Abstract
Background
Findings from observational clinical studies examining the relationship between biomarker expression and theranosis in colorectal cancer (CRC) have been conflicting.
Objective
We conducted this systematic review and meta-analysis to summarise the existing evidence to demonstrate the involvement of microRNAs (miRNAs) in chemoresistance and sensitivity in CRC through drug genetic pathways.
Methods
Using PRISMA guidelines, we systematically searched PubMed and Science Direct for relevant studies that took place between 2012 and 2017. A random-effects model of meta-analysis was applied to evaluate the pooled effect size of hazard ratios (HRs) across the included studies. Cochran’s Q test and the I2 statistic were used to detect heterogeneity. A funnel plot was used to assess potential publication bias.
Results
Of the 4700 studies found, 39 studies comprising 2822 patients with CRC met the inclusion criteria. The included studies used one or a combination of 14 chemotherapy drugs, including 5-fluorouracil and oxaliplatin. Of the 60 miRNAs, 28 were associated with chemosensitivity, 20 with chemoresistance, and one with differential expression and radiosensitivity; ten miRNAs were not associated with any impact on chemotherapy. The results outline the importance of 34 drug–regulatory pathways of chemoresistance and sensitivity in CRC. The mean effect size was 0.689 (95% confidence interval 0.428–1.110), indicating that the expression of miRNAs decreased the likelihood of death by about 32%.
Conclusion
Studies have consistently shown that multiple miRNAs could act as clinical predictors of chemoresistance and sensitivity. An inclusion of supplementary miRNA estimation in CRC routine practice needs to be considered to evaluate the efficacy of chemotherapy after confirming our findings with large-scale prospective cohort studies.
PROSPERO registration number
CRD42017082196.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
This is the first comprehensive systematic review to define the impact of microRNAs (miRNAs) in chemoresistance in colorectal cancer. |
Our results aid in comparing the association of drug-related genetic pathways with chemoresistance in colorectal cancer. |
This review highlights the critical role of biomarkers involved in colorectal cancer and will help determine their possible role in diagnosis and prognosis. |
1 Introduction
Colorectal cancer (CRC) is the third most common cancer in men, with 746,000 cases annually, and is the second most common in women, with 614,000 cases per year worldwide [1]. The conventional modalities of treatment for CRC include surgery [2], chemotherapy [3], radiation therapy [4], immunotherapy [5], targeted therapy [6], and precision medicine [7]. The commonly used chemotherapy drugs and monoclonal antibodies (mAbs) to treat CRC are 5-fluorouracil (5-FU) [8], oxaliplatin [9], cisplatin [10], doxorubicin [11], leucovorin [12], paclitaxel [13], mitomycin C (MMC) [14], tumour necrosis factor-related apoptosis-inducing ligand (TRAIL) [15], deoxycholic acid (DCA) [16], thapsigargin (Tg) and trichostatin A (TSA) [17], irinotecan [18], cetuximab [19], panitumumab, and bevacizumab [20]. However, chemoresistance is a significant hindrance to successful treatment in many CRC cases [21,22,23], and acquired drug resistance occurs with 90% of metastatic cancer [24]. Despite advances in treatment methods, the 5-year survival rate is 12.5% [24].
The involvement of microRNAs (miRNAs) in chemoresistance is associated with poor prognosis in several cancers [25,26,27,28,29,30,31]. Therefore, identification of biomarkers to detect possible chemoresistance in individual cases is a significant step towards specialised or personalised cancer treatment [32]. Emerging evidence has revealed that miRNAs can be considered as minimally invasive biomarkers useful for prognosis and as theranostic targets for monitoring treatment response (theranosis) [33]. Chemoresistance in CRC is mediated by the expression of a few specific miRNAs through drug-regulatory pathways [34, 35]. Both miRNA-19b and -21 were found to influence chemoresistance to 5-FU in human colon cells (DLD-1 and KM12C) [36]. 5-FU triggers A-disintegrin and metalloprotease domain-17 (ADAM-17), which causes growth factor shedding and growth factor receptor activation, leading to chemoresistance in CRC, which was found to be profoundly influenced by miRNA-222 [37, 38]. These drug-regulatory genes have been found to regulate cellular transformation and are influenced by miRNA expression [39].
Huang et al. [40] analysed 12 miRNAs (134, 146a, 17-3p, 181d, 191, 221, 222, 223, 25, 29a, 320a, and 92a) in plasma samples of both patients with CRC and healthy patients and found that levels of miRNA-29a and miRNA-92a were significantly higher in cancer tissues. miRNA was consistently upregulated in patients with CRC, with 21 of 30 patients expressing high levels throughout the 50 months of the post-treatment follow-up period [40]. The investigation of 5-FU resistance in 88 patients with CRC revealed that miRNA-10b expression was significantly higher in cancer tissues than in normal tissues and was connected to lymphatic invasion and poor prognosis, thus indicating miRNA-10b expression as a potential indicator of chemoresistance [41].
Cancer invasiveness and an increase in resistance to oxaliplatin and cisplatin were observed in both in vitro and in vivo studies; this mechanism was mediated by forkhead transcription factor forkhead box O3a (FOXO3a) and miRNA-induced metalloprotease enzyme, which indirectly promotes invasion [42]. Preclinical and clinical observational studies demonstrated that miRNA expression profiling could help to identify high-risk patients with CRC who may develop chemoresistance. Therefore, a comprehensive systematic review and meta-analysis was sought to review the published studies on miRNA-mediated chemoresistance in CRC (refer to the Electronic Supplementary Material [ESM] for the rationale of the study).
2 Methods
This systematic review and meta-analysis followed the 2015 PRISMA (Preferred Reporting Items for Systematic Review and Meta-analysis) guidelines [43] and was conducted following a previously established protocol (PROSPERO registration number: CRD42017082196).
2.1 Search Strategy and Selection Criteria
We searched the PubMed and Science Direct databases on October 2017 from 1 January 2012 to 25 October 2017, restricting the search to papers in the English language and to the last 5 years so the use of chemotherapeutic drugs was relatively current. The literature search was performed using the Medical Subject Heading (MeSH) search terms miRNA or microRNA, chemoresistance/sensitivity, and colorectal cancer in combination. The search strategy is presented in Table 1 in the ESM. We also manually searched the reference lists of all included publications for additional relevant studies. The titles and abstracts of all relevant studies were carefully examined and screened before full-text articles were retrieved. All search results were collated in a reference manager database (EndNote) to avoid duplication. Eligible studies had to meet the following inclusion criteria: involved miRNA and CRC, involved both clinical samples and in vitro preclinical analyses for patients with CRC, focused on resistance to CRC therapy, reported miRNA profiling platforms, and reported the genes or pathways involved in chemoresistance or sensitivity. We excluded the following: studies in languages other than English; reviews, editorials, opinions, case studies, and reports; unpublished materials, uninterpretable data, conference proceedings, or thesis; studies focusing only on long non-coding (lnc) RNA, and studies involving Fusobacterium nucleatum and its association with CRC chemoresistance.
2.2 Data Extraction and Quality Assessment
Two authors (RJ and MRM) independently evaluated and extracted the data from the screened articles using the selection criteria. Corresponding authors were contacted for supplementary materials if any necessary data were unavailable from the full text. Any disagreements between reviewers were resolved through discussion between the authors or by team decision or by consultation with the third reviewer (CK). The following data were collected and recorded for each study: first author and year of publication, patient information, location of the study, sex, ethnicity, tumour stage, number of samples, lymph node metastasis/nodal status, cell lines used, miRNAs involved, miRNA profiling platform, chemotherapy drugs used, drug-regulatory pathways, and associated genes. We described the effect size of the prognosis using the hazard ratio (HRs) and 95% confidence interval (CI) of survival of patients with CRC.
Two investigators (RJ and MRM) independently assessed each study for methodological quality using the MOOSE (Meta-analysis Of Observational Studies in Epidemiology) checklist by the Dutch Cochrane Centre [44]. A study checklist with predefined criteria prepared from the MOOSE study criteria list was used to assess the methodological quality of the studies included in the systematic review.
2.3 Statistical Analysis
Reporting of the sections, meta-analysis, subgroup analysis, and publication bias follows guidelines from the Meta-analysis concepts and applications workshop manual by Michael Borenstein. We used the Comprehensive Meta-Analysis (CMA) software (version 3.0, USA) to analyse the HRs and 95% CIs. The survival data, in the form of Kaplan–Meier curves, were transformed into HRs and 95% CIs. The forest plot was generated with combined outcome data to elucidate the clinical outcome effects of patient survival in CRC. Heterogeneity was obtained using Cochran’s Q test and Higgins I2 statistic [45]. Z-statistics were generated to analyse the standard deviations from the mean of all included studies if the pooled study results deviated. We used the subgroup analysis to compare the effect size in studies that employed a high expression and low expression of miRNAs.
2.4 Publication Bias
The inverted funnel plot depicts the level of publication bias. Publication bias was quantified using Egger’s bias indicator test, the Orwin and classic fail-safe N test, the Begg and Mazumdar rank collection test, and Duval and Tweedie’s trim-and-fill calculation.
3 Results
3.1 Study Search and Characteristics
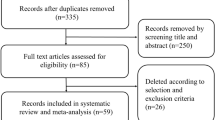
The initial search yielded 4700 studies. By implementing the search strategy, we identified a total of 2450 studies from PubMed (n = 200) and Science Direct (n = 2250) (Fig. 1). After removing duplicates, 163 potentially eligible studies were scrutinised for selection criteria. Crosschecking the existing reference lists of narrative and systematic reviews revealed no further relevant articles. Careful review of the 163 articles against the PRISMA guidelines identified 43 full-text studies that contained available data items. Of these 43, four were excluded because three studies evaluated lnc RNA expression in CRC and one investigated the association between F. nucleatum and CRC chemoresistance. We identified 39 studies involving 2822 patients with CRC, eligible for our systematic review. Seven studies were ultimately included in the meta-analysis.
Table 1 provides the main characteristics of the 39 included studies. The study period of the included studies was between 1999 and 2015. The most commonly used chemotherapy agents were 5-FU and oxaliplatin. Frozen CRC tissue samples were used in 23 studies, and four studies used formalin-fixed paraffin-embedded (FFPE) tissues; 17 studies did not specify the sampling type. A total of 3868 CRC tissue samples and 231 blood samples were included for analysis, 94 in stage I, 312 in stage II, 548 in stage III, and 114 samples in stage IV. Furthermore, 228 samples were observed from stage I to II, and 342 samples from stage III to IV. miRNA expression was analysed via microarray in a few studies, and all 39 studies used reverse transcription polymerase chain reaction (RT-PCR) for miRNA expression profiling.
3.2 Preclinical Investigation of MicroRNA (miRNA) Expression
In total, 39 studies reported a total of 40 cell lines utilised in the in vitro analysis to evaluate miRNA expression and its association with drug-regulatory genetic pathways. Figure 1 in the ESM presents the number of studies illustrating the most common assays. The most common cell lines were HCT-116, HCT-29, LoVo, SW480, and SW620. HCT-116 was used in 27 studies. The highest number of cell lines used in a single study was 13 [46]. The in vivo and in vitro assays from the studies included in our systematic review were the MTT/cell viability assay, luciferase assay, cell proliferation, western blotting, chemotherapy sensitivity assay, cell migration, cell invasion, apoptotic assay, clonogenic assay, tumorigenesis, colony formation, caspase-3 assay, BrdU assay, and radiosensitivity assay.
3.3 Clinical Investigation of miRNA Expression
3.3.1 Association Between miRNA Expression and Chemoresistance/Chemosensitivity
Of 60 miRNAs reported in the systematic review, 34 were downregulated and 24 were upregulated in patients with CRC (Fig. 2). Five upregulated miRNAs were associated with chemosensitivity, and 13 upregulated miRNAs were associated with chemoresistance. Similarly, 22 downregulated miRNAs were associated with chemosensitivity, and six downregulated miRNAs were associated with chemoresistance. Our report showed that miRNA-224 was differentially expressed and not related to either chemoresistance or sensitivity. Overall, the role of these miRNAs was investigated individually, as enhancers of chemoresistance (n = 20 miRNAs) or chemosensitivity (n = 28 miRNAs). We observed five miRNAs that were predominantly studied in ten different studies, with all being downregulated in patients with CRC. The internal control used was glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (ten studies) and U6 small nuclear RNA (U6 snRNA) (27 studies) for normalising the expression of miRNAs in the pooled studies. Some studies used both GAPDH and U6 snRNA.
Nine hallmarks of colorectal cancer chemotherapy BCL-2, GRP-78, EGFR, ADAM, cyclin, IGFR, AKT/PI3K, RAS, and FOX. Each hallmark shows specific miRNA that influences particular cellular function in CRC; some miRNAs control more than one hallmark, indicating multiple pathways regulated by them. Orange colour refers upregulated miRNAs; green colour indicates downregulated miRNAs. BCL-2 B-cell lymphoma-2, GRP-78 glucose-regulated protein 78 kDa, EGFR epidermal growth factor receptor, ADAM A disintegrin and metalloproteinase domain, IGFR insulin-like growth factor 1 receptor, AKT/PI3K protein kinase B/phosphoinositide 3-kinase, FOX forkhead box
3.4 Colorectal Cancer (CRC) Chemotherapy
Information about the CRC chemotherapy was available for 2822 patients across 37 studies. The included studies used a total of 14 drugs and their combinations, including 5-FU (1404 patients) [47], oxaliplatin (890 patients) [48], mitomycin C (180 patients) [49], cisplatin (314 patients) [50], doxorubicin (number of patients not reported) [51], leucovorin (137 patients) [52], paclitaxel (67 patients) [46], TRAIL (257 patients) [53], DCA (40 patients) [54], irinotecan (173 patients) [55], capecitabine (74 patients) [52], cetuximab (117 patients) [56], and thapsigargin (Tg) and TSA (ten patients) [17]. Of the 14 chemotherapy drugs studied individually and in combinations, 5-FU [57,58,59] was the most studied, followed by oxaliplatin [60, 61]. 5-FU is a non-specific drug treatment for all types of cancers [47, 62,63,64] and is also used in combination with oxaliplatin [65], leucovorin, and irinotecan [66]. The four studies investigated miRNA-139-5p [67, 68] and -497 [47, 50] twice in 5-FU treatment, whereas the remaining miRNAs were studied with other chemotherapy only once. Studies using cohort populations in the USA and China indicated a correlation between chemoresistance to 5-FU and increased miRNA-21 expression [69].
3.5 CRC Chemoresistance and Drug-Regulated Genetic Pathways
In the 39 studies, 34 unique miRNA-mediated drug-regulatory pathway-associated genes were reported (Fig. 2). We collated the drug-regulated gene pathways in CRC, with epidermal growth factor receptor (EGFR) (n = 3), phosphatidylinositol 3-kinase/protein kinase B (PI3K/AKT) (n = 2), ADAM (n = 2), B-cell lymphoma 2 (BCL-2) (n = 3), and insulin like growth factor 1 receptor (IGF1R; n = 7) being the most common pathways explored.
Tables 2 and 3 present the upregulation and downregulation of miRNAs contributing to chemosensitivity and resistance in patients with CRC through drug-regulated genetic pathways.
3.6 Meta-Analysis
3.6.1 Does miRNA Expression Affect Survival of Patients with CRC?
HRs and 95% CIs were explicitly reported in only four [42, 70,71,72] of the 39 studies and could be estimated from three studies [66, 71, 73] covering a total of 697 patients with CRC (Fig. 3). The mean effect size was 0.689, indicating that the expression of miRNAs decreased the likelihood of death by about 32%. The 95% CI for the HR was 0.428–1.110, which tells us the mean HR in the universe of studies could fall anywhere in this range.
Forest plot of pooled hazard ratio values from studies correlating the overall patient survival and miRNA expression with regards to chemotherapy. The pooled hazard ratios of hazard ratio values for colorectal cancer prognostic data were calculated and analysed using CMA software (version 3.3.070, USA). The red diamond represents the pooled effect estimate of survival for patients with colorectal cancer randomly assigned to miRNA evaluation. The black square line indicates the effect size of miRNA of the included studies with 95% confidence interval. The hazard ratio of 1 suggests no difference in survival risk of patients with colorectal cancer. A hazard ratio > 1 indicates an increased risk of patients’ survival, whereas a hazard ratio < 1 suggests a reduced risk of patients’ survival
Similarly, the Z value for test null hypothesis (that the mean risk ratio is 1.0) was − 1.531, p = 0.126. Therefore, we can reject the null that the risk of an event is the same in both upregulated and downregulated groups and conclude that the risk is higher in the upregulated group.
3.6.2 How Much Does the Effect Size Vary Across Studies?
The Q value was 34.640, df = 6, p = 0.000. Since the observed variance falls within the range that can be attributed to sampling error, we cannot reject the null that the true effect size is the same in all studies. Here, I2 was 82.679%. T2 is the variance of true effect sizes (in log units). Here, T2 was 0.360. T is the standard deviation of true effects (in log units). Here, T was 0.600.
3.6.3 Does the Effect Size Vary by Subgroup?
While the mean effect size across all studies is modest (HR 0.689), it is possible that the mean HR varies by subgroup. The mean HR in the upregulated and downregulated groups was 1.812 and 0.515, respectively. The Q value for the differences was 4.916, df = 1, p = 0.027. Therefore, there was no evidence that the HR varied according to survival of patients with CRC.
3.7 Publication Bias
Figure 4 presents the funnel plot correlating overall patient survival and miRNA expression with regards to chemotherapy.
Funnel plot of studies correlating the overall patient survival and miRNA expression with regards to chemotherapy. Dots represent the individual study; two studies on the bottom and three studies on the left-hand side of the plot. Given most of this area contains regions of high significance, publication bias would be unlikely to cause that asymmetry. This would reflect the fact that smaller studies (which appear toward the bottom) are more likely to be published if they have larger than average effects, which makes them more likely to meet the criterion for statistical significance
3.7.1 Classic Fail-Safe N
This meta-analysis includes data from seven CRC studies, which yield a Z value of − 1.70300 and a corresponding 2-tailed p = 0.08857. Since the combined result was not statistically significant, the fail-safe N was irrelevant.
3.7.2 Orwin Fail-Safe N
The criterion value must be set between the other two values for the Orwin fail-safe N to be computed. Here, the HR in observed studies was 0.689, which did not fall between the mean HR in the missing studies, so we could not calculate the Orwin fail-safe N.
3.7.3 Begg and Mazumdar Rank Correlation Test
In this case, Kendall’s tau b (corrected for ties, if any) was 0.09524, with a 1-tailed p value (recommended) of 0.38195 or a 2-tailed p value of 0.76389 (based on continuity-corrected normal approximation).
3.7.4 Egger’s Test of the Intercept
In this case, the intercept (B0) was 1.11103 (95% CI − 2.98430 to 5.20636), with t=0.69738, df = 5. The 1-tailed p value (recommended) was 0.25833, and the 2-tailed p value was 0.51665.
3.7.5 Duval and Tweedie’s Trim and Fill
This method suggests that two studies are missing (Fig. 2 in the ESM). Under the fixed-effects model, the point estimate for the combined studies was 0.76243 (95% CI 0.65272–0.89057). Using trim and fill, the imputed point estimate was 0.72164 (95% CI 0.61924–0.84096). Under the random-effects model, the point estimate for the combined studies was 0.92213 (95% CI 0.54705–1.55438). Using trim and fill, the imputed point estimate was 0.67177 (95% CI 0.39161–1.15234).
4 Discussion
Recent studies have demonstrated that specific miRNA expressions in CRC modulate chemosensitivity and resistance through regulation of drug-related genetic pathways [47, 58, 70, 74]. miRNA-mediated chemoresistance mechanisms in CRC have been explored in individual studies but have not been comprehensively characterised. Therefore, this systematic review and meta-analysis aimed to provide insights into miRNA expression patterns in the chemotherapy–drug mechanistic relationship as well as the regulation of genes associated with chemoresistance/sensitivity. This is the first systematic review to include different ethnic groups in various clinical settings.
Numerous studies have focused on the effect of miRNAs on chemoresistance, including in breast [75], cervical [76], colorectal [77], gastric [78], lung [79], oral [80], ovarian [81], pancreatic [82], prostate [83], and skin [84] cancers. Our systematic review showed that 60 miRNAs were upregulated as well as downregulated in CRC cell lines and tissues. Most of the studies investigated only one miRNA [47, 68, 85], whereas only seven studies focused on two or more miRNAs.
Previous reports have demonstrated crucial clinical functions of miRNAs that were consistent with our findings, particularly miRNA-21, which is used as a diagnostic and prognostic marker for several cancers, such as lung [86], breast [87] pancreas [88], CRC [89], and prostate [90]; miRNA-10b, -141, and -155 are used as diagnostic markers for lung cancer [91]; miRNA-143 is used as a diagnostic marker for CRC [92]; and, more importantly, miRNA-21 [93], -22 [94], -23a [95], -27b [96], -34a [97], -124 [98], and -135b [99] are being proposed as diagnostic markers in CRC. However, reports have also demonstrated conflicting expression patterns for miRNAs: miRNA-27a was found to be downregulated in one study [100] but upregulated in another [101].
Our systematic review highlights the importance of 34 drug-regulatory pathways, including the EGFR, IGF1R, and AKT/PI3K pathways, in CRC chemoresistance and susceptibility. Research has revealed that EGFR is involved in the prediction of overall survival and prognosis of cancers such as gastric [102], lung [103], head and neck cancer (HNC) [104], and CRC [105]. IGF1R plays an essential role in the regulation of cell proliferation, differentiation, and survival of tumour development [106] and has been well-studied in breast cancer [107], CRC [108], and prostate cancer [109]; it directly promotes angiogenesis via the PI3K/AKT pathway. Alternation in the PI3K pathway helps in identification of clinical outcomes in breast cancer [110], gastric cancer [111], CRC, and HNC [112]. The AKT pathway is a frequent target for lung cancer [113], breast cancer [110], CRC [114], and gastric cancer [115]. Our results highlighted the involvement of EGFR in increased chemosensitivity through miRNA-7, -20b, and -133b. Our results are consistent with another study on EGFR-targeted therapy [116]. Furthermore, miRNA-34a was observed as the direct target of Wnt signalling pathways, similar to other reports [39].
Our meta-analysis showed an overall pooled effect size could be a good predictor of patient survival. However, it is essential to note that we used only seven studies because insufficient data were reported in 32 studies. We noticed that several factors, including study strategy, inadequate information, and sample size might be responsible for the high level of heterogeneity.
4.1 Limitation and Strengths
Lack of statistical data in many included studies, including clinicopathological parameters, odds ratios (ORs), HR values, and quantitative data for various assays, limited our quantitative data synthesis. As the HR and CI values were retrieved from Kaplan–Meier curves, there could be some marginal errors, as values were not reported explicitly in the articles. The heterogeneity and differences in study design between different studies could have restricted both the analysis and a clinical hypothesis. A solution to this issue in future studies evaluating miRNA as theragnostic biomarkers would be to perform large collaborative studies in patients with CRC in established clinical settings.
One of the strengths of our study is the detailed correlation of the specific miRNAs with the regulation of chemoresistance in CRC. The clinical sources for miRNA profiling were investigated in our study using different clinical samples, including tissue and plasma. This study may be useful as a repository tabulating the miRNA gene regulatory pathways and its associations with chemotherapy in CRC. Furthermore, this study will provide lists of potential miRNA targets that could help to detect early chemoresistance and sensitivity in patients during treatment, encouraging individualised treatment.
5 Conclusions
This comprehensive systematic review and meta-analysis of published studies from around the world indicates the associations between the molecular mechanisms of chemoresistance and specific miRNAs in CRC. We anticipate that the interpretation of the molecular mechanisms of miRNAs in CRC will lead to improvements in the theranosis-based cancer therapy and oligonucleotide drugs currently under development.
References
World Health Organization. Colorectal Cancer: Estimated Incidence, Mortality and Prevalence Worldwide in 2012. 2012. http://globocan.iarc.fr/Pages/fact_sheets_cancer.aspx?cancer=colorectal. Accessed July 2018.
Devon KM, Vergara-Fernandez O, Victor JC, McLeod RS. Colorectal cancer surgery in elderly patients: presentation, treatment, and outcomes. Dis Colon Rectum. 2009;52(7):1272–7.
Scappaticci FA, Fehrenbacher L, Cartwright T, Hainsworth JD, Heim W, Berlin J, et al. Surgical wound healing complications in metastatic colorectal cancer patients treated with bevacizumab. J Surg Oncol. 2005;91(3):173–80.
Ayanian JZ, Zaslavsky AM, Fuchs CS, Guadagnoli E, Creech CM, Cress RD, et al. Use of adjuvant chemotherapy and radiation therapy for colorectal cancer in a population-based cohort. J Surg Oncol. 2003;21(7):1293–300.
Maisonial-Besset A, Witkowski T, Navarro-Teulon I, Berthier-Vergnes O, Fois G, Zhu Y, et al. Tetraspanin 8 (TSPAN 8) as a potential target for radio-immunotherapy of colorectal cancer. Oncotarget. 2017;8(13):22034.
Khan SA, Zeng Z, Shia J, Paty PB. EGFR gene amplification and KRAS mutation predict response to combination targeted therapy in metastatic colorectal cancer. Pathol Oncol Res. 2017;23(3):673–7.
Dienstmann R, Vermeulen L, Guinney J, Kopetz S, Tejpar S, Tabernero J. Consensus molecular subtypes and the evolution of precision medicine in colorectal cancer. Nat Rev Cancer. 2017;17(2):79–92.
Wiegering A, Matthes N, Mühling B, Koospal M, Quenzer A, Peter S, et al. Reactivating p53 and inducing tumor apoptosis (RITA) enhances the response of RITA-sensitive colorectal cancer cells to chemotherapeutic agents 5-fluorouracil and oxaliplatin. Neoplasia. 2017;19(4):301–9.
Costa T, Nuñez J, Felismino T, Boente L, Mello C. REOX: evaluation of the efficacy of retreatment with an oxaliplatin-containing regimen in metastatic colorectal cancer: a retrospective single-center study. Clin Colorectal Cancer. 2017;16(4):316–23.
Akerfeldt MC, Tran CMN, Shen C, Hambley TW, New EJ. Interactions of cisplatin and the copper transporter CTR1 in human colon cancer cells. J Biol Inorg Chem. 2017;22(5):765–74.
Potočnik N, Perše M, Cerar A, Injac R, Finderle Ž. Cardiac autonomic modulation induced by doxorubicin in a rodent model of colorectal cancer and the influence of fullerenol pretreatment. PLoS One. 2017;12(7):e0181632.
Odin E, Sondén A, Saksena P, Edsjö A, Carlsson G, Vedin A, Gustavsson B, Wettergren Y. P-281 Expression of folate pathway genes with putative impact on leucovorin metabolism and outcome of patients with advanced colorectal cancer. Ann Oncol. 2017. https://doi.org/10.1093/annonc/mdx261.279.
Lv C, Qu H, Zhu W, Xu K, Xu A, Jia B, Qing Y, Li H, Wei HJ, Zhao HY. Low-dose paclitaxel inhibits tumor cell growth by regulating glutaminolysis in colorectal carcinoma cells. Front Pharmacol. 2017;8:244.
Lev A, Deihimi S, Shagisultanova E, Xiu J, Lulla AR, Dicker DT, El-Deiry WS. Preclinical rationale for combination of crizotinib with mitomycin C for the treatment of advanced colorectal cancer. Cancer Biol Ther. 2017;18(9):694–704.
Zhang R, Xu J, Zhao J, Bai J. Knockdown of miR-27a sensitizes colorectal cancer stem cells to TRAIL by promoting the formation of Apaf-1-caspase-9 complex. Oncotarget. 2017;8(28):45213.
Kim E-K, Cho JH, Kim E, Kim YJ. Ursodeoxycholic acid inhibits the proliferation of colon cancer cells by regulating oxidative stress and cancer stem-like cell growth. PLoS One. 2017;12(7):e0181183.
Su S, Chang Y, Andreu-Vieyra C, Fang J, Yang Z, Han B, et al. miR-30d, miR-181a and miR-199a-5p cooperatively suppress the endoplasmic reticulum chaperone and signaling regulator GRP78 in cancer. Oncogene. 2013;32(39):4694–701.
Cohen DJ, Grabocka E, Bar-Sagi D, Godin R, Leichman LP. A phase Ib study combining irinotecan with AZD1775, a selective WEE 1 kinase inhibitor, in RAS/RAF mutated metastatic colorectal cancer patients who progressed on first line therapy. J Clin Oncol; 2017. https://doi.org/10.1200/JCO.2017.35.15_suppl.TPS3627.
Khelwatty S, Essapen S, Bagwan I, Green M, Seddon A, Modjtahedi H. The impact of co-expression of wild-type EGFR and its ligands determined by immunohistochemistry for response to treatment with cetuximab in patients with metastatic colorectal cancer. Oncotarget. 2017;8(5):7666.
Wainberg Z, Hecht JR. A phase III randomized, open-label, controlled trial of chemotherapy and bevacizumab with or without panitumumab in the first-line treatment of patients with metastatic colorectal cancer. Clin Colorectal Cancer. 2006;5(5):363–7.
Misale S, Yaeger R, Hobor S, Scala E, Janakiraman M, Liska D, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012;486(7404):532–6.
Russo M, Misale S, Wei G, Siravegna G, Crisafulli G, Lazzari L, et al. Acquired resistance to the TRK inhibitor entrectinib in colorectal cancer. Cancer Discov. 2016;6(1):36–44.
Diaz LA Jr, Williams RT, Wu J, Kinde I, Hecht JR, Berlin J, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 2012;486(7404):537–40.
Longley D, Johnston P. Molecular mechanisms of drug resistance. J Pathol. 2005;205(2):275–92.
Hollis M, Nair K, Vyas A, Chaturvedi LS, Gambhir S, Vyas D. MicroRNAs potential utility in colon cancer: early detection, prognosis, and chemosensitivity. World J Gastroenterol. 2015;21(27):8284.
Merhautova J, Demlova R, Slaby O. MicroRNA-based therapy in animal models of selected gastrointestinal cancers. Front Pharmacol. 2016;7:329.
Zhu L, Fang J. The structure and clinical roles of microRNA in colorectal cancer. Gastroenterol Res Pract. 2016;2016:1360348. https://doi.org/10.1155/2016/1360348.
Xie T, Huang M, Wang Y, Wang L, Chen C, Chu X. MicroRNAs as regulators, biomarkers and therapeutic targets in the drug resistance of colorectal cancer. Cell Physiol Biochem. 2016;40(1–2):62–76.
Liu X, Fu Q, Du Y, Yang Y, Cho WC. MicroRNA as regulators of cancer stem cells and chemoresistance in colorectal cancer. Curr Cancer Drug Targets. 2016;16(9):738–54.
Fanale D, Castiglia M, Bazan V, Russo A. Involvement of non-coding RNAs in chemo-and radioresistance of colorectal cancer. In: Non-coding RNAs in Colorectal Cancer. Springer, Berlin; 2016. p. 207–28.
Yi R, Li Y, Wang F-L, Miao G, Qi R-M, Zhao Y-Y. MicroRNAs as diagnostic and prognostic biomarkers in colorectal cancer. World J Gastrointest Oncol. 2016;8(4):330.
Amirkhah R, Farazmand A, Irfan-Maqsood M, Wolkenhauer O, Schmitz U. The role of microRNAs in the resistance to colorectal cancer treatments. Cell Mol Biol. 2015;61(6):17–23.
Vosa U, Vooder T, Kolde R, Vilo J, Metspalu A, Annilo T. Meta-analysis of microRNA expression in lung cancer. Int J Cancer. 2013;132(12):2884–93.
Khorrami S, Zavaran Hosseini A, Mowla SJ, Soleimani M, Rakhshani N, Malekzadeh R. MicroRNA-146a induces immune suppression and drug-resistant colorectal cancer cells. Tumor Biol. 2017;39(5):1010428317698365.
Xu K, Chen G, Qiu Y, Yuan Z, Li H, Yuan X, et al. miR-503-5p confers drug resistance by targeting PUMA in colorectal carcinoma. Oncotarget. 2017;8(13):21719.
Kurokawa K, Tanahashi T, Iima T, Yamamoto Y, Akaike Y, Nishida K, et al. Role of miR-19b and its target mRNAs in 5-fluorouracil resistance in colon cancer cells. J Gastroenterol. 2012;47(8):883–95.
Kyula JN, Van Schaeybroeck S, Doherty J, Fenning CS, Longley DB, Johnston PG. Chemotherapy-induced activation of ADAM-17: a novel mechanism of drug resistance in colorectal cancer. Clin Cancer Res. 2010;16(13):3378–89.
Xu K, Liang X, Shen K, Sun L, Cui D, Zhao Y, et al. MiR-222 modulates multidrug resistance in human colorectal carcinoma by down-regulating ADAM-17. Exp Cell Res. 2012;318(17):2168–77.
Strubberg AM, Madison BB. MicroRNAs in the etiology of colorectal cancer: pathways and clinical implications. Dis Models Mechan. 2017;10(3):197–214.
Huang Z, Huang D, Ni S, Peng Z, Sheng W, Du X. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer. 2010;127(1):118–26.
Nishida N, Yamashita S, Mimori K, Sudo T, Tanaka F, Shibata K, et al. MicroRNA-10b is a prognostic indicator in colorectal cancer and confers resistance to the chemotherapeutic agent 5-fluorouracil in colorectal cancer cells. Ann Surg Oncol. 2012;19(9):3065–71.
Zhang L, Pickard K, Jenei V, Bullock MD, Bruce A, Mitter R, et al. miR-153 supports colorectal cancer progression via pleiotropic effects that enhance invasion and chemotherapeutic resistance. Cancer Res. 2013;73(21):6435–47.
Moher D, Liberati A, Tetzlaff J, Altman DG, Group P. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6(7):e1000097.
Stroup DF, Berlin JA, Morton SC, Olkin I, Williamson GD, Rennie D, et al. Meta-analysis of observational studies in epidemiology: a proposal for reporting. Jama. 2000;283(15):2008–12.
Higgins J, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21(11):1539–58.
Liu Y, Gao S, Chen X, Liu M, Mao C, Fang X. Overexpression of miR-203 sensitizes paclitaxel (Taxol)-resistant colorectal cancer cells through targeting the salt-inducible kinase 2 (SIK2). Tumor Biol. 2016;37(9):12231–9.
Wang L, Jiang C-F, Li D-M, Ge X, Shi Z-M, Li C-Y, et al. MicroRNA-497 inhibits tumor growth and increases chemosensitivity to 5-fluorouracil treatment by targeting KSR1. Oncotarget. 2016;7(3):2660.
Qian X, Yu J, Yin Y, He J, Wang L, Li Q, et al. MicroRNA-143 inhibits tumor growth and angiogenesis and sensitizes chemosensitivity to oxaliplatin in colorectal cancers. Cell Cycle. 2013;12(9):1385–94.
Sui H, Cai G-X, Pan S-F, Deng W-L, Wang Y-W, Chen Z-S, et al. miR200c attenuates P-gp–mediated MDR and metastasis by targeting JNK2/c-jun signaling pathway in colorectal cancer. Mol Cancer Ther. 2014;13(12):3137–51.
Guo S, Jiang C, Wang G, Li Y, Wang C, Guo X, et al. MicroRNA-497 targets insulin-like growth factor 1 receptor and has a tumour suppressive role in human colorectal cancer. Oncogene. 2013;32(15):1910–20.
Qu J, Zhao L, Zhang P, Wang J, Xu N, Mi W, et al. MicroRNA-195 chemosensitizes colon cancer cells to the chemotherapeutic drug doxorubicin by targeting the first binding site of BCL2L2 mRNA. J Cell Physiol. 2015;230(3):535–45.
Zhou H, Lin C, Zhang Y, Zhang X, Zhang C, Zhang P, Xie X, Ren Z. miR‐506 enhances the sensitivity of human colorectal cancer cells to oxaliplatin by suppressing MDR1/P‐gp expression. Cell Prolif. 2017;50(3):e12341.
Huang G, Chen X, Cai Y, Wang X, Xing C. miR-20a-directed regulation of BID is associated with the TRAIL sensitivity in colorectal cancer. Oncol Rep. 2017;37(1):571–8.
Kong Y, Bai P-S, Sun H, Nan K-J, Chen N-Z, Qi X-G. The deoxycholic acid targets miRNA-dependent CAC1 gene expression in multidrug resistance of human colorectal cancer. Int J Biochem Cell Biol. 2012;44(12):2321–32.
To KK, Leung W, Ng SS. Exploiting a novel miR-519c–HuR–ABCG2 regulatory pathway to overcome chemoresistance in colorectal cancer. Exp Cell Res. 2015;338(2):222–31.
Zhou J, Lv L, Lin C, Hu G, Guo Y, Wu M, et al. Combinational treatment with microRNA-133b and cetuximab has increased inhibitory effects on the growth and invasion of colorectal cancer cells by regulating EGFR. Mol Med Rep. 2015;12(4):5407–14.
Fu Q, Cheng J, Zhang J, Zhang Y, Chen X, Luo S, et al. miR-20b reduces 5-FU resistance by suppressing the ADAM9/EGFR signaling pathway in colon cancer. Oncol Rep. 2017;37(1):123–30.
Liu B, Liu Y, Zhao L, Pan Y, Shan Y, Li Y, Jia L. Upregulation of microRNA‐135b and microRNA‐182 promotes chemoresistance of colorectal cancer by targeting ST6GALNAC2 via PI3K/AKT pathway. Mol Carcinog. 2017;56(12):2669–80.
Liu N, Li J, Zhao Z, Han J, Jiang T, Chen Y, et al. MicroRNA-302a enhances 5-fluorouracil-induced cell death in human colon cancer cells. Oncol Rep. 2017;37(1):631–9.
Fu Q, Cheng J, Zhang J, Zhang Y, Chen X, Xie J, et al. Downregulation of YEATS4 by miR-218 sensitizes colorectal cancer cells to L-OHP-induced cell apoptosis by inhibiting cytoprotective autophagy. Oncol Rep. 2016;36(6):3682–90.
Sun C, Wang F-J, Zhang H-G, Xu X-Z, Jia R-C, Yao L, et al. miR-34a mediates oxaliplatin resistance of colorectal cancer cells by inhibiting macroautophagy via transforming growth factor-β/Smad4 pathway. World J Gastroenterol. 2017;23(10):1816.
Wei X, Wang W, Wang L, Zhang Y, Zhang X, Chen M, et al. MicroRNA-21 induces 5-fluorouracil resistance in human pancreatic cancer cells by regulating PTEN and PDCD4. Cancer Med. 2016;5(4):693–702.
Kim NK, Park YS, Heo DS, Suh C, Kim SY, Park KC, et al. A phase III randomized study of 5-fluorouracil and cisplatin versus 5-fluorouracil, doxorubicin, and mitomycin C versus 5-fluorouracil alone in the treatment of advanced gastric cancer. Cancer. 1993;71(12):3813–8.
Khuri FR, Nemunaitis J, Ganly I, Arseneau J, Tannock IF, Romel L, et al. A controlled trial of intratumoral ONYX-015, a selectively-replicating adenovirus, in combination with cisplatin and 5-fluorouracil in patients with recurrent head and neck cancer. Nat Med. 2000;6(8):879–85.
Shang J, Yang F, Wang Y, Wang Y, Xue G, Mei Q, et al. MicroRNA-23a antisense enhances 5-fluorouracil chemosensitivity through APAF-1/caspase-9 apoptotic pathway in colorectal cancer cells. J Cell Biochem. 2014;115(4):772–84.
Li P-L, Zhang X, Wang L-L, Du L-T, Yang Y-M, Li J, et al. MicroRNA-218 is a prognostic indicator in colorectal cancer and enhances 5-fluorouracil-induced apoptosis by targeting BIRC5. Carcinogenesis. 2015;36(12):1484–93.
Li Q, Liang X, Wang Y, Meng X, Xu Y, Cai S, et al. miR-139-5p inhibits the epithelial-mesenchymal transition and enhances the chemotherapeutic sensitivity of colorectal cancer cells by downregulating BCL2. Sci Rep. 2016;6:27157.
Liu H, Yin Y, Hu Y, Feng Y, Bian Z, Yao S, et al. miR-139-5p sensitizes colorectal cancer cells to 5-fluorouracil by targeting NOTCH-1. Pathol Res Pract. 2016;212(7):643–9.
Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. Jama. 2008;299(4):425–36.
Fang L, Li H, Wang L, Hu J, Jin T, Wang J, et al. MicroRNA-17-5p promotes chemotherapeutic drug resistance and tumour metastasis of colorectal cancer by repressing PTEN expression. Oncotarget. 2014;5(10):2974.
Rasmussen MH, Jensen NF, Tarpgaard LS, Qvortrup C, Rømer MU, Stenvang J, et al. High expression of microRNA-625-3p is associated with poor response to first-line oxaliplatin based treatment of metastatic colorectal cancer. Mol Oncol. 2013;7(3):637–46.
Yin Y, Zhang B, Wang W, Fei B, Quan C, Zhang J, et al. miR-204-5p inhibits proliferation and invasion and enhances chemotherapeutic sensitivity of colorectal cancer cells by downregulating RAB22A. Clin Cancer Res. 2014;20(23):6187–99.
Suto T, Yokobori T, Yajima R, Morita H, Fujii T, Yamaguchi S, et al. MicroRNA-7 expression in colorectal cancer is associated with poor prognosis and regulates cetuximab sensitivity via EGFR regulation. Carcinogenesis. 2015;36(3):338–45.
Han J, Liu Z, Wang N, Pan W. MicroRNA-874 inhibits growth, induces apoptosis and reverses chemoresistance in colorectal cancer by targeting X-linked inhibitor of apoptosis protein. Oncol Rep. 2016;36(1):542–50.
Lin Teoh S, Das S. The role of MicroRNAs in diagnosis, prognosis, metastasis and resistant cases in breast cancer. Curr Pharm Des. 2017;23(12):1845–59.
Zhang W, Zhou J, Zhu X, Yuan H. MiR-126 reverses drug resistance to TRAIL through inhibiting the expression of c-FLIP in cervical cancer. Gene. 2017;627:420–7.
Zhang Y, Wang J. MicroRNAs are important regulators of drug resistance in colorectal cancer. Biol Chem. 2017;398(8):929–38.
Yang W, Ma J, Zhou W, Cao B, Zhou X, Yang Z, Zhang H, Zhao Q, Fan D, Hong L. Molecular mechanisms and theranostic potential of miRNAs in drug resistance of gastric cancer. Expert Opin Ther Targets. 2017;21(11):1063–75.
Haefliger S, Hudson A, Hayes S, Pavlakis N, Howell V. P2. 01-012 acquired chemotherapy resistance in vitro miRNA profiles of chemotherapy resistant squamous lung cancer cell lines. J Thorac Oncol. 2017;12(1):S790-S1.
Zhuang Z, Hu F, Hu J, Wang C, Hou J, Yu Z, et al. MicroRNA-218 promotes cisplatin resistance in oral cancer via the PPP2R5A/Wnt signaling pathway. Oncol Rep. 2017;38(4):2051–61.
Tung S, Huang W, Hsu F, Yang Z, Jang T, Chang J, et al. miRNA-34c-5p inhibits amphiregulin-induced ovarian cancer stemness and drug resistance via downregulation of the AREG-EGFR-ERK pathway. Oncogenesis. 2017;6(5):e326.
Amponsah PS, Fan P, Bauer N, Zhao Z, Gladkich J, Fellenberg J, et al. microRNA-210 overexpression inhibits tumor growth and potentially reverses gemcitabine resistance in pancreatic cancer. Cancer Lett. 2017;388:107–17.
Armstrong CM, Liu C, Lou W, Lombard AP, Evans CP, Gao AC. MicroRNA-181a promotes docetaxel resistance in prostate cancer cells. Prostate. 2017;77(9):1020–8.
Fattore L, Sacconi A, Mancini R, Ciliberto G. MicroRNA-driven deregulation of cytokine expression helps development of drug resistance in metastatic melanoma. Cytokine Growth Factor Rev. 2017;36:39–48.
Cheng J, Deng R, Zhang P, Wu C, Wu K, Shi L, et al. miR-219-5p plays a tumor suppressive role in colon cancer by targeting oncogene Sall4. Oncol Rep. 2015;34(4):1923–32.
Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9(3):189–98.
Yan L-X, Huang X-F, Shao Q, Huang M-Y, Deng L, Wu Q-L, et al. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. 2008;14(11):2348–60.
Wang P, Zhuang L, Zhang J, Fan J, Luo J, Chen H, et al. The serum miR-21 level serves as a predictor for the chemosensitivity of advanced pancreatic cancer, and miR-21 expression confers chemoresistance by targeting FasL. Mol Oncol. 2013;7(3):334–45.
Toiyama Y, Takahashi M, Hur K, Nagasaka T, Tanaka K, Inoue Y, et al. Serum miR-21 as a diagnostic and prognostic biomarker in colorectal cancer. J Natl Cancer Inst. 2013;105(12):849–59.
Zhang HL, Yang LF, Zhu Y, Yao XD, Zhang SL, Dai B, et al. Serum miRNA-21: elevated levels in patients with metastatic hormone-refractory prostate cancer and potential predictive factor for the efficacy of docetaxel-based chemotherapy. Prostate. 2011;71(3):326–31.
Roth C, Kasimir-Bauer S, Pantel K, Schwarzenbach H. Screening for circulating nucleic acids and caspase activity in the peripheral blood as potential diagnostic tools in lung cancer. Mol Oncol. 2011;5(3):281–91.
Ng E, Tsang W, Ng S, Jin H, Yu J, Li J, et al. MicroRNA-143 targets DNA methyltransferases 3A in colorectal cancer. Br J Cancer. 2009;101(4):699–706.
Valeri N, Gasparini P, Braconi C, Paone A, Lovat F, Fabbri M, et al. MicroRNA-21 induces resistance to 5-fluorouracil by down-regulating human DNA MutS homolog 2 (hMSH2). Proc Natl Acad Sci. 2010;107(49):21098–103.
Zhang H, Tang J, Li C, Kong J, Wang J, Wu Y, et al. MiR-22 regulates 5-FU sensitivity by inhibiting autophagy and promoting apoptosis in colorectal cancer cells. Cancer Lett. 2015;356(2):781–90.
Li X, Li X, Liao D, Wang X, Wu Z, Nie J, et al. Elevated microRNA-23a expression enhances the chemoresistance of colorectal cancer cells with microsatellite instability to 5-fluorouracil by directly targeting ABCF1. Curr Protein Pept Sci. 2015;16(4):301–9.
Ye J, Wu X, Wu D, Wu P, Ni C, Zhang Z, et al. miRNA-27b targets vascular endothelial growth factor C to inhibit tumor progression and angiogenesis in colorectal cancer. PLoS One. 2013;8(4):e60687.
Vogt M, Munding J, Grüner M, Liffers S-T, Verdoodt B, Hauk J, et al. Frequent concomitant inactivation of miR-34a and miR-34b/c by CpG methylation in colorectal, pancreatic, mammary, ovarian, urothelial, and renal cell carcinomas and soft tissue sarcomas. Virchows Archiv. 2011;458(3):313–22.
Zhang Y, Zheng L, Huang J, Gao F, Lin X, He L, et al. MiR-124 Radiosensitizes human colorectal cancer cells by targeting PRRX1. PLoS One. 2014;9(4):e93917.
Valeri N, Braconi C, Gasparini P, Murgia C, Lampis A, Paulus-Hock V, et al. MicroRNA-135b promotes cancer progression by acting as a downstream effector of oncogenic pathways in colon cancer. Cancer Cell. 2014;25(4):469–83.
Bao Y, Guo Y, Li Z, Fang W, Yang Y, Li X, et al. MicroRNA profiling in Muc2 knockout mice of colitis-associated cancer model reveals epigenetic alterations during chronic colitis malignant transformation. PLoS One. 2014;9(6):e99132.
Chintharlapalli S, Papineni S, Abdelrahim M, Abudayyeh A, Jutooru I, Chadalapaka G, et al. Oncogenic microRNA-27a is a target for anticancer agent methyl 2-cyano-3, 11-dioxo-18β-olean-1, 12-dien-30-oate in colon cancer cells. Int J Cancer. 2009;125(8):1965–74.
Fuse N, Kuboki Y, Kuwata T, Nishina T, Kadowaki S, Shinozaki E, et al. Prognostic impact of HER2, EGFR, and c-MET status on overall survival of advanced gastric cancer patients. Gastric Cancer. 2016;19(1):183–91.
Gainor JF, Shaw AT, Sequist LV, Fu X, Azzoli CG, Piotrowska Z, Huynh TG, Zhao L, Fulton L, Schultz KR, Howe E, Farago AF, Sullivan RJ, Stone JR, Digumarthy S, Moran T, Hata AN, Yagi Y, Yeap BY, Engelman JA, Mino-Kenudson M. EGFR mutations and ALK rearrangements are associated with low response rates to PD-1 pathway blockade in non-small cell lung cancer (NSCLC): a retrospective analysis. Clin Cancer Res. 2016;22(18):4585–93.
Concha-Benavente F, Srivastava RM, Trivedi S, Lei Y, Chandran U, Seethala RR, et al. Identification of the cell-intrinsic and-extrinsic pathways downstream of EGFR and IFNγ that induce PD-L1 expression in head and neck cancer. Cancer Res. 2016;76(5):1031–43.
Arnold D, Lueza B, Douillard J-Y, Peeters M, Lenz H-J, Venook A, et al. Prognostic and predictive value of primary tumour side in patients with RAS wild-type metastatic colorectal cancer treated with chemotherapy and EGFR directed antibodies in six randomized trials. Ann Oncol. 2017;28(8):1713–29.
He Q, Xu F, Li X, Chang C. 167-IGF-IR promoted clonal cell proliferation in myelodysplastic syndromes through PI3K and MAPK pathways. Leukemia Res. 2017;55:S103–5.
Elbaz M, Ahirwar D, Ravi J, Nasser MW, Ganju RK. Novel role of cannabinoid receptor 2 in inhibiting EGF/EGFR and IGF-I/IGF-IR pathways in breast cancer. Oncotarget. 2017;8(18):29668.
Jin M, Long Z-W, Yang J, Lin X. Correlations of IGF-1R and COX-2 expressions with Ras and BRAF genetic mutations, clinicopathological features and prognosis of colorectal cancer patients. Pathol Oncol Res. 2018;24(1):45–57.
DeRita RM, Zerlanko B, Singh A, Lu H, Iozzo RV, Benovic JL, et al. c-Src, insulin-like growth factor I receptor, G-protein-coupled receptor kinases and focal adhesion kinase are enriched into prostate cancer cell exosomes. J Cell Biochem. 2017;118(1):66–73.
Yang SX, Polley E, Lipkowitz S. New insights on PI3K/AKT pathway alterations and clinical outcomes in breast cancer. Cancer Treat Rev. 2016;45:87–96.
Fang W-L, Huang K-H, Lan Y-T, Lin C-H, Chang S-C, Chen M-H, et al. Mutations in PI3K/AKT pathway genes and amplifications of PIK3CA are associated with patterns of recurrence in gastric cancers. Oncotarget. 2016;7(5):6201.
Fang X, Yang D, Luo H, Wu S, Dong W, Xiao J, et al. SNORD126 promotes HCC and CRC cell growth by activating the PI3K–AKT pathway through FGFR2. J Mol Cell Biol. 2016;9(3):243–55.
Lastwika KJ, Wilson W, Li QK, Norris J, Xu H, Ghazarian SR, et al. Control of PD-L1 expression by oncogenic activation of the AKT–mTOR pathway in non–small cell lung cancer. Cancer Res. 2016;76(2):227–38.
Arqués O, Chicote I, Puig I, Tenbaum SP, Argilés G, Dienstmann R, et al. Tankyrase inhibition blocks Wnt/β-catenin pathway and reverts resistance to PI3K and AKT inhibitors in the treatment of colorectal cancer. Clin Cancer Res. 2016;22(3):644–56.
Riquelme I, Tapia O, Leal P, Sandoval A, Varga MG, Letelier P, et al. miR-101-2, miR-125b-2 and miR-451a act as potential tumor suppressors in gastric cancer through regulation of the PI3K/AKT/mTOR pathway. Cell Oncol. 2016;39(1):23–33.
Masuda T, Hayashi N, Kuroda Y, Ito S, Eguchi H, Mimori K. MicroRNAs as biomarkers in colorectal cancer. Cancers. 2017;9(9):124.
Wang J, Li H, Wang Y, Wang L, Yan X, Zhang D, et al. MicroRNA-552 enhances metastatic capacity of colorectal cancer cells by targeting a disintegrin and metalloprotease 28. Oncotarget. 2016;7(43):70194.
Wang T, Han P, He Y, Zhao C, Wang G, Yang W, et al. Lin28A enhances chemosensitivity of colon cancer cells to 5-FU by promoting apoptosis in a let-7 independent manner. Tumor Biol. 2016;37(6):7657–65.
Lu W, Wang J, Yang G, Yu N, Huang Z, Xu H, et al. Posttranscriptional regulation of Galectin-3 by miR-128 contributes to colorectal cancer progression. Oncotarget. 2017;8(9):15242.
Karaayvaz M, Zhai H, Ju J. miR-129 promotes apoptosis and enhances chemosensitivity to 5-fluorouracil in colorectal cancer. Cell Death Dis. 2013;4(6):e659.
Ye J, Wang Z, Zhao J, Chen W, Wu D, Wu P, et al. MicroRNA-141 inhibits tumor growth and minimizes therapy resistance in colorectal cancer. Mol Med Rep. 2017;15(3):1037–42.
Han P, Li J-W, Zhang B-M, Lv J-C, Li Y-M, Gu X-Y, et al. The lncRNA CRNDE promotes colorectal cancer cell proliferation and chemoresistance via miR-181a-5p-mediated regulation of Wnt/β-catenin signaling. Mol cancer. 2017;16(1):9.
Wan L-Y, Deng J, Xiang X-J, Zhang L, Yu F, Chen J, et al. miR-320 enhances the sensitivity of human colon cancer cells to chemoradiotherapy in vitro by targeting FOXM1. Biochem Biophys Res Commun. 2015;457(2):125–32.
Tan S, Shi H, Ba M, Lin S, Tang H, Zeng X, et al. miR-409-3p sensitizes colon cancer cells to oxaliplatin by inhibiting Beclin-1-mediated autophagy. Int J Mol Med. 2016;37(4):1030–8.
Jin Y, Jiang Z, Guan X, Chen Y, Tang Q, Wang G, et al. miR-450b-5p suppresses stemness and the development of chemoresistance by targeting SOX2 in colorectal cancer. DNA and cell Biol. 2016;35(5):249–56.
Amankwatia E, Chakravarty P, Carey F, Weidlich S, Steele R, Munro A, et al. MicroRNA-224 is associated with colorectal cancer progression and response to 5-fluorouracil-based chemotherapy by KRAS-dependent and-independent mechanisms. Br J Cancer. 2015;112(9):1480–90.
Acknowledgements
The authors acknowledge the Meta-analysis Concepts and Applications workshop manual by Michael Borenstein for his guidelines on reporting meta-analysis, subgroup analysis and publication bias, (www.meta-analysis-workshops.com).
Author information
Authors and Affiliations
Contributions
RJ contributed to the conceptualisation, study design, search strategy, protocol development, and review by revising different versions. RJ, MRM, CK, SB, AG, NR and KMG provided input into the study design, supervision, ensured the absence of errors, and arbitrated in case of disagreement. MRM and CK engaged in initial searches to determine the feasibility and in the data collection, analysis, and drafting of the manuscript. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Funding
No sources of funding were used to conduct this study or prepare this manuscript.
Conflict of interest
RJ, MRM, CK, SB, AG, NR and KMG have no conflicts of interest that are directly relevant to the content of this article.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Madurantakam Royam, M., Kumarasamy, C., Baxi, S. et al. Current Evidence on miRNAs as Potential Theranostic Markers for Detecting Chemoresistance in Colorectal Cancer: A Systematic Review and Meta-Analysis of Preclinical and Clinical Studies. Mol Diagn Ther 23, 65–82 (2019). https://doi.org/10.1007/s40291-019-00381-6
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40291-019-00381-6