Abstract
Fluorescent pseudomonads are one of the most important microbial communities which play a key role in rhizosphere to enhance plant growth-promotion and protection. The diverse groups of antibiotics viz. 2,4-diacetylphloroglucinol (DAPG), phenazine-1-carboxylic acid (PCA) and pyoluteorin (PLT) are produced by fluorescent pseudomonads inhibiting growth of fungal pathogens which results in health upliftment of plants. The present study, discusses about frequency and diversity of 138 antibiotic-producing fluorescent pseudomonads isolated from eight genotypes of rapeseed mustard rhizosphere (Brassica juncea L. Czern.). The plant growth promoting traits and antibiotics (DAPG, PCA and PLT) production of isolates were examined by using polymerase chain reaction (PCR), thin layer chromatography (TLC) and dot blot-hybridization. Among 138 isolates, 47, 25 and 9 % of isolates were positive in indole production, phosphate solubilization and antagonism potential against Sclerotinia sclerotiorum (causal agent of white mold disease in rapeseed mustard), respectively. PCR amplifications showed that none of the isolates had phlD (DAPG) and phzC (PCA) genes, but four isolates (UKA-2, UKA-8, UKA-11, UKA-66) had pltB (PLT) gene, which was further confirmed by TLC and DNA dot-blot hybridization. BOX profiles of pltB positive isolates were distinct, showing unique genetic diversity in the small population. The four pltB positive fluorescent pseudomonad isolates could be used as promising bio-control and plant growth-promoting inoculants for Indian rapeseed mustard.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
During the last three decades it has been demonstrated that secondary metabolites produced by antagonistic bacteria play a key role in the suppression of various soil-borne plant pathogens. Among the members of plant growth-promoting rhizobacteria, fluorescent pseudomonads (FP) are frequently found to produce antibiotics like 2,4-diacetyl phloroglucinol (DAPG), phenazine-1-carboxylic acid (PCA), pyoluteorin (PLT) and pyrrolnitrin (PRN) etc. which have been recovered from rhizosphere of many crops. These bacteria have been studied extensively because they are highly effective biocontrol agents of a wide genotype of plant diseases when applied as seed or soil treatments [1].
It was reported that there was considerable genetic diversity which exists amongst DAPG [2], PLT [3, 4] and PCA [5] producing FP isolates. It has been shown by amplified ribosomal DNA restriction analysis (ARDRA), whole cell repetitive sequence based PCR (rep-PCR) and restriction fragment length polymorphism (RFLP) analysis in many crops. The genetic diversity analysis for DAPG, PCA and PLT-producing FP have not been studied so far in Indian rapeseed mustard (IRM). Similarly, the relationship between plant genotype and the PGPR rhizosphere colonization is also not known in IRM.
Rape-seed mustard is a member of family Brassicaceae, an economically important edible oil seed crop in India which has been cultivated for a long time especially in the northern plains of the country. The presence of sulphur compounds (glucosinolate 2-phenylethylglucosinolate) in the root exudates of these plants influences the rhizospheric resident population [6], which exhibits negative influence on Arbuscular mycorrhiza (AM) fungi [7]. These information hypothesized that the plant may have a great influence on the fluorescent pseudomonads which may be reflected on population density and antibiotic production. Most of the work on antibiotic-producing fluorescent pseudomonads has been carried out on flax [8], pea [9], tomato [10], wheat [11, 12] and green pepper [3] rhizospheres. As there was not much information with mustard rhizosphere, the present investigation was carried out with special emphasize on antibiotic-producing plant growth promoting fluorescent pseudomonads of mustard.
Material and Methods
Isolation of Fluorescent Pseudomonads (FP)
Rhizosphere of eight genotypes of IRM (Pusa Bold, Pusa Carinata, Pusa Mahak, Pusa Jaganath, Pusa Karishma, Pusa Vijay, NPJ-114 and Bio-3401) were collected from two locations (Entomology and Gentetics fields) of Indian Agricultural Research Institute, New Delhi (28°38′N, 77°09′E; 228.61 m above mean sea level), India. Their serial dilutions were made and plated on King’s B medium (KBM) [13] and incubated at 30 ± 2 °C for 48 h. FP colonies were screened under ultra-violet transilluminator. These colonies were sub-cultured on KBM and pure cultures of 138 isolates were maintained on nutrient agar (NA) slants.
Indole Acetic Acid (IAA) Production
FP isolates were inoculated in 5 mL Luria–Bertani (LB) broth and incubated at 30 ± 2 °C at 180 rpm for 48 h. Two microlitre of log phase cultures were spot inoculated on both LB and LB agar supplemented with 5 mM l-tryptophan. Sterilized nylon membranes were overlaid on those spots after complete drying and incubated at 30 ± 2 °C for 48 h. The grown cultures on nylon membrane were soaked in Salkowski reagent (2 % of 0.5 M FeCl3 in 35 % HClO4) and pink color development around the colonies was observed and compared to reference strain P. fluorescence Pf-5 [14].
Phosphate Solubilization
The phosphate solubilization test was done on a solid medium described by Pikovaskya [15]. Agar plates were prepared and the FP isolates were spot inoculated on the grids and incubated for 5–6 days. A clear zone around the colony was taken as positive for P-solubilization.
Dual Plate Assay Against Sclerotinia sclerotiorum
Dual plate assay was carried out of FP isolates against S. sclerotiorum, a causal organism of white mold disease in rapeseed mustard. All screenings were carried out on potato dextrose agar (PDA) plates. An actively growing fungal agar plug (3 mm diameter) was placed at the centre of PDA plates. FP isolates were streaked 2 cm away from the fungal disk and incubated at 28 ± 2 °C for 7 days.
Bacterial Genomic DNA Extraction
FP isolates were grown in 5 mL LB broth and incubated at 30 ± 2 °C for 24 h. Genomic DNA was extracted by phenol–chloroform method [16].
Polymerase Chain Reaction (PCR) of Antibiotics Production Genes
Polymerase chain reactions were carried out to detect the presence of three antibiotic production genes viz. 2,4-diacetylphloroglucinol (DAPG: phlD gene), phenazine (PCA: phzC and phzD genes) and pyoluteorin (PLT: pltB gene). The detailed specification of primer, respective reference strains and source of these antibiotics are mentioned in Tables 1 and 2. Amplification of phzD and phlC genes was carried out by PCR using a thermal cycler (Eppendorf, Master cycler gradient). The amplification reactions were performed in a 25 μL volume by mixing 4 ng μL−1 of template DNA with polymerase reaction buffer (10×), 1.5 mM MgCl2, 200 μM dNTPs, 20 pmol each primers of DAPG (Phl2a and Phl2b) and PCA (PCA2a and PCA3b) and 1.5 U Taq polymerase [17]. The following programme was used for thermocycling conditions, 94 °C denaturation for 90 s, followed by 35 cycles at 94 °C denaturation for 35 s, 53 °C annealing for 30 s and 72 °C extension for 45 s and final extension at 72 °C for 30 s.
Similarly, the amplification of pltB gene was performed with primers pltBf and pltBR (20 pmol each). The PCR program consisted of a denaturation step at 94 °C for 120 s followed by 29 cycles at 94 °C for 60 s, 58 °C for 30 s, 72 °C for 60 s and a final extension at 72 °C for 120 s [19]. For every PCR reaction, a negative control (no template DNA) and a positive control (corresponding reference stains Pf-5, Ps-Q2-87 and Ps-2-79) were invariably maintained (Table 1). The amplified product was run on 1.2 % agarose gel (containg 0.5 μL mL−1 EtBr) along with 100 bp and 1 kb ladders at a constant voltage (5 V cm−1) for an hour and band was visualized under ultra-violet transilluminator.
Thin Layer Chromatography (TLC) of Pyoluteorin Antibiotic
Inocula were prepared from cells harvested from 3 days old cultures of fluorescent pseudomonads grown on KBM broth of 30 ± 2 °C on a rotary shaker at 180 rpm. The supernatant was collected by centrifugation at 3500 rpm for 5 min and transferred to micro-centrifuge tubes, vortexed for 30 s with 500 μL of ethyl acetate and again centrifuged. The ethyl acetate phase was collected and dried. The dried residue was dissolved in 10 μL methanol and proceeded for TLC analysis along with reference strain Pf-5 [19].
Dot-Blot Hybridization for Pyoluteorin Antibiotic
Dot-blots were probed with a α-32P labeled pltB (~773 bp) purified fragment from reference strain Pf-5. Five microliter of purified DNA samples of 138 FP isolates along with 2 μL of the purified PCR product (~773 bp fragment) of pyoluteorin-producing reference strain Pf-5 were put onto nylon membrane and dot-blot hybridization was performed following the protocol of Udo and Dashti [20].
BOX A1R-PCR for Genetic Diversity of Pyoluteorin-Producers
Amplification of 156 bp box element was carried out by PCR using a thermal cycle (Eppendorf, Master cycler gradient) with BOX-A1R Primer (5′CTACGGCAAGGCGACGCTGACG3′) [21]. The amplification reactions were performed in a 50 μL volume by mixing template DNA (40 ng μL−1) with polymerase reaction buffer (10×), 1.5 mM MgCl2, 200 μM dNTPs, BOXA1R-primer (15 pmol μL−1) and 3 U Taq polymerase. The program consisting of a denaturation step at 95 °C for 7 min followed by 30 cycles at 94 °C for 60 s, 53 °C for 60 s and 65 °C for 8 min was used to eliminate additional fragments resulting from final extension at 65 °C for 16 min. The amplified product was run on 1.5 % agarose gel (containing 0.5 μL mL−1 Etbr) along with 100 bp and 1 kb ladder at a constant voltage (5 V cm−1) for 6 h and bands were visualized under ultra-violet transilluminator.
Data Analysis
Pyoluteorin producing fluorescent pseudomonads isolates were identified using biochemical-based PibWin software [22]. For genetic diversity, BOX-PCR fingerprints were converted into two-dimensional binary matrix (1, presence of a given band; 0 absence of a given band) and analyzed with NTSYS-PC (Applied Biostatistics Inc. NY). Dendrogram was generated by using the SIMQUAL (Similarity for Qualitative data) and SAHN (Sequential Agglomerative Hierarchial and Nested) clustering subroutines of NTSYS-PC [23].
Results and Discussion
One hundred and thirty eight FP were isolated from rhizosphere of eight different genotypes of IRM. Among them, 71.4 % of isolates of genotype BIO-3401 were non-functional in growth-promoting traits viz. indole, P-solubilization, antagonistic against S. sclerotiorum, antibiotics production (DAPG, PCA and PLT) whereas it was 57.1, 55.8, 38.4, 35.7, 28.5 and 22.7 % in NPJ 114, Pusa Bold, Pusa Carinata, Pusa Jaganath, Pusa Mahak and Pusa Karishma, respectively. The frequency of FP from rhizosphere of Pusa Vijay was found to be predominant (data not shown). It clearly indicated the varietal effect. This was amply demonstrated by the lower frequencies of the isolates in genotype BIO-3401 rhizosphere which was strongly inhibitory towards the rhizospheric FP. This might be due to root exudates containing sulphur compounds causing allelopathic effect against rhizospheric microbes [6] and also variation of root exudation in different plant species [24]. The above reasons were also corroborated with the earlier findings [8, 25].
Plant-Growth Promoting Traits
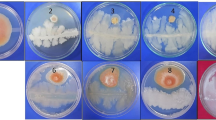
A qualitative test of IAA production indicated that out of 138 FP, 47 % were found to be positive with variable IAA production. Most of the isolates were found to produce IAA constitutively in the absence of the precursor tryptophan (Figs. 1a, b, 2). Twenty five percent of isolates showed positive in P-solubilization (Fig. 2). Against S. sclerotiorum, 9 % of isolates showed antagonistic activity (Fig. 2). Among the isolates, there was a wide variation in functional diversity of 6 growth promoting traits. Only 4 isolates possessed three functional traits (IAA, antagonistic activity and antibiotic production), 29 had two functions (IAA and antagonistic activity) and others possessed only a single function (IAA) (Fig. 2).
FP often predominates among the bacteria of plant rhizosphere, and some can have beneficial effects on plants, either by direct stimulation of plant growth or by exerting antagonism towards soil borne pathogens [26, 27]. Rhizobacteria from the rhizosphere of different Brassica species varied greatly in their auxin production [28]. It has been shown that host genotype influences the enhancement of plant growth by auxin producing strains. In the present study, 47 % of isolates were positive for IAA production. Similarly, the host genotype had a drastic influence in selecting phosphate (P)-solubilizers. Only 35 of the 138 isolates were found to solubilize insoluble P, corresponding to a frequency of 25.3 %. The P requirement of rapeseed mustard is low or may be the root exudates do not stimulate P-solubilizers in the vicinity of roots of mustard.
Only 13 isolates, corresponding to a frequency of 9.4 % were able to inhibit S. sclerotiorum which is an important soil-borne fungal pathogen of mustard causing white mold disease in the crop. Since the three reference strains were not able to inhibit this fungus in vitro, it could be possible that the three antibiotics, phenazine, pyoluteorin and DAPG have no effect. This finding indicates that 13 isolates may produce some other metabolite which could suppress the pathogen [29]. The low frequency of antibiotic producing fluorescent pseudomonad isolates in rapeseed mustard rhizosphere is gaining much importance as no chemical control measures are so effective against white mold disease.
PCR Screening of FP for DAPG, PCA and PLT
The three reference strains Pf-5, Ps-Q2-87 and Ps-2-79 were PCR amplified for their respective antibiotic genes corresponding to phlD (DAPG) with 745 bp, phzC (phenazine) with 1150 bp and pltB (pyoluteorin) with 773 bp (Fig. 3a). None of the isolates showed the presence of the phlD and phzC codes for DAPG and phenazine genes (Fig. 3b, c). However, 4 isolates i.e. UKA-2, UKA-8, UKA-11, and UKA-66 showed the pltB gene amplification (~773 bp) (Fig. 3d). These four isolates were identified by biochemical profiles and the data was computed with PibWin software. All isolates were 95–99 % similar to fluorescent pseudomonads (Table 3).
PCR amplification of antibiotic genes for of 2,4-diacetyl phloroglucinol (DAPG), phenazine-1-carboxylic acid (PCA) and pyoluteorin (PLT). a Amplification of reference strains. Lane 1 100 bp DNA ladder; Lane 2 Pf-5 (~745 bp); Lane 3 Ps-Q2-87 (~745 bp); Lane 4 Pf-5 (~773 bp); Lane 5 Ps-2-79 (~1150 bp); Lane 6 1 kb DNA ladder. b Gel picture of DAPG with reference strain Ps-Q2-87. c Gel picture of phenazine with reference strain Ps-2-79. d Amplification of pltB gene (pyoluteorin)with reference strain Pf-5
It is generally accepted that pathogen inhibition by antibiotic metabolites is one of the primary mechanisms of biocontrol provided by these root colonizing bacteria [30]. In the rhizosphere of lily (Lilium candidum L.), phlD+ pseudomonads were not detected, although it supported in an average the highest population densities of fluorescent Pseudomonas [31]. It is also reported that host plant species has significant influence on the composition and activity of specific indigenous antagonistic Pseudomonas. Raaijmakers et al. [17] could not detect any phenazine producer in roots of wheat grown in three soils. Similarly, in the present study, only four fluorescent isolates amplified the pltB gene, which indicated the lower frequency for this antibiotic in mustard rhizosphere.
TLC and Dot-Blot Hybridization of PLT-Producing Isolates
The four positive pyoluteorin producing isolates were further confirmed through TLC with reference strains Pf-5. Visualization of TLC plates under ultra violet light and spraying with DSA revealed only one isolate, UKA-66 showing the brown spot corresponding to PLT (Fig. 4a) and other three were undetectable by TLC.
To validate the negative PCR results a DNA dot blot experiment was carried out with all 138 FP onto nylon membranes. Dot blots were probed with a α-32P labeled pltB (~773 bp) purified fragment from reference strain Pf-5. Strong signals were observed in PCR positive strains along with reference strain Pf-5; however 7 more isolates, UKA-32, UKA-33, UKA-68, UKA-71, UKA-83, UKA-124 and UKA-128 gave good detectable signals (Fig. 4b, c). DNA blotting experiments increased this number to 11, confirming that negative PCR is not always confirmatory. Eight blots were scored to have strong detectable signal. Two PCR positive isolates UKA-2 and UKA-11 gave faint signals, quite possibly because of heterologous sequences. However, TLC of the 4 PCR positive isolates gave the appropriate band only in one isolate (UKA-66) probably due to little concentration of the antibiotics produced by other isolates (since extraction was carried out with 5 mL of culture) so as to be undetectable by TLC.
Genetic Diversity of Pyoluteorin Producing Isolates
In the present study DNA fingerprints were generated for total chromosomal DNA for all the four isolates positive for the antibiotic pyoluteorin using BOX A1R primer. Distinct banding patterns were generated using the 22-mer oligonucleotide primer in combination with PCR conditions that favored the simultaneous amplification of multiple different sized DNA fragments. The range of amplified bands were found between 300 bp and >10 kb. Total number of bands scored were 31. No single band was uniformly present in these isolates showing a high degree of heterogeneity (Fig. 5a). Dendrogram generated of the BOX profiles confirmed the presence of polymorphism. Two clusters were generated with the similarity coefficient being less than 40 %. UKA-66 and UKA-2 were more similar to each other and UKA-8 was closer to UKA-11 (Fig. 5b).
It has been postulated that the genotypic diversity within a group of microorganisms that share the same antagonistic trait provides a largely untapped resource for improving biological control of soil borne pathogens [32]. Studies have supported the hypothesis that certain indigenous phlD+ genotypes preferentially colonize the roots of specific crop plants. Similar type of findings were reported by other researchers [17, 18, 33]. In the present study, BOX-PCR distinguished 4 isolates amplifying pltB gene. The result showed that the isolate UKA-8 was positive for pltB, IAA production and antagonistic activity. Isolate UKA-66 was positive for pltB, IAA production and P-solubilization, isolate UKA-2 was positive for IAA and pltB, but UKA-11was positive only for pltB. However, the BOX profiles of each of these were unique and different indicating the genetic heterogeneity existing in these isolates. The present findings will further help to identify the efficient PGPR strains from the mixed heterogenous rhizosphere population.
Conclusion
Overall, the present study revealed the lower frequency and diversity of plant growth promoting traits of FP in IRM. It was also demonstrated that out of three antibiotics (DAPG, phenazine and pyoluteorin), none of the isolates produced DAPG and phenazine and only 4 isolates produced pyoluteorin which further confirmed the lower frequency of antibiotic producing FP in IRM rhizosphere.
Abbreviations
- FP:
-
Fluorescent pseudomonads
- IRM:
-
Indian rapeseed mustard
- DAPG:
-
2,4-Diacetyl phloroglucinol
- PCA:
-
Phenazine-1-carboxylic acid
- PLT:
-
Pyoluteorin
References
de La Fuente L, Mavrodi DV, Landa BB, Thomashow LS (2006) phlD-based genetic diversity and detection of genotype of 2,4-diacytylphloroglucinol-producing Pseudomonas fluorescens. FEMS Microbiol Ecol 50:64–78
Weller DM, Mavrodi DV, van Pelt JA, Pieterse CM, van Loon LC, Bakker PA (2012) Induced systemic resistance in Arabidopsis thaliana against Pseudomonas syringae pv. tomato by 2,4-diacetyl phloroglucinol producing Pseudomonas fluorescens. Phytopathology 102:403–412
Liu H, Dong D, Peng H, Zhang X, Xu Y (2006) Genetic diversity of phenazine-and pyoluteorin-producing pseudomonads isolated from green pepper rhizosphere. Arch Microbiol 185(2):91–98
Ramette A, Frapolli M, Fischer-Le Saux M, Gruffaz C, Meyer JM, Défago G, Sutra L, Moenne-Loccoz Y (2011) Pseudomonas protegens sp. nov., widespread plant-protecting bacteria producing the biocontrol compounds 2,4-diacetyl phloroglucinol and pyoluteorin. Syst Appl Microbiol 34:180–188
Mavrodi DV, Mavrodi OV, Parejko JA, Bonsall RF, Kwak YS, Paulitz TC, Thomashow LS, Weller DM (2012) Accumulation of the antibiotic phenazine-1-carboxylic acid in the rhizosphere of dryland cereals. Appl Environ Microbiol 78:804–812
Barto EK, Antunes PM, Stinson K, Koch AM (2011) Differences in arbuscular mycorrhizal fungal communities associated with sugar maple seedlings in and outside of invaded garlic mustard forest patches. Biol Invasions 13(12):2755–2762
Vierheilig H, Bennett R, Kiddle G, Kaldorf M, Ludwig-Müller J (2000) Differences in glucosinolate patterns and arbuscular mycorrhizal status of glucosinolate-containing plant species. New Phytol 146(2):343–352
Lemanceau P, Corberand T, Gardan L, Latour X (1995) Effect of two plant species, flax (Linum usitatissimum L.) and tomato (Lycopersicon esculentum Mill.) on the diversity of soil-borne populations of fluorescent pseudomonads. Appl Environ Microbiol 61:1004–1012
Landa BB, Mavrodi OV, Raaijmakers JS, Gardener BBM (2002) Differential ability of genotype of 2,4-diacetyl phloroglucinol producing Pseudomonas fluorescens strains to colonize the roots of pea plants. Appl Environ Microbiol 68:3226–3237
Rao MS, Dwivedi MK, Tiwari SP, Kumar RM, Rajinikanth R, Chaya MK, Grace GN, Priti K, Ratnamma M, Kamalnath M, Prabu P, Vidyashree N, Gopalkrishna C, Shivananda TN (2014) Documentation of bio-efficacy of DAPG producing Pseudomonas fluorescens-1% as on Meloidogyne incognita and Ralstonia solanacearum infecting tomato in different agro-climatic regions. Pest Manag Horticult Ecosyst 20(2):222–226
de Souza JT, Weller DM, Raaijmakers JM (2003) Frequency, diversity and activity of 2,4-diacetylphloroglucinol-producing fluorescent Pseudomonas spp. in Dutch take-all decline soils. Phytopathology 93:54–63
Weller DM (2015) Take-all decline and beneficial pseudomonads. In: Principles of plant–microbe interactions. Springer, New York, pp 363–370
King EO, Ward MK, Raney DE (1954) Two simple media for demonstration of pyocyanin and fluroscein. J Lab Clin Med 44:301–307
Bric JM, Bostock RM, Silverstone SE (1991) Rapid in situ assay for Indoleacetic acid production by bacteria immobilized on a nitrocellulose membrane. Appl Environ Microbiol 57:535–538
Pikovskya RI (1948) Mobilization of phosphorus and soil in connection with the vital activity of some microbial species. Microbiology 17:362–370
Masterson RV, Prakash RK, Amerly AG (1985) Conservation of symbiotic nitrogen fixation gene sequence in R. japonicum. J Bacteriol 163:21–26
Raaijmakers JM, Weller DM, Thomashow LS (1997) Frequency of antibiotic producing Pseudomonas spp. in natural environments. Appl Environ Microbiol 63:881–887
Mavrodi OJ, Gardener BBM, Mavrodi DN, Bansall RF (2001) Genetic diversity of phlD from 2,4-diacetylphloroglucinol producing fluorescent Pseudomonas spp. Phytopathology 91:35–43
Udo EE, Dashti AA (2000) Detection of genes encoding aminoglycoside-modifying enzymes in staphylococci by polymerase chain reaction and dot blot hybridization. Int J Antimicrob Agents 13(4):273–279
Picard C, Bosco M (2005) Maize heterosis affects the structure and dynamics of indigenous rhizospheric auxins-producing Pseudomonas populations. FEMS Microbiol Ecol 53:349–357
Ottaviani D, Masini L, Bacchiocchi S (2003) A biochemical protocol for the isolation and identification of current species of Vibrio in seafood. J Appl Microbiol 95(6):1277–1284
Rohlf FJ (2000) NTSYS-pc: numerical taxonomy system. Ver. 2.1. Exeter Publishing, Ltd., Setauket
Costa R, Gotz M, Mrotzek N, Lottmann J (2006) Effects of site and plant species on rhizosphere community structure as revealed by molecular analysis of microbial guilds. FEMS Microbiol Ecol 56:236–249
Berg G, Zachow C, Lottmann J, Gotz M (2005) Impact of plant species and site on rhizosphere associated fungi antagonistic to Verticillum dahliae kleb. Appl Environ Microbiol 71:4203–4213
Kumar U, Lakkineni VK, Kannepalli K (2013) Antagonistic potential and functional diversity of endo- and rhizospheric bacteria of basmati rice. Oryza 50(2):162–168
Kumar U, Mishra S (2014) Functional and genetic diversity of primary and secondary- metabolites producing fluorescent pseudomonads from rhizosphere of rice (Oryza sativa L.). J Appl Zool Res 25:83–93
Asghar H, Zahir Z, Arshad M, Khaliq A (2002) Relationship between in vitro production of auxins by rhizobacteria and their growth-promoting activities in Brassica juncea L. Biol Fert Soils 35:231–237
Mazurier S, Corberand T, Lemanceau P, Raaijmakers JM (2009) Phenazine antibiotics produced by fluorescent pseudomonads contribute to natural soil suppressiveness to Fusarium wilt. ISME J 3(8):977–991
Haas G, Keel C (2003) Regulation of antibiotic production in root-colonizing Pseudomonas spp. and relevance for biological control of plant disease. Annu Rev Microbiol 41:117–153
Bergsma-Vlami M, Prins ME, Raaijmakers JM (2005) Influence of plant species on populations dynamics genotype diversity and antibiotic production in the rhizosphere by indigenous Pseudomonas spp. FEMS Microbiol Ecol 52:59–69
Raaijmakers JM, Vlami M, de Souza JT (2002) Antibiotic production by bacterial biocontrol agents. Ant Van Leewen 81:537–547
Landa BB, Mavrodi OV, Raaijmakers JS, Gardener BBM (2002) Differential ability of genotype of 2,4-diacetyl phloroglucinol producing Pseudomonas fluorescens strains to colonize the roots of pea plants. Appl Environ Microbiol 68:3226–3237
Raaijmakers JM, Bonsall RF, Weller DM (1999) Effect of population diversity of Pseudomonas fluorescens on production of 2,4-diacetyl phloroglucinol in the rhizosphere of wheat. Phytopathology 89:470–475
Acknowledgments
The first author thanks to Indian Council of Agricultural Research (ICAR) and Indian Agricultural Research Institute, New Delhi, India for providing financial assistance in the form of Junior Research Fellowship (JRF) and laboratory facility of conducting research work, respectively.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declares that there are no conflicts of interest.
Rights and permissions
About this article
Cite this article
Kumar, U., Panneerselvam, P., Banik, A. et al. Lower Frequency and Diversity of Antibiotic-Producing Fluorescent Pseudomonads in Rhizosphere of Indian Rapeseed–Mustard (Brassica juncea L. Czern.). Proc. Natl. Acad. Sci., India, Sect. B Biol. Sci. 88, 579–586 (2018). https://doi.org/10.1007/s40011-016-0792-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40011-016-0792-1