doi.org/10.1007/s11248-016-9943-7
Hammad SAR, Ali OAM (2014) Physiological and biochemical studies on drought tolerance of wheat plants by application of amino acids and yeast extract. Annals of Agricultural Sci 59:133–145. https://doi.org/10.1016/j.aoas.2014.06.018
Han Y, Shi Q, Jiang J (2015) Multisite interaction with Sufu regulates Ci/Gli activity through distinct mechanisms in Hh signal transduction. Proc Natl Acad Sci USA 112(20):6383–6388. https://doi.org/10.1073/pnas.1421628112
Hasunuma T, Miyazawa S, Yoshimura S, Shinzaki Y, Tomizawa K, Shindo K, Choi SK, Misawa N, Miyake C (2008) Biosynthesis of astaxanthin in tobacco leaves by transplastomic engineering. Plant J 55(5):857–868. https://doi.org/10.1111/j.1365-313X.2008.03559.x
He J, Li P, Huo H, Liu L, Tang T, He M, Huang J, Liu L (2019) Heterologous expression of HpBHY and CrBKT increases heat tolerance in Physcomitrella patens. Plant Divers 41:266e274. https://doi.org/10.1016/j.pld.2019.04.001
Huang JC, Zhong YJ, Liu J, Sandmann G, Chen F (2013) Metabolic engineering of tomato for high-yield production of astaxanthin. Metab Eng 17:59–67. https://doi.org/10.1016/j.ymben.2013.02.005
Jackson M (2015) Elevated temperature effects on carotenoid biosynthesis in the diploid strawberry Fragaria vesca. Dissertation, University of Maryland, Maryland
Johnson MG, Malley C, Goffinet B, Shaw AJ, Wickett NJ (2016) A phylotranscriptomic analysis of gene family expansion and evolution in the largest order of pleurocarpous mosses (Hypnales, Bryophyta). Mol Phylogenet Evol 98:29–40. https://doi.org/10.1016/j.ympev.2016.01.008
Juneja A, Ceballos RM, Murthy GS (2013) Effects of environmental factors and nutrient availability on the biochemical composition of algae for biofuels production: a review. Energies 6(9):4607–4638. https://doi.org/10.3390/en6094607
Kang C, Zhai H, Xue L, Zhao N, He S, Liu Q (2018) A lycopene β-cyclase gene, IbLCYB2, enhances carotenoid contents and abiotic stress tolerance in transgenic sweetpotato. Plant Sci 272:243–254. https://doi.org/10.1016/j.plantsci.2018.05.005
Kato M, Ikoma Y, Matsumoto H, Sugiura M, Hyodo H, Yano M (2004) Accumulation of carotenoids and expression of carotenoid biosynthetic genes during maturation in citrus fruit. Plant Physiol 134:824–837. https://doi.org/10.1104/pp.103.031104
Kim J, DellaPenna D (2006) Defining the primary route for lutein synthesis in plants: the role of Arabidopsis carotenoid beta-ring hydroxylase CYP97A3. Proc Natl Acad Sci USA 103(9):3474–3479. https://doi.org/10.1073/pnas.0511207103
Kim SH, Jeong JC, Park S, Bae JY, Ahn MJ, Lee HS, Kwak SS (2014) Down-regulation of sweetpotato lycopene β-cyclase gene enhances tolerance to abiotic stress in transgenic calli. Mol Biol Rep 41(12):8137–8148. https://doi.org/10.1007/s11033-014-3714-4
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Leuenberger M, Morris JM, Chan AM, Leonelli L, Niyogi KK, Fleming GR (2017) Dissecting and modeling zeaxanthin- and lutein-dependent nonphotochemical quenching in Arabidopsis thaliana. Proc Natl Acad Sci USA 114(33):E7009–E7017. https://doi.org/10.1073/pnas.1704502114
Li F, Vallabhaneni R, Yu J (2008) The maize phytoene synthase gene family: overlapping roles for carotenogenesis in endosperm, photomorphogenesis, and thermal stress tolerance. Plant Physiol 147:1334–1346. https://doi.org/10.1104/pp.108.122119
Li R, Kang C, Song X, Yu L, Liu D, He S, Zhai H, Liu Q (2017) A ζ-carotene desaturase gene, IbZDS, increases β-carotene and lutein contents and enhances salt tolerance in transgenic sweetpotato. Plant Sci 262:39–51. https://doi.org/10.1016/j.plantsci.2017.05.014
Li C, Ji J, Wang G, Li ZD, Wang YR, Fan YJ (2020) Overexpression of LcPDS LcZDS and LcCRTISO genes from wolfberry for carotenoid biosynthesis enhanced carotenoid accumulation and salt tolerance in tobacco. Front Plant Sci 11:119. https://doi.org/10.3389/fpls.2020.00119
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25(4):402–408. https://doi.org/10.1006/meth.2001.1262
López-Emparán A, Quezada-Martinez D, Zúñiga-Bustos M, Cifuentes V, Iñiguez-Luy F, Federico ML (2014) Functional analysis of the Brassica napus L. phytoene synthase (PSY) gene family. PLoS ONE 9(12):e114878. https://doi.org/10.1371/journal.pone.0114878
Maestri E, Klueva N, Perrotta C, Gulli M, Nguyen HT, Marmiroli N (2002) Molecular genetics of heat tolerance and heat shock proteins in cereals. Plant Mol Biol 48:67e681. https://doi.org/10.1023/A:1014826730024
Manivannan P, Rabert GA, Rajasekar M, Somasundaram R (2014) Drought stress-induced modification on growth and pigments composition in different genotypes of Helianthus annuus L. Curr Bot 5:7–13. http://scienceflora.org/journals/index.php/jp/
Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ, Lu S, Chitsaz F et al (2017) CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucl Acids Res 45(D1):D200–D203. https://doi.org/10.1093/nar/gkw1129
Merget B, Wolf M (2010) A molecular phylogeny of Hypnales (Bryophyta) inferred from ITS2 sequence-structure data. BMC Res Notes 3:320. https://doi.org/10.1186/1756-0500-3-320
Moeller RE, Gilroy S, Williamson CE, Grad G, Sommaruga R (2005) Dietary acquisition of photoprotective compounds (mycosporine-like amino acids, carotenoids) and acclimation to ultraviolet radiation in a freshwater copepod. Limnol Oceanogr 50:427–439. https://doi.org/10.4319/lo.2005.50.2.0427
Morris WL, Ducreux LJ, Fraser PD, Millam S, Taylor MA (2006) Engineering ketocarotenoid biosynthesis in potato tubers. Metab Eng 8:253e263. https://doi.org/10.1016/j.ymben.2006.01.001
Onele AO, Mazina AB, Leksin IY, Minibayeva FV (2022) DsDBF1, a type A-5 DREB gene, identified and characterized in the moss Dicranum scoparium. Life (Basel) 13(1):90. https://doi.org/10.3390/life13010090
Park CH, Park YE, Yeo HJ, Yoon JS, Park S-Y, Kim JK, Park SU (2021) Comparative analysis of secondary metabolites and metabolic profiling between diploid and tetraploid Morus alba L. J Agric Food Chem 69:1300–1307. https://doi.org/10.1021/acs.jafc.0c06863
Ramel F, Birtic S, Cuiné S, Triantaphylidès C, Ravanat J-L, Havaux M (2012) Chemical quenching of singlet oxygen by carotenoids in plants. Plant Physiol 158(3):1267–1278. http://www.jstor.org/stable/41435324
Reddy CS, Lee SH, Yoon JS, Kim JK, Lee SW, Hur M, Koo SC, Meilan J, Lee WM, Jang JK, Hur Y, Park SU, Kim YB (2017) Molecular cloning and characterization of carotenoid pathway genes and carotenoid content in Ixeris dentata var. Albif lora. Molecules 22:1449. https://doi.org/10.3390/molecules22091449
Ruban AV (2016) Nonphotochemical chlorophyll fluorescence quenching: mechanism and effectiveness in protecting plants from photodamage. Plant Physiol 170(4):1903–1916. https://doi.org/10.1104/pp.15.01935
Ruiz-Sola M, Rodríguez-Concepción M (2012) Carotenoid biosynthesis in Arabidopsis: a colorful pathway. Arab Book 10:e0158. https://doi.org/10.1199/tab.0158
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Sánchez JF, Fernández-Sevilla JM, Acién FG, Cerón MC, Pérez-Parra J, Molina-Grima E (2008) Biomass and lutein productivity of Scenedesmus almeriensis: influence of irradiance, dilution rate and temperature. Appl Microbiol Biotechnol 79(5):719–729. https://doi.org/10.1007/s00253-008-1494-2
Sathasivam R, Radhakrishnan R, Kim JK, Park SU (2021) An update on biosynthesis and regulation of carotenoids in plants. S Afr J Bot 140:290–302. https://doi.org/10.1016/j.jare.2021.03.011
Stanley L, Yuan YW (2019) Transcriptional regulation of carotenoid biosynthesis in plants: so many regulators, so little consensus. Front Plant Sci 10:1017. https://doi.org/10.3389/fpls.2019.01017
Sudrajat DJ, Siregar IZ, Khumaida N, Siregar UJ, Mansur I (2015) Adaptability of white jabon (Anthocephalus cadamba Miq.) Seedling from 12 populations to drought and waterlogging. Agrivita J Agric Sci 37(2):130–143. https://doi.org/10.17503/agrivita.v37i2.455
Syed NH, Kalyna M, Marquez Y, Barta A, Brown JW (2012) Alternative splicing in plants-coming of age. Trends Plant Sci 17(10):616–623. https://doi.org/10.1016/j.tplants.2012.06.001
Takemura M, Maoka T, Misawa N (2014) Carotenoid analysis of a liverwort marchantia polymorpha and functional identification of its lycopene b- and e-cyclase genes. Plant Cell Physiol 55(1):194–200. https://doi.org/10.1093/pcp/pct170
Tan BC, Joseph LM, Deng WT, Liu LJ, Li QB, Cline K, McCarty DR (2003) Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J 35:44–56. https://doi.org/10.1046/j.1365-313X.2003.01786.x
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting position-specific gap penalties and weight matrix choice. Nucl Acids Res 22:4673–4680. https://doi.org/10.1093/nar/22.22.4673
Truscott TG (1990) The photophysics and photochemistry of the carotenoids. J Photochem Photobiol B Biol 6:359–371. https://doi.org/10.1016/1011-1344(90)85110-I
Tuan PA, Kim JK, Lee J, Park WT, Kwon do Y, Kim YB, Kim HH, Kim HR, Park SU (2012) Analysis of carotenoid accumulation and expression of carotenoid biosynthesis genes in different organs of Chinese cabbage (Brassica rapa Subsp pekinensis). EXCLI J 11:508–516 PMID: 27540344; PMCID: PMC4983711
Tuan PA, Kim YB, Kim JK, Arasu MV, Al-Dhabi NA, Park SU (2014) Molecular characterization of carotenoid biosynthetic genes and carotenoid accumulation in Scutellaria baicalensis Georgi. EXCLI J 14:146–157. http://creativecommons.org/licenses/by/4.0/
Wahid A, Gelani S, Ashraf M, Foolad MR (2007) Heat tolerance in plants: an overview. Environ Exp Bot 61:199e223. https://doi.org/10.1016/j.envexpbot.2007.05.011
Wang JW, Czech B, Weigel D (2009) miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138(4):738–749. https://doi.org/10.1016/j.cell.2009.06.014
Welsch R, Wüst F, Bär C, Al-Babili S, Beyer P (2008) A third phytoene synthase is devoted to abiotic stress-induced abscisic acid formation in rice and defines functional diversification of phytoene synthase genes. Plant Physiol 147(1):367–380. https://doi.org/10.1104/pp.108.117028
Zhu YH, Jiang JG, Chen Q (2008) Characterization of cDNA of lycopene beta-cyclase responsible for a high level of beta-carotene accumulation in Dunaliella salina. Biochem Cell Biol 86(3):285–292. https://doi.org/10.1139/O08-012
Zhu HS, Chen MD, Wen QF, Li YP (2015) Isolation and characterization of the carotenoid biosynthetic genes LCYB LCYE and CHXB from Strawberry and their relation to carotenoid accumulation. Sci Hortic 182:134–144. https://doi.org/10.1016/j.scienta.2014.12.007
Funding
This study was supported by the Russian Science Foundation grant No. 22-24-00595 (for AR, MK, AM).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Renkova, A.G., Koulintchenko, M.V., Mazina, A.B. et al. Genes of carotenoid biosynthesis pathway in the moss Hylocomium splendens: identification and differential expression during abiotic stresses. Theor. Exp. Plant Physiol. 36, 83–96 (2024). https://doi.org/10.1007/s40626-024-00309-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40626-024-00309-4